Abstract
Loss or mutation of the TP53 tumor suppressor gene is not commonly observed in acute myeloid leukemia (AML), suggesting that there is an alternate route for cell transformation. We investigated the hypothesis that previously observed Bcl-2 family member overexpression suppresses wild-type p53 activity in AML. We demonstrate that wild-type p53 protein is expressed in primary leukemic blasts from patients with de novo AML using 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE) and phospho-specific flow cytometry. We found that p53 was heterogeneously expressed and phosphorylated in AML patient samples and could accumulate following DNA damage. Overexpression of antiapoptosis protein Bcl-2 in AML cells was directly correlated with p53 expression and phosphorylation on serine residues 15, 46, and 392. Within those patients with the highest levels of Bcl-2 expression, we identified a mutation in FLT3 that duplicated phosphorylation site Y591. The presence of this mutation correlated with greater than normal Bcl-2 expression and with previously observed profiles of potentiated STAT and MAPK signaling. These results support the hypothesis that Flt3-mediated signaling in AML enables accumulation of Bcl-2 and maintains a downstream block to p53 pathway apoptosis. Bcl-2 inhibition might therefore improve the efficacy of existing AML therapies by inactivating this suppression of wild-type p53 activity.
Introduction
We have previously reported that signaling profiles of acute myeloid leukemia (AML) cells identify patients with a poor response to course one of chemotherapy.1 In that study we showed that mutation of fms-like tyrosine kinase 3 (Flt3) was associated with increased activity of signal transduction and activator of transcription (STAT) family members Stat5 and Stat3, but we did not explore how this signaling might contribute to the observed therapy resistance. Here we examine the connection between a target of altered Stat5 signaling in AML—the Bcl-2 protein—and the suppression of normal apoptotic responses to DNA damage in leukemia cells. Evasion of apoptosis contributes to the formation and continued survival of cancer cells2 and insight into signaling mechanisms that suppress apoptosis in AML cells could be used to improve the efficacy of existing therapies.3
AML is particularly relevant to the study of suppressed apoptotic pathways in cancer because p53, a central driver of apoptosis and guardian of genomic integrity, is not generally lost or mutated in this disease.4,5 p53 is a sequence-specific transcription factor that can halt progression through the cell cycle or initiate apoptosis.6 Furthermore, p53 is a key tumor suppressor protein often lost or mutated in human cancers; however, TP53 has been reported to be mutant in only 7% of AML cases.4 Subsequent research has also shown that downstream p53 effector genes, such as p21, are rarely lost in AML and that wild-type p53 protein is expressed in newly established AML derived cell lines.7
For this study, we measured several properties of p53 in primary AML cells, including TP53 gene sequence, per-cell abundance of p53 protein, and p53 phosphorylation at 5 residues.8 Following DNA damage and cell stresses, p53 protein is rapidly phosphorylated at several residues,9 including those we measured here (serines 15, 20, 37, 46, and 392). Phosphorylation of p53 is thought to regulate p53 localization, conformation, and activity,10 but the precise contribution of each phosphorylation site is controversial. We treated each of these 5 p53 phosphorylations as independent signaling events to determine associations with each other and with other properties of AML cells, such as mutational status of Flt3 and expression of apoptosis protein Bcl-2.
Bcl-2 is a potent oncogene that is heterogeneously expressed in AML and associated with a poor response to chemotherapy.11–13 In AML originating from immature CD34+ myeloid cells, Bcl-2 expression is correlated with lowered rates of complete remission14 and the ratio of Bax to Bcl-2 is highly predictive of long-term survival.15,16 Despite these reports, expression of Bcl-2 has not been found to correlate significantly with other prognostic indicators in AML, such as cytogenetic alterations13 or FLT3 mutation. However, Bcl-2 family members are transcriptional targets of STAT protein transcription factors Stat5 and Stat3 in AML cells,17 suggesting that observed alterations in leukemic cell signaling via STATs1 might cause the overexpression of Bcl-2 and allow AML blasts to evade apoptosis.
Signaling pathways upstream of Bcl-2 can be activated by a number of different mechanisms, including genetic changes that result in improper activation of signaling pathways,18 changes in STAT gene sequence and expression,19 and abnormalities of upstream activators of signaling (eg, Flt3, c-kit, platelet-derived growth factor receptor).20 FLT3 mutations are observed in approximately 30% of adult patients with AML.21 FLT3 mutations correlate with increased signaling activity of STAT proteins Stat3 and Stat5 in primary AML cells1 and are an accurate, independent predictor of disease relapse after chemotherapy.22 The majority of mutations in FLT3 are internal tandem duplications situated in the juxtamembrane region. In this juxtamembrane region 3 tyrosine phosphorylation sites have been identified (Y589, Y591, Y599).23,24 The region duplicated is highly variable among patients but nearly always involves at least one of these phosphorylation sites. Approximately half of our patients with Flt3 length mutations (LMs) have a duplication of Y591 (R.H., unpublished observations, June 2005). Changes to signaling1 and poor response to chemotherapy22 associated with Flt3-LM suggest that a key outcome of Flt3 signaling in AML may be evasion of apoptosis mediated by Bcl-2 expression. These changes to signaling might be related to Flt3 mutations that duplicate specific phosphorylation sites. Consistent with this hypothesis is recent evidence that Flt3-LM enables activation of Stat5 through binding to Flt3 phosphorylation sites Y589 and Y591.24
Given the importance of p53 in mediating cell death following DNA damage, we wanted to determine whether wild-type p53 protein was expressed and functional in AML and, if so, understand why DNA damage induction therapies are resisted by AML blast cells. Because of the known role of STAT proteins in transcription of antiapoptotic Bcl-2 family members, we further examined relationships between Flt3 mutations, Bcl-2 protein expression, and levels of phosphorylated p53.
Patients, materials, and methods
Patients and preparation of cells
The study was approved by the local ethics committee (REK Vest) and the Data Inspectorate, Norway. REK Vest is affiliated with the University of Bergen and Haukeland University Hospital. Samples were collected after informed consent was obtained. Samples were selected from a large group of consecutive patients with de novo AML and high peripheral blood blast counts.21 These patients were admitted to the hospital from April 1999 to August 2003. The median age was 60 years (range, 29-84 years). Because these patients were selected for high blast counts, enriched AML-cell populations containing more than 95% tumor cells were prepared using a simple density gradient separation of peripheral blood samples (Ficoll-Hypaque; NyCoMed, Oslo, Norway; specific density 1.077) before standardized cryopreservation according to previously developed techniques.25 These patients represent a portion of a group studied previously for Flt3 signaling and mutation in AML21 and were previously characterized for signaling profiles associated with chemotherapy response and Flt3 mutation.1 Peripheral blood mononuclear cells (PBMCs) were collected from healthy donors, separated as described for AML cells, and used fresh. Cell lines with wild-type p53, the AML line Molm-13, and the prostate cancer line LNCaP, were from American Type Culture Collection (Manassas, VA; www.atcc.org) and were cultured in RPMI (Sigma-Aldrich, Poole, United Kingdom) with 10% fetal calf serum (HyClone, South Logan, UT). A list of patient characteristics is included (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Sequencing of p53
Sequencing of TP53 was performed by temporal temperature gradient gel electrophoresis (TTGE) as previously described.26
Treatment of cells
Enriched AML cells were thawed, counted, pelleted, and resuspended at 2 × 106 cells/mL in StemSpan H3000 defined, serum free-medium (StemCell Technologies, Vancouver, BC, Canada). Leukemia samples were transferred to individual wells or cytometry tubes (Falcon 2052, BD Biosciences, San Jose, CA) and allowed to rest at 37°C for a total of 2 hours. For induction of DNA double-strand breaks, samples were irradiated by 25 Gy from a 137Ce source and analyzed 2 hours later by 2-dimensional polyacrylamide gel electrophoresis (2D-PAGE). For an in vitro model of course one of AML chemotherapy, samples were cultured in StemSpan H3000 media containing 360 nM idarubicin (Pfizer, New York, NY) or vehicle for 2 hours or 4 hours before harvesting cells. Following treatment, samples were returned to a 37°C tissue culture incubator for indicated times. For 2D-PAGE analysis, cells were lysed and protein extract analyzed as described (see “2D SDS-PAGE”). For flow cytometry, paraformaldehyde (PFA; Electron Microscopy Services, Fort Washington, PA) was added to each tube to a final concentration of 1.6% to stop signaling. Cells were fixed for 15 minutes at room temperature, pelleted, permeabilized by resuspension in 2 mL methanol for 10 minutes, and stored at 4 °C for less than 1 week before being stained for flow cytometry.
2D SDS-PAGE
Cells were washed twice in 100 volumes of 0.9% NaCl, lysed in 7% trichloroacetic acid, desalted with 3 washes in water-saturated ether (1 mL, 14 000g for 20 minutes, 4°C) and resolved in 2D sample buffer (7 M urea, 2 M thiourea, 100 mM DTT, 1.5% ampholyte 3-10, 0.5% ampholyte 5-6, 0.5% CHAPS). 2D SDS-PAGE was performed using 7 cm pH 3-10 (Zoom Strip, Invitrogen, Carlsbad, CA) isoelectric focusing (IEF) gel strips. Focusing strips were incubated with sample rehydration buffer (8 M urea, 1% CHAPS, 20 mM DTT, 1.5% ampholyte 3-10, 0.5% ampholyte 5-6, bromophenol blue) containing the protein sample to rehydrate the strips. Rehydration was performed overnight at room temperature. IEF was performed at 200 V for 40 minutes, 450 V for 30 minutes, 750 V for 30 minutes, and 2000 V for 60 minutes. Following IEF, the strips were either stored at −80°C or equilibrated directly for 15 min in LDS sample buffer (Invitrogen) containing 100 mM DTT and then 15 minutes in LDS sample buffer containing 125 mM iodoacetamide. For the second dimension the ZOOM strip was aligned in 0.5% agarose solution added into the IEF well of a NuPAGE Novex 4 to 12% bis-Tris ZOOM Gel (Invitrogen). Electrophoresis was performed at 200 V for 60 minutes and proteins were transferred to polyvinylidene fluoride (PVDF) membrane by standard electroblotting.
Immunoblotting
p53 protein was detected by BP53-12 antibody (Santa Cruz Biotechnology, Santa Cruz, CA) or phospho-specific antibodies (Cell Signaling Technology, Beverly, MA) that recognize phosphorylated S37, S46, or S392. Primary antibodies were used in combination with secondary alkaline phosphatase-conjugated mouse antibody (Applied Biosystems, Foster City, CA) and visualized using the Western-star Immunodetection system (Applied Biosystems–Tropix, Foster City, CA) or secondary horseradish peroxidase-conjugated mouse or rabbit antibody (Jackson ImmunoResearch, West Grove, PA) and visualized using the SuperSignal West Pico system (Pierce, Rockford, IL). Additional description of antibodies is included in Table S2.
Intracellular phospho-specific flow cytometry
Flow cytometry analysis was performed as previously described for AML patient samples.1 PFA fixed, methanol-permeabilized cells were rehydrated by addition of 2 mL phosphate buffered saline (PBS), resuspension by vortexing, and then centrifugation. The cell pellet was washed once with 2 mL PBS + 1% BSA (Sigma, St Louis, MO), resuspended in 50 μL PBS + 1% BSA, and then split evenly into new cytometry tubes for staining. Then, 50 μL of an antibody mix containing 0.13 μg primary conjugated phospho-specific antibody per sample was added to each tube of AML blasts and staining proceeded for 20 minutes at room temperature. Antibodies for p53 included monoclonal α-phospho-p53-S15 clone 16G8 and polyclonal α-phospho-p53-S20, α-phospho-p53-S37, α-phospho-p53-S46, and α-phospho-p53-S392 (Cell Signaling Technology). These antibodies were conjugated to biotin, bound to streptavidin (SA)–conjugated fluorophores SA-Alexa 488, SA-PE, and SA-Alexa 647 (Molecular Probes, Eugene, OR), and used in combination with α-Bcl-2-FITC or α-p53-FITC clone DO-7 (BD PharMingen, San Diego, CA). Stained AML blasts were washed by adding 2 mL PBS + 1% BSA and resuspended in 200 μL PBS. At least 30 000 ungated live cell events were collected for each sample on a FACSCalibur dual-laser cytometer (Becton Dickinson, Franklin Lakes, NJ). Additional description of antibodies is included in Table S2.
Statistical analysis and relationship mapping
Normalization of flow cytometry data was performed by comparing median fluorescence intensity (MFI) values to either an unstimulated control (Figure 3) or the median in AML (Figures 4–5) using the log10 ratio of a sample divided by the control. With this scale, +1 corresponds to a 10-fold increase in MFI. MFI normalization allowed examination of p53 phosphorylation, p53 expression, and Bcl-2 on comparable scales. Significance of differences in averages for 2 populations (eg, expression of Bcl-2 in Flt3 LM groups) was determined using a Student t test. Linear regression analysis of correlation between graded parameters was determined using the square of the Pearson product momentum correlation coefficient (r). Correlations where r2 was greater than 0.6 were included as solid lines in a map of relationships between detected parameters (Figure 6A). In this map, each related parameter was placed at approximately 1 − r2 units from the nodes to which it was related. For example, the correlation between Bcl-2 expression and p53 phosphorylation at serine 46 across 30 AML patients was r2 = 0.64, so the nodes “Bcl-2” and “ser46” were placed 1 − 0.64 = 0.46 units apart on the map. Therefore, the more closely related 2 parameters, the closer they were placed on the map.
Results
p53 is wild type and expressed at high levels in AML
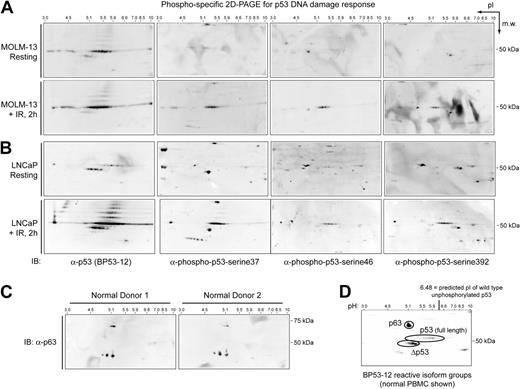
AML blast cells have been reported to contain 2 wild-type TP53 alleles.4 We sequenced TP53 of blast cells from 18 AML patients (Table S1) and all sequenced patients had wild-type TP53. We further asked whether wild-type p53 protein was expressed in AML blast cells. Normal cells express high levels of p53 in response to cell stress (eg, DNA damage), but express low levels of p53 in the resting state. To obtain normal, primary cells with wild-type p53, PBMCs were isolated from the blood of healthy donors by the same method used for isolation of AML blasts. It was especially important to examine p53 in primary cells, because perturbation of p53 pathway function is likely required for generation of a cell line.1 We used 2D-PAGE immunoblotting with the BP53-12 α-p53 antibody to examine expression level, molecular weight, and isoelectric point (pI; measured using pH) of proteins from cell extracts of normal PBMCs and AML blasts (Figure 1). The BP53-12 antibody recognizes many forms of p53, including wild type, most common mutants, N-phosphorylated, and cytoplasmic p53 (Table S2). AML blasts from 6 patients (Figure 1A) and normal PBMCs (Figure 1B) were analyzed and 3 primary isoform groups reactive with BP53-12 were detected. The molecular masses of the isoforms were 63 kDa, 53 kDa, and 47 kDa. The level and presence of individual spots within these isoform groups, especially for the 53-kDa band corresponding to full-length p53, varied significantly among AML patients. In contrast, isoform groups varied little among normal PBMCs from different donors (Figure 1B). In normal cells, full-length p53 isoforms were expressed at much lower levels than in most AML blast samples. The 47-kDa isoform group, termed Δp53, was expressed at high levels in all resting cells. Δp53 was abundant in all AML patient samples tested and was generally restricted to a smaller range in pI than full-length p53 (pH range of 0.5, from 4.8 to 5.3). In contrast, full-length p53 expression and isoform status varied widely in AML patients (Figure 1A), and the range in pI of full-length p53 was wider (pH range of 1.48, from 5.00 to 6.48). In resting normal cells, Δp53 was expressed at approximately a 10-fold higher level than that of full-length p53 (Figure 1B).
Expressed, wild-type p53 in primary AML blasts exists in several acidic isoforms not seen in resting normal cells. (A) Expression of wild-type p53 was detected at high levels in AML blast samples. However, the acidity and expression level of p53 isoforms was variable among AML patients, such that the 2D pattern of p53 created a signature that distinguished AML patients from one another. (B) In contrast, full-length wild-type p53 was expressed at low levels (53-kDa band) and with a consistent 2D pattern in PBMCs from healthy donors.
Expressed, wild-type p53 in primary AML blasts exists in several acidic isoforms not seen in resting normal cells. (A) Expression of wild-type p53 was detected at high levels in AML blast samples. However, the acidity and expression level of p53 isoforms was variable among AML patients, such that the 2D pattern of p53 created a signature that distinguished AML patients from one another. (B) In contrast, full-length wild-type p53 was expressed at low levels (53-kDa band) and with a consistent 2D pattern in PBMCs from healthy donors.
p63, phosphorylated p53, full-length p53, and Δp53 isoforms are expressed in AML cells
We asked whether phosphorylated p53 was present within the identified isoform groups of p53. We performed 2D-PAGE immunoblotting for total p53 or p53 phosphorylated at serines 37, 46, or 392 from unstimulated and irradiated Molm-13 (AML derived; Figure 2A) and LNCaP (prostate cancer derived; Figure 2B) cells that both express wild-type p53.27,28 Equivalent results were obtained for both cell types. In resting Molm-13 and LNCaP cells, full-length p53 and Δp53 isoforms were detected at the same pI as in normal PBMCs and AML blast cells. Phosphorylation of p53 was low or not detected in unstimulated cells. At 2 hours following irradiation (IR; 25 Gy), phosphorylation of full-length p53 was detected. Phosphorylation of serines 37 and 392 was detected in more than 5 spots of the 53-kDa band, whereas phosphorylation of serine 46 was observed in fewer spots (Figure 2A-B). Phosphorylation of Δp53 was not observed at 2 hours following irradiation at any of the residues tested.
Characterization of p53 isoforms by 2D-PAGE immunoblotting. Phosphorylated p53 was detected by 2D-PAGE in resting and irradiated (A) Molm-13 and (B) LNCaP cells that express wild-type p53. Following IR, expression of total full-length p53 increased, a characteristic laddering was detected, and phosphorylation occurred at serines 37, 46, and 392, primarily in the full-length p53 isoform group. Following IR, no change in reactivity with phospho-specific antibodies was observed for p63 or Δp53 isoforms. (C) The 63-kDa isoform reactive with the BP53-12 antibody in normal PBMCs was p63. Also seen was a triplet of low-molecular-weight α-p63–reactive spots that were not detected by BP53-12. (D) p63, full-length p53, and the putative p53 truncation (designated Δp53) were the 3 major isoform groups reactive with BP53-12.
Characterization of p53 isoforms by 2D-PAGE immunoblotting. Phosphorylated p53 was detected by 2D-PAGE in resting and irradiated (A) Molm-13 and (B) LNCaP cells that express wild-type p53. Following IR, expression of total full-length p53 increased, a characteristic laddering was detected, and phosphorylation occurred at serines 37, 46, and 392, primarily in the full-length p53 isoform group. Following IR, no change in reactivity with phospho-specific antibodies was observed for p63 or Δp53 isoforms. (C) The 63-kDa isoform reactive with the BP53-12 antibody in normal PBMCs was p63. Also seen was a triplet of low-molecular-weight α-p63–reactive spots that were not detected by BP53-12. (D) p63, full-length p53, and the putative p53 truncation (designated Δp53) were the 3 major isoform groups reactive with BP53-12.
2D-PAGE analysis of cell extracts from normal PBMCs using antibodies against p63 indicated the isoform group detected by BP53-12 at 63 kDa was likely p63 protein (Figure 2C). An additional p63 isoform group of low molecular weight (spot triplet of approximately 35-kDa molecular mass) was specifically detected by α-p63 and not by BP53-13. An example 2D-PAGE immunoblot of normal PBMCs (Figure 2D) shows the 3 major isoform groups designated p63 (63 kDa), p53 full-length (53 kDa), and Δp53 (47 kDa).
Wild-type p53 is phosphorylated and accumulates following DNA damage in AML blasts
Following cell stress, wild-type p53 in normal cells is rapidly phosphorylated, binds DNA, and activates expression or repression of target genes.8–10 We asked how p53 was regulated following DNA damage in AML using an in vitro model of idarubicin chemotherapy. AML blast cells from each of 6 patients were treated with idarubicin for 0 hours, 2 hours, or 4 hours prior to 2D-PAGE mapping of p53 (representative immunoblots shown in Figure 3A). Following idarubicin treatment, full-length p53 accumulated and shifted to a more acidic pI. We quantitated changes in the pI of full-length p53 detected in treated AML blasts as the shift of full-length p53 toward a lower, more acidic pI relative to p53 in untreated cells (n = 6; Figure 3B). The density of full-length p53 increased following DNA damage and the density of Δp53 decreased following DNA damage (Figure 3A).
Phosphorylation of wild-type p53 is induced by DNA damage in AML. (A) Idarubicin treatment of AML blasts led to rapid accumulation of numerous full-length p53 isoforms of high acidity. The blue line indicates the resting pI of the most acidic full-length p53 isoform for this representative AML patient sample (AML-P18). (B) The change in pI of full-length p53 following idarubicin treatment was measured in AML blast samples from 6 patients. The acidic movement of full-length p53 relative to that in untreated cells was statistically significant (n = 6). Similar results were obtained for normal PBMCs. (C) Phosphorylation of p53 at 5 residues (serines 15, 20, 37, 46, and 392) and total per-cell level of p53 were detected by phospho-specific flow cytometry in AML blast samples from 6 patients. Phosphorylation and accumulation of p53 in AML blast samples following DNA damage was measured as the log10-fold increase in MFI at 4 hours following idarubicin treatment, as compared to cells treated with idarubicin and fixed immediately (0 hours). Black indicates a comparable MFI was observed at 4 hours and 0 hours.
Phosphorylation of wild-type p53 is induced by DNA damage in AML. (A) Idarubicin treatment of AML blasts led to rapid accumulation of numerous full-length p53 isoforms of high acidity. The blue line indicates the resting pI of the most acidic full-length p53 isoform for this representative AML patient sample (AML-P18). (B) The change in pI of full-length p53 following idarubicin treatment was measured in AML blast samples from 6 patients. The acidic movement of full-length p53 relative to that in untreated cells was statistically significant (n = 6). Similar results were obtained for normal PBMCs. (C) Phosphorylation of p53 at 5 residues (serines 15, 20, 37, 46, and 392) and total per-cell level of p53 were detected by phospho-specific flow cytometry in AML blast samples from 6 patients. Phosphorylation and accumulation of p53 in AML blast samples following DNA damage was measured as the log10-fold increase in MFI at 4 hours following idarubicin treatment, as compared to cells treated with idarubicin and fixed immediately (0 hours). Black indicates a comparable MFI was observed at 4 hours and 0 hours.
We measured the phosphorylation of p53 at serines 15, 20, 37, 46, and 392 using phospho-specific flow cytometry in primary AML blast cells treated for 0 hours or 4 hours with idarubicin (Figure 3C). The degree of p53 phosphorylation was quantitated as the log10 ratio of 4 hours idarubicin treatment compared with control AML patient samples exposed to idarubicin and then immediately fixed (0 hours treatment). Levels of total p53 protein were also determined in these cells (Figure 3C) using the DO7 α-p53 antibody (Table S2). As controls, cell size and autofluorescence were also measured and showed no change over time following idarubicin treatment (Figure 3C). Following idarubicin treatment, increases in p53 phosphorylation varied among AML patient samples, but followed a general pattern of increased phosphorylation and accumulation of p53. In no patient did p53 phosphorylation decrease; however, the level of phosphorylation was low for some residues in certain samples (eg, serine 392 in AML-P01, AML-P02, and AML-P05).
High levels of Bcl-2 protein expression are detected in AML patients expressing phosphorylated p53
Phospho-specific flow cytometry was used to detect p53 phosphorylation in resting AML blasts from 30 patients. For each of the measured parameters (Bcl-2, p53, p-p53-S15, p-p53-S20, p-p53-S37, p-p53-S46, and p-p53-S392) we calculated the MFI of AML blasts from each patient (Figure 4A). To normalize the fluorescent scale, the MFI of each parameter for each sample was compared to the median MFI for the group. For example, Bcl-2 expression in a particular patient sample that was above the median for AML was colored yellow, whereas Bcl-2 expression below the median was colored blue.
A coordinate profile of Bcl-2 expression, p53 expression, and p53 phosphorylation is observed in primary AML samples. (A) Expression of Bcl-2, p53, p-p53-ser15, p-p53-ser20, p-p53-ser37, p-p53-ser46, and p-p53-ser392 was measured for all 30 patient samples. Three clusters of p-p53 patterns were detected: low-phospho-p53; phosphorylation at serine 20 or 37; or phosphorylation at serine 15, 46, or 392. (B) Correlations between p53 phosphorylation and Bcl-2 expression were analyzed by linear regression. Data for each of 30 AML patient samples were plotted in 2 dimensions to determine dependence. Bcl-2 was significantly correlated with p53 phosphorylation at serines 15, 46, and 392.
A coordinate profile of Bcl-2 expression, p53 expression, and p53 phosphorylation is observed in primary AML samples. (A) Expression of Bcl-2, p53, p-p53-ser15, p-p53-ser20, p-p53-ser37, p-p53-ser46, and p-p53-ser392 was measured for all 30 patient samples. Three clusters of p-p53 patterns were detected: low-phospho-p53; phosphorylation at serine 20 or 37; or phosphorylation at serine 15, 46, or 392. (B) Correlations between p53 phosphorylation and Bcl-2 expression were analyzed by linear regression. Data for each of 30 AML patient samples were plotted in 2 dimensions to determine dependence. Bcl-2 was significantly correlated with p53 phosphorylation at serines 15, 46, and 392.
Data from AML patient samples were clustered according to p53 phosphorylation at serines 15, 20, 37, 46, and 392 and characterized as having one of 3 patterns of p53 phosphorylation, designated low phospho-p53, phosphorylation at serine 20 or 37, or phosphorylation at serine 15, 46, or 392 (Figure 4A). Patients with a pattern of high p53 phosphorylation at serines 15, 46, and 392 tended to express a high amount of Bcl-2 (Figure 4A); total p53 protein expression was also very high in these patients (Figure 4A). Patients with a pattern of p53 phosphorylation at serines 20 or 37 (and lacking other p53 phosphorylations) did not consistently have a particular level of Bcl-2 expression. AML patients with low p53 phosphorylation typically expressed lower than the median amount of Bcl-2 (Figure 4A).
The relationship between Bcl-2 level and p53 phosphorylation was plotted for 30 AML patients and the correlation coefficients determined to gauge whether the association had a specific threshold for association or was the result of a scalable relationship. No correlation of Bcl-2 expression with serines 20 and 37 was observed (Figure 4B). A scalable correlation between Bcl-2 level and phosphorylation of serines 15, 46, and 392 was observed over a wide range of Bcl-2 expression (Figure 4B). The observed correlations between p53 phosphorylation and Bcl-2 protein expression were quantitated by linear regression (Table S3) and are reported as r2 values.
A high level of Bcl-2 expression in AML blasts is associated with LM of Flt3 and specific p53 phosphorylations
LM of the receptor tyrosine kinase Flt3 has been observed to drive transcription of antiapoptotic members of the Bcl-2 family.29 Thirteen of the 30 patients in our sampling had a Flt3 LM. The FLT3 genes from 11 of these 13 patient samples had been sequenced as part of another study (R.H., unpublished observations, June 2005) and were classified into 2 groups depending on the type of mutation: (1) Flt3-LM-SPY591, containing a single Y591 phosphorylation site and (2) Flt3-LM-DupY591, containing 2 Y591 phosphorylation sites. We used phospho-specific flow cytometry to evaluate p53 phosphorylation and Bcl-2 protein expression at the single-cell level in these groups (Figure 5A). A higher MFI for Bcl-2 expression was observed in cells from patients with the Flt3-LM-DupY591 sequence than in cells with the Flt3-LM-SPY591 mutation (P < .001; Figure 5B). Primary AML blast cells expressing high amounts of Bcl-2 protein generally expressed high amounts of p53 phosphorylated at serines 15, 46, and 392 (eg, AML-P28). No correlation between p53 phosphorylation at serine 20 or serine 37 with Bcl-2 levels was observed (Figures S2–S3). Additional comparisons of p-p53-S46, p-p53-S392, and total p53 expression at the single-cell level were investigated and are included (Figures S4–S6). The MFI of each AML sample (as shown in Figure 4) was therefore a sufficient representation of these unimodal data.
Identification of Y591 mutation of Flt3 in patients sharing a profile of p53 phosphorylation and Bcl-2 overexpression. (A) Phosphorylation of p53 at serine 15 and Bcl-2 expression were measured in PBMCs from healthy donors or in AML blast cells. Four samples with Flt3 mutations had duplication of Y591 (DupY591). Seven samples had Flt3 mutations that did not duplicate Y591 (SPY591; 6 representatives shown). The remaining samples displayed other Flt3 mutations or were considered wild type (6 representatives shown). (B) Bcl-2 expression, relative to the median in AML blast cells, is shown for normal PBMCs or AML patient samples with different Flt3 mutational status. Significantly greater Bcl-2 expression was observed in samples from AML patients with Flt3 LM that duplicated Y591 than in cells from patients with Flt3 LM that did not (DupY591 versus SPY591; P < .001). Wild-type Flt3 AML patient samples displayed a spread of Bcl-2 values centered at the median observed for all 30 AML samples (a value of 0 on the log10 scale). The median Bcl-2 expression for each group is marked with a black line. (C) A percentage of STAT and MAPK signaling we previously analyzed in this cohort of AML patients3 was plotted for each patient against the Bcl-2 expression in patients without (left plot) or with (right plot) a Flt3 LM. Among patients with a Flt3 LM, potentiated STAT and MAPK signaling was associated with Bcl-2 expression (r2 = 0.51). In contrast, significantly higher levels of aggressive signaling were observed in patients whose Flt3 mutation duplicated Y591, as compared to patients with other Flt3 LMs (P < .01).
Identification of Y591 mutation of Flt3 in patients sharing a profile of p53 phosphorylation and Bcl-2 overexpression. (A) Phosphorylation of p53 at serine 15 and Bcl-2 expression were measured in PBMCs from healthy donors or in AML blast cells. Four samples with Flt3 mutations had duplication of Y591 (DupY591). Seven samples had Flt3 mutations that did not duplicate Y591 (SPY591; 6 representatives shown). The remaining samples displayed other Flt3 mutations or were considered wild type (6 representatives shown). (B) Bcl-2 expression, relative to the median in AML blast cells, is shown for normal PBMCs or AML patient samples with different Flt3 mutational status. Significantly greater Bcl-2 expression was observed in samples from AML patients with Flt3 LM that duplicated Y591 than in cells from patients with Flt3 LM that did not (DupY591 versus SPY591; P < .001). Wild-type Flt3 AML patient samples displayed a spread of Bcl-2 values centered at the median observed for all 30 AML samples (a value of 0 on the log10 scale). The median Bcl-2 expression for each group is marked with a black line. (C) A percentage of STAT and MAPK signaling we previously analyzed in this cohort of AML patients3 was plotted for each patient against the Bcl-2 expression in patients without (left plot) or with (right plot) a Flt3 LM. Among patients with a Flt3 LM, potentiated STAT and MAPK signaling was associated with Bcl-2 expression (r2 = 0.51). In contrast, significantly higher levels of aggressive signaling were observed in patients whose Flt3 mutation duplicated Y591, as compared to patients with other Flt3 LMs (P < .01).
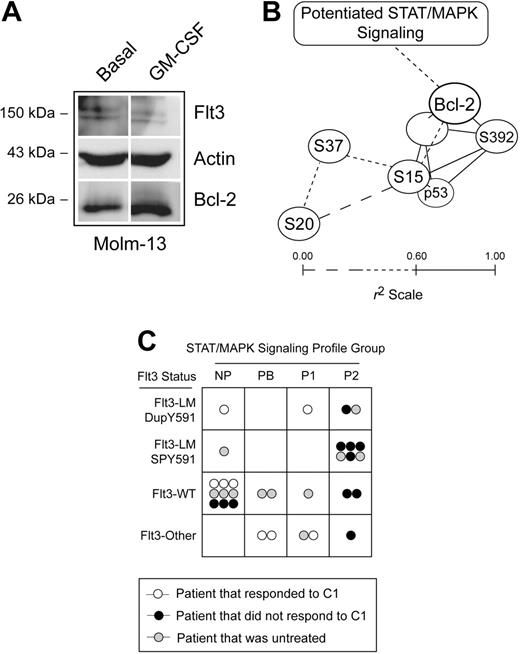
In a previous study we identified profiles of STAT and MAPK signaling responses that correlated with poor responses to clinical outcome in the same patients here examined for Bcl-2 expression and p53 phosphorylation.1 The total strength of STAT and MAPK pathway signaling observed in this study was summarized in a single statistic by adding the proliferative signaling in these patients (gain of Stat3, Stat5, and Erk1/2 signaling responses and loss of Stat1 signaling response) and calculating the percentage of the total observed in AML. Among patients with Flt3 LMs, greater signaling was associated with greater Bcl-2 expression (Figure 5C, P < .01). In contrast, in patients where Flt3 was not detected to be mutant there was no association between signaling profile and Bcl-2 expression (Figure 5C). This connection between previously observed profiles of signaling and Bcl-2 expression suggested that factors observed to trigger potentiated STAT and MAPK signaling, such as GM-CSF, Flt3 ligand, G-CSF, and IL-3 in the previous study,1 might directly increase Bcl-2 expression. To test this idea, we stimulated the AML cell line Molm-13 with GM-CSF and observed an increase in Bcl-2 expression by Western blot (Figure 6A).
Flt3 mutation, potentiated STAT and MAPK signaling, and Bcl-2 expression are part of a common pathway opposing p53 activity in AML patients. (A) Western blots of the Molm-13 cell line left untreated or stimulated with 20 ng/mL GM-CSF were used to determine whether upstream signaling observed to be important in profiles of STAT and MAPK signaling increases the level of Bcl-2 expression. Actin and Flt3 are shown as controls. (B) Relationships between p53 phosphorylation, p53 expression, and Bcl-2 expression were plotted in 3 dimensions based on correlation coefficient (r2) for the 30 AML patient samples. All relationships where r2 was more than 0.60 (solid lines) were plotted. Lines shown are of length 1 − r2 units; the closer the correlation between 2 biomarkers in AML blasts, the closer the lines on this graph. Patients with Flt3-LM-DupY591 expressed higher levels of Bcl-2 and phosphorylated p53, relative to other AML samples, as indicated by brackets. Refer to Figure S1 and Table S3 for more information. (C) A graph summarizes chemotherapy response, AML signaling cluster,1 and Flt3 mutational status. No AML patient with Flt3-LM-SPY591 responded to course one of chemotherapy and these patients primarily displayed a potentiated type 2 signaling profile (SC-P2), as was commonly observed in patients with Flt3-LM. SC-P2 was characterized primarily by potentiated Stat5 and Stat3 signaling responses and an attenuated Stat1 response to IFN-γ. Refer to Table S1 for more information.
Flt3 mutation, potentiated STAT and MAPK signaling, and Bcl-2 expression are part of a common pathway opposing p53 activity in AML patients. (A) Western blots of the Molm-13 cell line left untreated or stimulated with 20 ng/mL GM-CSF were used to determine whether upstream signaling observed to be important in profiles of STAT and MAPK signaling increases the level of Bcl-2 expression. Actin and Flt3 are shown as controls. (B) Relationships between p53 phosphorylation, p53 expression, and Bcl-2 expression were plotted in 3 dimensions based on correlation coefficient (r2) for the 30 AML patient samples. All relationships where r2 was more than 0.60 (solid lines) were plotted. Lines shown are of length 1 − r2 units; the closer the correlation between 2 biomarkers in AML blasts, the closer the lines on this graph. Patients with Flt3-LM-DupY591 expressed higher levels of Bcl-2 and phosphorylated p53, relative to other AML samples, as indicated by brackets. Refer to Figure S1 and Table S3 for more information. (C) A graph summarizes chemotherapy response, AML signaling cluster,1 and Flt3 mutational status. No AML patient with Flt3-LM-SPY591 responded to course one of chemotherapy and these patients primarily displayed a potentiated type 2 signaling profile (SC-P2), as was commonly observed in patients with Flt3-LM. SC-P2 was characterized primarily by potentiated Stat5 and Stat3 signaling responses and an attenuated Stat1 response to IFN-γ. Refer to Table S1 for more information.
A graphical summary of the observed relationships between Flt3 mutation, Bcl-2 expression, and specific p53 phosphorylations is shown in Figure 6B. In this map of the correlations observed in this study, the closer 2 nodes are plotted, the greater their observed correlation (r2) across AML patients. The values for Bcl-2 expression, p53 expression, and p53 phosphorylation are included in full with other patient characteristics, such as signaling profiles,1 in Table S1. The previously reported response to course one of chemotherapy, Flt3 LM group, and signaling cluster for each patient, has been summarized graphically in Figure 6C.
Discussion
We showed here that p53 was not only wild type in AML blasts, but was expressed, diversely phosphorylated in resting cells, and able to respond to DNA damage. In a defined subset of AML patients with extremely high levels of phosphorylated p53, we detected very high levels of Bcl-2 expression associated with a specific mutation in Flt3 (Flt3-LM-DupY591). This mutation in Flt3 was associated with a pattern of signaling including potentiated Stat5 activity previously linked to poor clinical outcome in AML.1 Together, these results suggest that Bcl-2 expression links altered signaling in AML blast cells to inactivation of the p53 pathway and enables continued survival of leukemic cells despite expression of wild-type p53 (Figure S7).
We identified several p53 isoforms in normal cells and AML blasts (Figure 2), and it is notable that most of the variation in p53 among AML patients (Figure 1) constituted differences in phosphorylation of full-length p53 (Figure 2). TP53 was wild type and expressed in all 18 samples from AML patients evaluated and the highest pI of the spot for full-length p53 was at a pH of approximately 6.5, in agreement with the computationally predicted pH of 6.48 for unphosphorylated, full-length p53 protein (Figure 1). These results suggest that differences in p53 levels among AML patients detected by flow cytometry (Figures 3,Figure 4–5) also represent differences in full-length, wild-type p53.
Following DNA damage, p53 protein is normally phosphorylated at several residues.9 Idarubicin treatment (Figure 3) and DNA damage by irradiation (Figure 2) led to accumulation of p53 in a highly phosphorylated form in both 2D-PAGE and flow cytometry assays. This increase in acidity of full-length p53 suggests that normal phosphorylation of p53 in response to DNA damage was intact in AML blast cells. The use of idarubicin in these experiments was relevant to AML therapy because most first-line therapies currently include an anthracycline. Phospho-specific 2D-PAGE for p53 indicated that phosphorylation of p53 in response to DNA damage occurred primarily within the full-length p53 isoform group (Figure 2A). We did not examine in detail p53 transcriptional activity in primary AML blast cells here; however, studies by our group have shown that expression of p53 target genes, including p21 and Bax, can be detected in AML blasts during in vivo therapy.30 Typically, the p53 response of malignant cells is disabled by loss of heterozygosity of p53 or by loss of p53 pathway effectors.6,31 The finding that wild-type p53 in AML blast cells was phosphorylated following DNA damage caused by a common chemotherapeutic agent is particularly striking (Figure 3). These data suggest that there exists a block to p53 pathway-driven apoptosis downstream of p53 transcriptional activity and that therapeutic removal of this block could increase the efficacy of chemotherapy for AML.
A key question raised by our results was why AML blast cells were able to undergo malignant transformation and accumulate genetic lesions, such as FLT3 LM, despite expression of wild-type, active p53. Our observation that Bcl-2 expression was greatest in AML blast cells expressing high levels of phosphorylated p53 (Figure 4) suggests that signaling-driven expression of antiapoptosis proteins inactivates the p53 pathway in AML. At both the individual cancer-cell level (Figure 4A) and in populations of malignant cells from individual AML patients (Figure 4B; Table S2), the level of p53 phosphorylation at serines 15, 46, and 392, but not at serines 20 and 37, was correlated with Bcl-2 protein expression. Flt3 drives transcription of antiapoptotic members of the Bcl-2 family (eg, Bcl-2 and Bcl-XL)29 and Flt3-LM might also increase Bcl-2 expression via activation of Stat5.24 Those patients with the highest levels of p53 phosphorylation and Bcl-2 expression were nearly all members of the same Flt3 LM sequence group, Flt3-DupY591 (P < .001, Figure 5B). We initially hypothesized that, despite expressing phosphorylated, wild-type p53, the duplication of Flt3 Y591 and associated overexpression of Bcl-2 in these patients might be indicators of a more chemoresistant disease. Instead, we found that AML patients with the worst chemotherapy response were those with SP-Y591 (Figure 6B). p53 phosphorylation and Bcl-2 expression was present in these samples, but not at a high level relative to other AML samples (Figure 5A), and these patients displayed a uniformly poor response to course one of chemotherapy (Figure 6B).
To further address the role of Bcl-2 expression in the survival of AML cells expressing wild-type Flt3, Flt3-LM-SPY591, and Flt3-LM-DupY591, we treated representative patient samples in vitro with idarubicin and the Bcl-2 inhibitor HA14-1 (data not shown). However, all groups had essentially equivalent levels of cell death in response to chemotherapy—both following idarubicin alone and following idarubicin combined with the Bcl-2 inhibitor HA14-1. In vitro chemotherapy response is established as a predictive factor for therapy response in childhood lymphoblastic leukemia,32,33 and may also reflect prognostic molecular features of these leukemias.34 In contrast, in AML the relation between in vitro chemotherapy response, prognosis, and molecular features is less clear.35,36 Our own experiences studying in vitro spontaneous and drug-induced apoptosis in primary AML cells, including patients with various Flt3 mutational status, have so far not convinced us that chemotherapy response reflects important prognostic features (data not shown, Bruserud et al,21 and Ryningen et al37 ). These results indicate that killing of AML cells in vitro (over a period of several hours) will likely be a less useful measurement than their immediate signaling biology, including basal phosphorylation and responses to stimulation (ie, a signaling profile including p53 phosphorylation).
Flt3-LM-DupY591 was associated with Bcl-2 overexpression (Figure 5B) and a profile of STAT/MAPK signaling that includes potentiated Stat5 signaling (Figure 5C). These results suggest that duplication of Flt3 Y591 is a particularly potent signaling amplifier able to overcome very high expression of p53 in AML blast cells. In contrast, apoptotic signaling in AML blasts from patients with Flt3-LM-SPY591 might be compromised by changes other than potentiated signaling responses and Bcl-2 overexpression, such as loss of Stat1 pathway activity or a loss of proapoptotic Bax.16 Consistent with our observations of clinical outcome (Figure 6C), we hypothesize that a loss of proapoptosis pathway activity coupled with potentiated signaling (as with Flt3-LM-SPY591) might compromise DNA damage-based chemotherapies more effectively than large gains in antiapoptosis signaling in the setting of intact DNA damage responses (as with Flt3-LM-DupY591). In both cases, we would expect blocks to antiapoptosis signaling pathways to improve the efficacy of DNA damage-based chemotherapies (Figure S7).
The results presented here support a model where altered signaling in AML cells, including potentiated Stat5 signaling responses associated with Flt3 duplication at Y591, contributes to a shift in the balance of proapotosis and antiapoptosis Bcl-2 family members and enables AML blasts to resist induction of cell death by chemotherapy despite an apparently intact p53 pathway. Treatment of AML by blocking Flt3 or its signaling effectors (eg, Stat5) and Bcl-2, combined with chemotherapy, might be more effective at reversing apoptosis suppression and inducing rapid death of AML blast cells than conventional chemotherapy alone.
Authorship
Conflict-of-interest disclosure: G.P.N. is a paid consultant of BD Biosciences, a vendor of antibodies and flow cytometry instrumentation.
J.M.I. and N.Å. contributed equally to the study.
Correspondence: Bjørn Tore Gjertsen, Department of Internal Medicine, Hematology Section, University of Bergen, Haukeland University Hospital, N5021 Bergen, Norway; e-mail: bjorn.gjertsen@med.uib.no; and Garry P. Nolan, Department of Microbiology and Immunology, Baxter Laboratories for Genetic Pharmacology, Stanford University School of Medicine, Stanford, CA 94035; e-mail: gnolan@stanford.edu.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
This work was supported by the Norwegian Research Council Functional Genomics Program (FUGE) grant number 151859 and by the National Heart, Lung and Blood Proteomics Center Contract N01-HV-28183I. B.T.G and Ø.B. were supported by the Norwegian Cancer Society (Kreftforeningen). R.H. was supported by a Helse Vest HF research grant. J.M.I was supported by the G. J. Lieberman Fellowship and is a Leukemia and Lymphoma Society Fellow.
We thank A. Ryningen for several useful discussions of Bcl-2 family members and R. Ihrie for review of this manuscript.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal