Abstract
The synthesis of interferon-β (IFNβ) and IFN-inducible factors elicited by lipopolysaccharide (LPS) depends on the transcriptional activity of interferon regulatory factor 3 (IRF-3) downstream of Toll-like receptor-4 (TLR4). To examine the ability of human newborns to mount TLR4-mediated IRF-3–dependent responses, we analyzed the pattern of genes expressed on the addition of LPS to cord blood or cord blood monocyte-derived dendritic cells (moDCs). Expression of IFNβ and IFN-inducible genes was selectively impaired in neonatal blood and moDCs as compared with their adult counterparts. This selective defect was confirmed by microarray experiments on moDCs. Altered expression of IFN-inducible genes was related to impaired IFNβ synthesis because IFNβ signaling was functional in neonatal moDCs. However, addition of exogenous IFNβ failed to restore LPS-induced IL-12p70 synthesis which was previously shown to be defective in neonatal moDCs. Although LPS-induced IRF-3 nuclear translocation was observed both in adult and neonatal moDCs, IRF-3 DNA-binding activity and association with the coactivator CREB-binding protein (CBP) were decreased in neonatal as compared with adult moDCs. We conclude that impaired IRF-3/CBP interaction in neonatal blood cells exposed to LPS is associated with impaired expression of IFNβ and IFN-inducible genes. Because IRF-3 activity is also required for IL-12p70 synthesis, our findings provide a molecular basis for the decreased ability of LPS-stimulated neonatal moDCs to elicit Th1-type responses.
Introduction
The immaturity of the immune system in early life results in increased susceptibility to infections and suboptimal responses to vaccines because of impaired B-cell and T-cell responses.1,2 Neonatal CD4+ T cells are indeed unable to mount efficient Th1-type responses to most stimuli with the exception of BCG vaccine.3,4 Intrinsic T-cell defects responsible for impaired Th1 differentiation in neonates include hypermethylation status of the ifng locus and deficient up-regulation of CD40 ligand expression at the T-cell membrane.5,6 In human newborns, we found that decreased production of interleukin (IL)–12p70 by myeloid dendritic cells (DCs) in response to several stimuli represents an additional contributing factor.7,8 However, there is clear evidence that IL-12 plays a redundant role in the induction of protective immune responses during natural exposure to most intracellular microorganisms.9 Type I interferons (IFNs) are also critically involved in such responses.10
LPS and its analogs are known to promote Th1 responses,11 and it is therefore important to decipher LPS-induced responses in neonatal cells. The critical receptor for LPS is Toll-like receptor-4 (TLR4) which is coupled to 2 major signaling pathways: (1) the “myeloid differentiation factor 88 (MyD88)–dependent” pathway involving MyD88 and TIR (Toll/IL-1R)–containing adaptor protein (TIRAP) and leading to early NF-κB activation and synthesis of several inflammatory cytokines, and (2) the “MyD88-independent pathway” involving TIR-containing adaptor inducing interferon IFNβ (TRIF) and TRIF-related adaptor molecule (TRAM) and leading to activation of interferon regulatory factor 3 (IRF-3), IFNβ synthesis, and expression of IFN-inducible genes.12 We observed that the expression of IFNβ and IFN-inducible genes is specifically impaired in LPS-stimulated cord blood or cord blood monocyte-derived dendritic cells (moDCs) as compared with their adult counterparts. This led us to explore IFNβ signaling and IRF-3 activation events in neonatal moDCs.
Material and methods
Cells and reagents
Human cord blood was obtained from the placentas of normal full-term deliveries at the obstetric departments of CHU Tivoli-La Louvière, CHU-Charleroi, and Clinique Notre Dame, Charleroi. Buffy coats and whole peripheral adult blood samples were obtained from local routine blood donations or from healthy volunteers, respectively. This study was approved by the ethical committee of the Hôpital Erasme, Brussels, and the animal welfare committee of the faculty of medicine of the Université Libre de Bruxelles, Belgium. Informed consent was obtained for all cord blood and adult blood samples. moDCs were generated from cord blood mononuclear cells or peripheral blood mononuclear cells, as previously described.7 Lipopolysaccharide (LPS) from Escherichia coli (0128:B12) was obtained from Sigma-Aldrich (Bornem, Belgium). Recombinant human IFNβ (Avonex) was purchased from Biogen Idec (Brussels, Belgium).
Whole blood and moDCs stimulation
Heparinized whole cord or adult blood (200 μL) was stimulated at 37°C with the indicated concentration of LPS. To assess the role of plasma factors, hemocytes were collected by centrifugation, washed 3 times, and resuspended in either neonatal or adult plasma. moDCs were cultured in RPMI 1640 (Cambrex, Verviers, Belgium) supplemented with 2 mM L-glutamine, 20 μg/mL gentamicin, 50 mM β2-Mercapto-ethanol, 1% nonessential amino acids, and 10% fetal calf serum (Hyclone, Perbio, Erembodegem, Belgium). moDCs were stimulated at 37°C with LPS (0.1-1 μg/mL).
RNA purification and real-time reverse transcriptase–polymerase chain reaction (RT-PCR)
mRNA levels from whole blood assays were quantified as described by Stordeur et al.13 Total RNA or mRNA was extracted on the MagnaPure instrument following the manufacturer's instruction (Roche Diagnostics, Brussels, Belgium). Primer sequences are listed in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Quantification of cytokine production
Human IFNβ (detection limit, 1 IU/mL) and IL-12p70 (detection limit, 15 pg/mL) levels were determined after 24 hours of incubation in cell-free supernatants by specific enzyme-linked immunoabsorbent assay (ELISA; Biosource, Nivelles, Belgium, and R&D Systems, Abingdon, United Kingdom, respectively). CXCL10 serum levels (detection limit, 15 pg/mL) were determined using Duoset ELISA (R&D Systems).
Microarray experiments
RNA preparation.
Adult and cord blood moDCs (3 donors in each group) were left untreated or stimulated with LPS (1 μg/mL) for 6 hours. Total RNA was isolated using Tripure (Roche Diagnostics).
RNA preparation and Affymetrix GeneChip hybridization.
The quality of the RNAs was checked with an Agilent 2100 bioanalyzer (Agilent Technologies, Palo Alto, CA). All the RNAs used in the experiment met the quality criteria defined by Agilent. Genes expressed in each sample were analyzed on a high-density oligonucleotide microarray (U133A; Affymetrix, Santa Clara, CA) containing 22 690 transcripts. Target preparation and microarray processing procedures were performed as described in the Affymetrix GeneChip Expression Analysis Manual (http://www.affymetrix.com/support/technical/manual/expression_manual.affx). Briefly, 3 μg total RNA was used to synthesize double-stranded cDNA with SuperScript II reverse transcriptase (Life Technologies, Rockville, MD) and a T7–(dT)24 primer (Proligo, Paris, France). Then, biotinylated cRNA was synthesized from the double-stranded cDNA with the RNA transcript labeling kit (Enzo Life Sciences, Farmingdale, NY) and was purified and fragmented. The fragmented cRNA was hybridized into the oligonucleotide microarray, which was washed and stained with streptavidin-phycoerythrin. Scanning was performed with an Agilent microarray scanner.
Data analysis.
All the “dat” files (corresponding to the scan image) and the “rpt” files (corresponding to each GeneChip) were first checked for the control of hybridization defined by the Affymetrix GeneChip manual (uniformity of the signal, good position of the grid, scale factor, background, percentage of presence, and presence of housekeeping genes and spike control). All experiments were also analyzed with the “deg” algorithm associated with Bioconductor 1.3 software (http://www.bioconductor.org/). This algorithm analyzes the quality of the mRNA by comparing the signal intensities in the 5′ and 3′ ends of the mRNA, which should be similar if no RNA degradation has occurred. All quality controls were positive. All data analyses were performed with the use of the R statistics software (available at www.r-project.org) and various Bioconductor packages for R (available at www.bioconductor.org). In particular, the simpleaffy package was used to run the Affymetrix QC tests, which were passed by all chips. The raw data were preprocessed for background correction, normalization, and probe summarization by the RMA method.14 The differential gene expression between any 2 groups was computed by 2 different statistical methods, the RankProduct15 and the CyberT16 methods. In each of them, a correction for multiple testing was done with the method of Benjamini and Hochberg,17 and the threshold of 5% was chosen for the false discovery rate; thus, the genes had to have a corrected P value less or equal to .05 to be considered as differentially expressed. For each 2-group comparison, the final set of differentially expressed genes was obtained as the intersection of the gene lists obtained by both methods.
Detection of IRF-3 and NF-κB by immunofluorescence
moDCs were adhered onto glass coverslips for at least 1 hour before stimulation. Cells were fixed with PBS containing 2% paraformaldehyde and stored in methanol. For the staining, cells were rehydratated in PBS and incubated with PBS-BSA 2%. Permeabilization with PBS containing 0.1% Triton X-100 and 2% BSA was followed by application of the following primary antibodies: IRF-3 mouse monoclonal (BD Biosciences, Erembodegem, Belgium) and p65/Rel A rabbit polyclonal (Santa Cruz Biotechnology, Santa Cruz, CA). moDCs were then fluorescently labeled with secondary antibodies (anti–mouse Alexa 488 and anti–rabbit Alexa 568; Molecular Probes, Invitrogen, Merelbeke, Belgium). The coverslips were washed and mounted with Vectashield antifade solution before being analyzed. Immunofluorescence was visualized by using an epi-fluorescence microscope (Nixon Eclipse 80i; Analis, Namur, Belgium) with a 20× Plan Apochromat objective and equipped with a digital camera.
Detection of IRF-3 DNA-binding activity
IRF-3 binding activity in nuclear extracts was measured with Trans-AM IRF transcription factor assay kit (Active Motif Europe, Rixensart, Belgium) according to the manufacturer's protocol. Briefly, nuclear extracts (5 μg) were incubated with plate-coated ISRE consensus oligonucleotide. Plates were washed and anti–IRF-3 antibody was added to the wells. Antibody binding was detected with a secondary HRP-conjugated antibody and developed with TMB substrate. The intensity of the reaction was measured at 450 nm.
Immunoblotting and immunoprecipitation
moDCs (106 cells) were lysed in Laemmli buffer and equal volumes of cell lysate from each condition were resolved by 8% to 10% sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). Analysis of protein phosphorylation was visualized by direct Western blotting using antibodies directed against phosphor–Stat-1 (Cell Signaling Technology, Leiden, The Netherlands). To control loading, immunoblots were reprobed with anti–Stat-1 (Santa Cruz Biotechnology). To analyze IRF-3 and p65 nuclear translocation, 15 μg nuclear extract from each condition was fractioned onto 8% SDS-PAGE. The immunoblots were analyzed using anti–IRF-3 or anti-p65 antibodies (Santa Cruz Biotechnology), and the loading was controlled by an anti–USF-2 antibody. For coimmunoprecipitation studies, whole-cell lysates (WCLs) from 3 × 106 moDCs were lysed in 400 μL buffer (50 mM Tris pH 7.4, 150 mM NaCl, 5 mM EDTA pH 8.0, 30 mM NaF, 1 mM Na3V04, 40 mM β-glycerophosphate, 0.1 mM PMSF in EtOH, protease inhibitors cocktail [Roche Diagnostics], 10% glycerol, and 1% Nonidet-P40) and then incubated overnight with 1 μg anti-CBP antibody (C-1; Santa Cruz Biotechnology). WCLs were incubated with 60 μL of 1:1 slurry of protein A and G sepharose beads (GE Healthcare, Diegem, Belgium) for 2 hours at 4°C. The beads were washed 5 times with lysis buffer and resuspended in equal volume of Laemmli buffer.
Results
IFNβ gene expression is defective in neonatal blood exposed to LPS
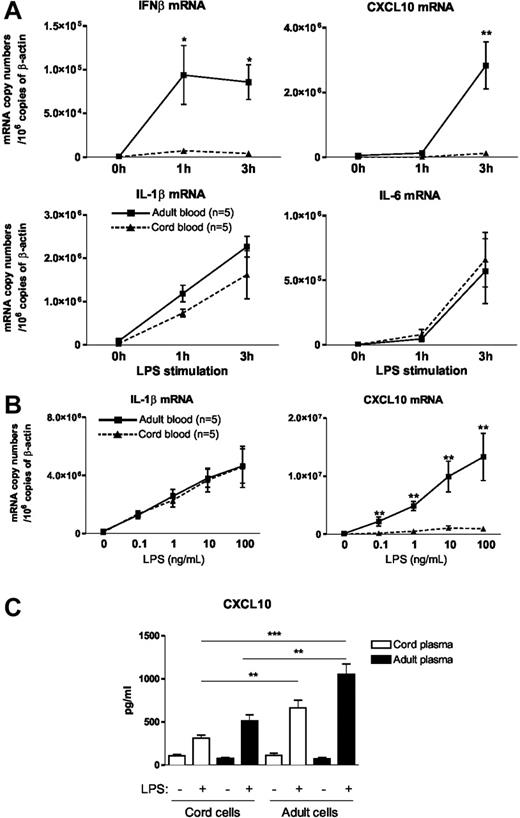
In a first set of experiments, we used real-time RT-PCR to measure IFNβ; gene expression in LPS-stimulated whole blood obtained from human placental cord and adults. As shown in Figure 1A, we found that the addition of LPS to cord blood failed to induce IFNβ mRNA expression that was clearly detected in adult blood. In line with this seminal observation, the induction of CXCL10, a prototypic IFN-dependent gene,18 was also defective in cord blood. In contrast, induction of IL-1β and IL-6 mRNA was comparable in cord and adult blood. Because IL-1β; and IL-6 gene expression merely depends on the MyD88-dependent signaling pathway whereas the expression of IFNβ; and CXCL10 is TRIF/TRAM dependent, these results suggest that the MyD88-independent TLR4 signaling pathway is preferentially affected in neonatal blood cells. To determine whether similar conclusions could be drawn under suboptimal conditions of stimulation, we performed dose-response curves of LPS. As shown in Figure 1B, we observed similar dose-dependent induction of IL-1β mRNA in adult and cord blood samples. In contrast, CXCL10 mRNA levels were strongly reduced in neonatal blood, whatever the LPS concentration used. These experiments indicate that the selectivity of the defect that we have observed in LPS-stimulated cord blood is not influenced by the strength of the stimulation. Next, to determine the impact of circulating factors on these differences, we examined CXCL10 release from cord and adult hemocytes resuspended in either adult or cord plasma. We first confirmed that CXCL10 protein levels released on LPS challenge were strongly reduced in cord blood (cord cells in cord plasma) as compared with adult blood (adult cells in adult plasma) (Figure 1C). When cord blood cells were resuspended in adult plasma, LPS-induced CXCL10 levels increased slightly. Conversely, the addition of neonatal plasma to adult cells reduced CXCL10 levels elicited by LPS. Although the differences did not reach statistical significance, these results suggest that plasma factors could contribute to impaired TLR4-mediated CXCL10 production. Importantly, the differences between adult and cord blood cells remained statistically significant when both cell populations were resuspended in the same plasma from the same origin (adult or newborn), confirming that intrinsic cellular factors contribute to the differences observed between adult and cord blood.
Impaired IFNβ synthesis in LPS-stimulated neonatal blood. (A) Heparinized cord or adult blood was incubated with LPS (10 ng/mL) for the indicated time. mRNA was then extracted and real-time RT-PCR was performed. Results are expressed in mRNA copy numbers per 106 β-actin mRNA copies. The mean and SEM obtained from 5 different samples are shown. *P < .05 and **P < .01 cord versus adult blood. (B) Whole cord and adult blood samples were incubated with the indicated LPS concentration for 3 hours. The mean and SEM obtained from 5 different samples are shown. (C) Cord and adult hemocytes were washed and resuspended in either adult or neonatal plasma. Blood cells were stimulated for 24 hours in the presence of LPS (10 ng/mL). CXCL10 levels were analyzed by specific ELISA. Mean and SEM obtained from 10 different blood samples are shown. **P < .01 and ***P < .001.
Impaired IFNβ synthesis in LPS-stimulated neonatal blood. (A) Heparinized cord or adult blood was incubated with LPS (10 ng/mL) for the indicated time. mRNA was then extracted and real-time RT-PCR was performed. Results are expressed in mRNA copy numbers per 106 β-actin mRNA copies. The mean and SEM obtained from 5 different samples are shown. *P < .05 and **P < .01 cord versus adult blood. (B) Whole cord and adult blood samples were incubated with the indicated LPS concentration for 3 hours. The mean and SEM obtained from 5 different samples are shown. (C) Cord and adult hemocytes were washed and resuspended in either adult or neonatal plasma. Blood cells were stimulated for 24 hours in the presence of LPS (10 ng/mL). CXCL10 levels were analyzed by specific ELISA. Mean and SEM obtained from 10 different blood samples are shown. **P < .01 and ***P < .001.
Impaired synthesis of IFNβ and defective expression of IFN-inducible genes in LPS-stimulated neonatal moDCs
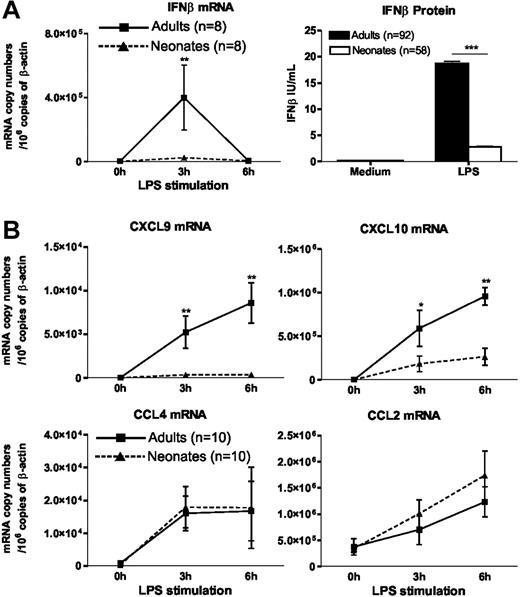
To explore the cellular and molecular mechanisms leading to impaired IFNβ synthesis in LPS-stimulated cord blood, we performed additional experiments on DCs differentiated from cord or adult blood monocytes (moDCs) under the same culture conditions. This approach allows the study of LPS-induced responses of neonatal cells in the absence of inhibitory factors present in cord blood (Figure 1C; Levy et al19 ). First, we confirmed the findings made in whole blood because IFNβ mRNA accumulation in response to LPS was profoundly depressed in neonatal moDCs as compared with their adult counterparts (Figure 2A). The profound defect in IFNβ synthesis was also apparent when we measured IFNβ protein levels in supernatants of LPS-stimulated moDCs (Figure 2A).
Impaired IFNβ synthesis and IFN-dependent gene expression in LPS-stimulated neonatal moDCs. Adult or cord blood moDCs were incubated with LPS (100 ng/mL) for the indicated times. Total RNA was extracted and real-time RT-PCR was performed for mRNA quantification. For determination of IFNβ levels in the supernatants, cells were stimulated for 24 hours. Histograms and error bars represent mean ± SEM. *P < .05, **P < .01, ***P < .001.
Impaired IFNβ synthesis and IFN-dependent gene expression in LPS-stimulated neonatal moDCs. Adult or cord blood moDCs were incubated with LPS (100 ng/mL) for the indicated times. Total RNA was extracted and real-time RT-PCR was performed for mRNA quantification. For determination of IFNβ levels in the supernatants, cells were stimulated for 24 hours. Histograms and error bars represent mean ± SEM. *P < .05, **P < .01, ***P < .001.
As shown in Figure 2B, the expression of CXCL9 and CXCL10 genes, which are known to be IFN-inducible, was also significantly decreased in LPS-stimulated moDCs, whereas the induction of CCL2 and CCL4 genes, which depend on NF-κB activation, was of similar magnitude in neonatal and adult moDCs. The preferential suppression of IFN-inducible genes in LPS-stimulated neonatal moDCs was confirmed using oligonucleotide microarrays. Among the 520 LPS-responsive genes identified in adult moDCs, 58 were found to be significantly less expressed in neonatal moDCs (list of genes available in Table S2). Thirty-three (57%) of these 58 genes were previously shown to be inducible by type I IFNs.20,21 Moreover, among the first 10 genes for which the difference between neonatal and adult cells was the most important, 8 are known to be IFN inducible (Table 1). However, no significant difference of expression was found between adult and neonatal moDCs in a set of 12 NF-κB-inducible genes (Table 2). The latter finding is consistent with our previous observation indicating that LPS-induced NF-κB p65 DNA binding is comparable in neonatal and adult moDCs.8
List of the 10 most differentially expressed genes in LPS-stimulated adult (Ad) versus neonatal (NB) moDCs
| Gene description . | Symbol . | Ad . | NB . | Ad LPS vs NB LPS, fold . | ||
|---|---|---|---|---|---|---|
| Basal* . | LPS* . | Basal* . | LPS* . | |||
| Chemokine (C-X-C motif) ligand 9† | CXCL9 | 27 ± 1 | 1982 ± 997 | 29 ± 3 | 50 ± 11 | 39.6 |
| Chemokine (C-X-C motif) ligand 11† | CXCL11 | 13 ± 1 | 2213 ± 192 | 13 ± 1 | 88 ± 57 | 25.1 |
| Tumor necrosis factor (ligand) superfamily, member 10† | TNFSF10 | 40 ± 13 | 1140 ± 359 | 24 ± 5 | 166 ± 83 | 6.9 |
| Claudin 1 | CLDN1 | 16 ± 1 | 108 ± 59 | 15 ± 1 | 18 ± 3 | 5.9 |
| Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A† | APOBEC3A | 27 ± 3 | 1351 ± 41 | 23 ± 2 | 231 ± 161 | 5.8 |
| Exostoses (multiple) 1† | EXT1 | 54 ± 3 | 468 ± 137 | 44 ± 5 | 82 ± 20 | 5.7 |
| Zinc finger CCCH type, antiviral 1† | ZAP | 54 ± 3 | 628 ± 79 | 35 ± 2 | 116 ± 38 | 5.4 |
| Neutrophil cytosolic factor 1 (47kDa) | NCF1 | 500 ± 219 | 2927 ± 629 | 148 ± 51 | 567 ± 115 | 5.2 |
| Homeo box (expressed in ES cells) 1† | HESX1 | 27 ± 5 | 334 ± 95 | 27 ± 2 | 65 ± 26 | 5.1 |
| CD38 antigen (p45)† | CD38 | 89 ± 8 | 1674 ± 242 | 74 ± 8 | 362 ± 145 | 4.6 |
| Sialoadhesin† | SN | 95 ± 7 | 562 ± 194 | 70 ± 11 | 125 ± 33 | 4.5 |
| Gene description . | Symbol . | Ad . | NB . | Ad LPS vs NB LPS, fold . | ||
|---|---|---|---|---|---|---|
| Basal* . | LPS* . | Basal* . | LPS* . | |||
| Chemokine (C-X-C motif) ligand 9† | CXCL9 | 27 ± 1 | 1982 ± 997 | 29 ± 3 | 50 ± 11 | 39.6 |
| Chemokine (C-X-C motif) ligand 11† | CXCL11 | 13 ± 1 | 2213 ± 192 | 13 ± 1 | 88 ± 57 | 25.1 |
| Tumor necrosis factor (ligand) superfamily, member 10† | TNFSF10 | 40 ± 13 | 1140 ± 359 | 24 ± 5 | 166 ± 83 | 6.9 |
| Claudin 1 | CLDN1 | 16 ± 1 | 108 ± 59 | 15 ± 1 | 18 ± 3 | 5.9 |
| Apolipoprotein B mRNA editing enzyme, catalytic polypeptide-like 3A† | APOBEC3A | 27 ± 3 | 1351 ± 41 | 23 ± 2 | 231 ± 161 | 5.8 |
| Exostoses (multiple) 1† | EXT1 | 54 ± 3 | 468 ± 137 | 44 ± 5 | 82 ± 20 | 5.7 |
| Zinc finger CCCH type, antiviral 1† | ZAP | 54 ± 3 | 628 ± 79 | 35 ± 2 | 116 ± 38 | 5.4 |
| Neutrophil cytosolic factor 1 (47kDa) | NCF1 | 500 ± 219 | 2927 ± 629 | 148 ± 51 | 567 ± 115 | 5.2 |
| Homeo box (expressed in ES cells) 1† | HESX1 | 27 ± 5 | 334 ± 95 | 27 ± 2 | 65 ± 26 | 5.1 |
| CD38 antigen (p45)† | CD38 | 89 ± 8 | 1674 ± 242 | 74 ± 8 | 362 ± 145 | 4.6 |
| Sialoadhesin† | SN | 95 ± 7 | 562 ± 194 | 70 ± 11 | 125 ± 33 | 4.5 |
Gene expression is described as mean intensity ± SEM obtained from 3 donors in each group.
IFN-dependent genes.
Expression of NF-κB-dependent genes in moDCs adult (Ad) and neonatal (NB)
| Gene description . | Symbol . | Ad, basal, mean ± SEM . | Adults, LPS, mean ± SEM . | NB, basal, mean ± SEM . | NB, LPS, mean ± SEM . | Ad LPS vs NB LPS, fold . |
|---|---|---|---|---|---|---|
| Chemokine (C-C motif) ligand 2 | CCL2 | 216 ± 110 | 3057 ± 536 | 1282 ± 364 | 6994 ± 862 | 0.4 |
| Chemokine (C-C motif) ligand 3 | CCL3 | 515 ± 168 | 5084 ± 419 | 361 ± 139 | 5630 ± 757 | 0.9 |
| Chemokine (C-C motif) ligand 4 | CCL4 | 599 ± 141 | 7644 ± 672 | 366 ± 132 | 7327 ± 930 | 1.0 |
| Intercellular adhesion molecule 1 (CD54) | ICAM1 | 539 ± 137 | 1732 ± 128 | 544 ± 110 | 1779 ± 145 | 1.0 |
| Interleukin 12B | IL12B | 14 ± 1 | 1857 ± 359 | 15 ± 0.3 | 1074 ± 411 | 1.7 |
| Interleukin 1 α | IL1A | 48 ± 2 | 1729 ± 684 | 95 ± 36 | 2444 ± 591 | 0.7 |
| Interleukin 1 β | IL1B | 251 ± 94 | 7787 ± 1639 | 174 ± 30 | 9972 ± 603 | 0.8 |
| Interleukin 6 | IL6 | 50 ± 4 | 3802 ± 1171 | 52 ± 10 | 2400 ± 185 | 1.6 |
| Interleukin 8 | IL8 | 277 ± 32 | 6141 ± 912 | 1637 ± 995 | 8623 ± 979 | 0.7 |
| IκBα | NFKB1A | 1698 ± 257 | 6672 ± 703 | 928 ± 179 | 3617 ± 139 | 1.8 |
| Prostaglandin-endoperoxide synthase 2 | PTGS2 | 16 ± 1 | 1237 ± 428 | 16 ± 2 | 998 ± 307 | 1.2 |
| Tumor necrosis factor α | TNF | 147 ± 45 | 1870 ± 320 | 77 ± 17 | 1175 ± 452 | 1.6 |
| Gene description . | Symbol . | Ad, basal, mean ± SEM . | Adults, LPS, mean ± SEM . | NB, basal, mean ± SEM . | NB, LPS, mean ± SEM . | Ad LPS vs NB LPS, fold . |
|---|---|---|---|---|---|---|
| Chemokine (C-C motif) ligand 2 | CCL2 | 216 ± 110 | 3057 ± 536 | 1282 ± 364 | 6994 ± 862 | 0.4 |
| Chemokine (C-C motif) ligand 3 | CCL3 | 515 ± 168 | 5084 ± 419 | 361 ± 139 | 5630 ± 757 | 0.9 |
| Chemokine (C-C motif) ligand 4 | CCL4 | 599 ± 141 | 7644 ± 672 | 366 ± 132 | 7327 ± 930 | 1.0 |
| Intercellular adhesion molecule 1 (CD54) | ICAM1 | 539 ± 137 | 1732 ± 128 | 544 ± 110 | 1779 ± 145 | 1.0 |
| Interleukin 12B | IL12B | 14 ± 1 | 1857 ± 359 | 15 ± 0.3 | 1074 ± 411 | 1.7 |
| Interleukin 1 α | IL1A | 48 ± 2 | 1729 ± 684 | 95 ± 36 | 2444 ± 591 | 0.7 |
| Interleukin 1 β | IL1B | 251 ± 94 | 7787 ± 1639 | 174 ± 30 | 9972 ± 603 | 0.8 |
| Interleukin 6 | IL6 | 50 ± 4 | 3802 ± 1171 | 52 ± 10 | 2400 ± 185 | 1.6 |
| Interleukin 8 | IL8 | 277 ± 32 | 6141 ± 912 | 1637 ± 995 | 8623 ± 979 | 0.7 |
| IκBα | NFKB1A | 1698 ± 257 | 6672 ± 703 | 928 ± 179 | 3617 ± 139 | 1.8 |
| Prostaglandin-endoperoxide synthase 2 | PTGS2 | 16 ± 1 | 1237 ± 428 | 16 ± 2 | 998 ± 307 | 1.2 |
| Tumor necrosis factor α | TNF | 147 ± 45 | 1870 ± 320 | 77 ± 17 | 1175 ± 452 | 1.6 |
Exogenous IFNβ restores expression of IFN-inducible genes but not IL-12 p35 synthesis in LPS-stimulated neonatal moDCs
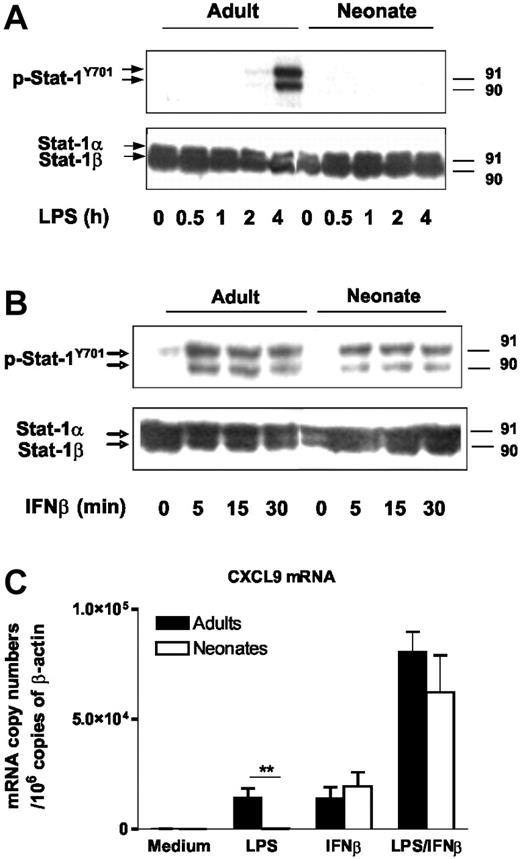
To determine whether the defective expression of IFN-inducible genes in LPS-stimulated moDCs is only related to deficient IFNβ synthesis or also involves a defect in IFNβ signaling, we analyzed Stat-1 phosphorylation at Y701, a critical step in the type I IFN-signaling pathway. As expected, phosphorylation of Stat-1 on tyrosine 701 was induced in LPS-stimulated adult moDCs but not in neonatal DC (Figure 3A). In contrast, Stat-1 phosphorylation in response to exogenous IFNβ was clearly induced in both cell types, suggesting that IFNβ signaling is functional in neonatal cells (Figure 3B). Indeed, exogenous IFNβ induced similar levels of CXCL9 mRNA in adult and cord blood moDCs (Figure 3C). Furthermore, when exogenous IFNβ was added to LPS, we observed synergistic activation of CXCL9 gene expression in both adult and neonatal cells at similar levels. These data establish that impaired expression of IFN-inducible genes in LPS-stimulated neonatal moDCs results from deficient transcriptional activation of the IFNβ; gene.
IFNβ signaling is functional in neonatal moDCs. (A-B) Stat-1 phosphorylation at Y701 in adult and cord blood moDCs was assessed by Western blotting. Cells were stimulated with LPS (100 ng/mL) (A) or recombinant IFNβ (10 U/mL) (B) for the indicated time. Total Stat-1 levels are shown as controls. One representative experiment of 3 is shown. (C) CXCL9 mRNA levels were determined in adult and cord blood moDCs by real-time RT-PCR. Cells were incubated with medium alone, LPS (100 ng/mL), and/or IFNβ (100 U/mL) for 6 hours. Mean and SEM from 5 different donors are shown. **P < .01 cord versus adult moDCs.
IFNβ signaling is functional in neonatal moDCs. (A-B) Stat-1 phosphorylation at Y701 in adult and cord blood moDCs was assessed by Western blotting. Cells were stimulated with LPS (100 ng/mL) (A) or recombinant IFNβ (10 U/mL) (B) for the indicated time. Total Stat-1 levels are shown as controls. One representative experiment of 3 is shown. (C) CXCL9 mRNA levels were determined in adult and cord blood moDCs by real-time RT-PCR. Cells were incubated with medium alone, LPS (100 ng/mL), and/or IFNβ (100 U/mL) for 6 hours. Mean and SEM from 5 different donors are shown. **P < .01 cord versus adult moDCs.
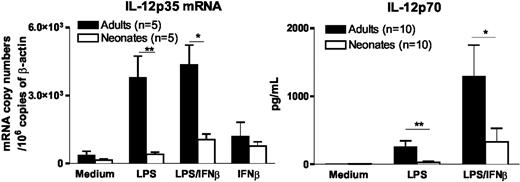
We previously demonstrated that defective IL-12p70 synthesis in LPS-stimulated neonatal moDCs is related to impaired activation of the IL-12p35 gene, whereas their expression of IL-12p40 gene is normal.7 Because a recent report by Gautier et al22 demonstrated a role for type I IFNs in TLR-induced IL-12p35 synthesis, we examined whether the addition of exogenous IFNβ to LPS would restore IL-12p35 gene expression and IL-12p70 synthesis in neonatal moDCs. As shown in Figure 4, IL-12p35 mRNA accumulation and IL-12p70 synthesis remained severely depressed in LPS-stimulated neonatal moDCs even in the presence of exogenous IFNβ. IFNβ alone induced low levels of IL-12p35 mRNA in adult moDCs, and a similar effect was observed in neonatal moDCs. These data indicate that the defect in IL-12p35 gene activation in LPS-stimulated neonatal moDCs is not exclusively caused by deficient IFNβ synthesis.
Impaired IL-12 synthesis in neonatal moDCs is not overcome by exogenous IFNβ. For quantification of IL-12p35 mRNA levels, adult and cord blood moDCs were incubated with medium alone, LPS (100 ng/mL), and/or IFNβ (100 U/mL) for 6 hours. For determination of bioactive IL-12p70 levels in the supernatants by ELISA, cells were stimulated for 24 hours. Mean and SEM from 5 (RT-PCR experiments) and 10 (ELISA experiments) different donors are shown. *P < .05 and **P < .01 cord versus adult moDCs.
Impaired IL-12 synthesis in neonatal moDCs is not overcome by exogenous IFNβ. For quantification of IL-12p35 mRNA levels, adult and cord blood moDCs were incubated with medium alone, LPS (100 ng/mL), and/or IFNβ (100 U/mL) for 6 hours. For determination of bioactive IL-12p70 levels in the supernatants by ELISA, cells were stimulated for 24 hours. Mean and SEM from 5 (RT-PCR experiments) and 10 (ELISA experiments) different donors are shown. *P < .05 and **P < .01 cord versus adult moDCs.
Impaired interaction of IRF-3 with CREB binding protein in neonatal moDCs
IFNβ synthesis on TLR4 stimulation involves interaction of TRIF and TRAM with the 2 noncanonical inhibitor κB kinases (IKKs), TANK-binding kinase 1 (TBK1) and IKKϵ.23 These kinases phosphorylate IRF-3 which undergoes dimerization, translocation, and association with the coactivators CREB-binding protein (CBP) and p300.24 Using Western blotting, we found comparable expression of TRIF, TRAM, IRF-3, IKKϵ, and TBK1 in adult and neonatal moDCs (Figure S1). We compared by immunofluorescence analysis LPS-induced nuclear translocation of NF-κB p65 and IRF-3, which is critically involved in IFNβ; gene activation. As shown in Figure 5A, NF-κB and IRF-3 translocated to the nucleus on LPS stimulation, both in adult and neonatal moDCs. Nuclear translocation of IRF-3 was confirmed by Western blot experiments on nuclear fractions (Figure 5B). We then assessed association of IRF-3 with the coactivator CBP in coimmunoprecipitation experiments. In adult moDCs, we observed the expected interaction of IRF-3 with CBP on LPS stimulation (Figure 5C). Remarkably, this IRF-3/CBP interaction was not detected in neonatal moDCs, whereas NF-κB p65 interaction with CBP was observed in both adult and neonatal cells (Figure 5C). Finally, using an ELISA-based technique which previously documented the induction of NF-κB p65-binding activity in neonatal moDCs,8 we found that LPS-induced IRF-3 DNA binding was significantly reduced in neonatal moDCs (Figure 5D).
Impaired interaction of IRF-3 and CBP in neonatal moDCs. (A-B) Analysis of IRF-3 and NF-κB p65 translocation in adult and cord blood moDCs. (A) Cells were incubated in the presence of medium (unstimulated) or LPS (100 ng/mL) for 2 hours, fixed, and stained for IRF-3 (Alexa 488, green), p65/RelA (Alexa 568, red). Representative fields of IRF-3 and p65 fluorescence from 5 adults and 6 neonates are shown. (B) Nuclear translocation of IRF-3 and p65 were analyzed by Western blotting as described in “Material and methods.” One representative adult and cord blood sample of 3 are shown. (C) Impaired interaction of IRF-3 with CBP in LPS-stimulated cord blood moDCs. Coimmunoprecipitation experiments were performed as described in “Material and methods.” The asterisk (*) denotes IgG. Two representative adult and cord blood samples of 6 are shown. (D) Assessment of IRF-3 DNA-binding capacity in adult and cord blood moDCs. Cells were left untreated or stimulated for 2 hours with LPS. Nuclear extracts were isolated, and IRF-3 DNA-binding activity was measured using TransAM transcription factor assay kit. Results are presented as mean and SEM from at least 5 different donors. *P <.05 as compared with adult moDC.
Impaired interaction of IRF-3 and CBP in neonatal moDCs. (A-B) Analysis of IRF-3 and NF-κB p65 translocation in adult and cord blood moDCs. (A) Cells were incubated in the presence of medium (unstimulated) or LPS (100 ng/mL) for 2 hours, fixed, and stained for IRF-3 (Alexa 488, green), p65/RelA (Alexa 568, red). Representative fields of IRF-3 and p65 fluorescence from 5 adults and 6 neonates are shown. (B) Nuclear translocation of IRF-3 and p65 were analyzed by Western blotting as described in “Material and methods.” One representative adult and cord blood sample of 3 are shown. (C) Impaired interaction of IRF-3 with CBP in LPS-stimulated cord blood moDCs. Coimmunoprecipitation experiments were performed as described in “Material and methods.” The asterisk (*) denotes IgG. Two representative adult and cord blood samples of 6 are shown. (D) Assessment of IRF-3 DNA-binding capacity in adult and cord blood moDCs. Cells were left untreated or stimulated for 2 hours with LPS. Nuclear extracts were isolated, and IRF-3 DNA-binding activity was measured using TransAM transcription factor assay kit. Results are presented as mean and SEM from at least 5 different donors. *P <.05 as compared with adult moDC.
Discussion
Type I IFNs, which include several IFNα subtypes and a single IFNβ subtype, were initially identified for their essential antiviral properties. Their broader immunoregulatory role has recently emerged in the context of TLR signaling.25 Herein, we first show that TLR4-triggered IFNβ synthesis is blunted in cord blood. Taken together with previous findings indicating that IFNα production in response to PolyI:C (TLR3 ligand) or CpG oligonucleotides (TLR9 ligand) is strongly decreased in cord blood,26,27 these data indicate that deficient type I IFN synthesis is a key feature of TLR-mediated responses in blood cells of human newborns.
Several soluble factors influence responses to LPS, including LPS-binding protein and soluble CD14,28 which has been shown to be present in reduced amounts in cord blood.19 In fact, LPS binding to cord blood monocytes was found to be reduced as compared with adult cells.29 Furthermore, IL-10 produced by B cells was recently shown to repress DC function in newborn mice.30 We show herein that differences in adult and cord plasma have some influence on LPS-triggered CXCL10 production. However, our experiments indicate that neonatal blood cells display reduced capacity to produce this IFN-inducible chemokine even when they are resuspended in adult plasma. Interestingly, Yan et al31 have previously reported that adherent cord blood mononuclear cells (CBMCs) display reduced expression of MyD88 as compared with their adult counterparts. This finding was correlated to impaired TNF-α production in response to suboptimal LPS stimulation. Herein, we observed that IRF-3–dependent responses are impaired whatever the LPS concentration used or the presence of adult or neonatal plasma. Furthermore this selective defect was apparent even after in vitro differentiation of monocytes in moDCs, a situation in which MyD88 expression was found to be comparable in adult and neonatal cells. To explore the molecular basis of deficient TLR4-triggered IFNβ synthesis in neonatal cells and its possible relationship with the previously reported decreased IL-12p35 production,7 we therefore focused our attention on the comparison of adult and neonatal moDCs generated and stimulated under the same culture conditions. Analysis of the profile of genes expressed in LPS-stimulated neonatal moDCs confirmed a defective IFNβ production that was associated with decreased expression of type I IFN-dependent genes but not of genes which depend on NF-κB activation. This observation indicates that TRIF-dependent signaling events leading to IFNβ synthesis are preferentially affected in neonatal cells. It is consistent with the deficient up-regulation of costimulatory molecules on LPS-stimulated neonatal DCs32 as this process was found to be dependent on type I IFN release.33,34 We show here that CXCL9, CXCL10, and CXCL11 are part of the array of IFN-dependent factors which are poorly expressed in neonatal moDCs. This might be relevant to the outcome of T-cell responses as these chemokines preferentially act on Th1 cells expressing CXCR3.35
Activation of the IFNβ; promoter critically depends on the recruitment of a holocomplex which includes IRF-3 dimers and the coactivator CBP.36,37 We related impaired IFNβ synthesis in LPS-stimulated neonatal moDCs to altered association of IRF-3 with CBP. Indeed, IRF-3 nuclear translocation occurred in LPS-stimulated neonatal moDCs, but IRF-3 binding to DNA and its interaction with the coactivator CBP were blunted. This dichotomy is compatible with the 2-step activation model recently proposed for IRF-3.38 In this model, phosphorylation of Ser396 by TBK1 pathway is sufficient for IRF-3 nuclear translocation, but further phosphorylation of Ser/Thr residues is required for CBP recruitment and IRF-3 binding to promoter regions. Alternatively, a more global perturbation in the assembly of the enhanceosome complex at the enhancer region of the IFNβ; gene39 could also account for our observations.
Beside its critical role in TLR4-mediated IFNβ synthesis,40 IRF-3 was recently found to be directly involved in IL-12p35 gene induction.41 Interestingly, decreased IL-12p70 synthesis in neonatal moDCs was previously shown to be related to altered chromatin remodeling in the IL-12p35 promoter region.8 Because IRF-3 is known to facilitate gene expression through the recruitment of chromatin remodeling complexes,39,42 impaired recruitment of IRF-3/CBP complex to the promoter region in neonatal moDCs could be responsible for defective IL-12p35 chain synthesis on LPS stimulation.8
In conclusion, we suggest that deficient IRF-3/CBP interaction in LPS-stimulated neonatal DCs reduces their ability to elicit Th1 responses by limiting their production of type I interferon, IL-12p70, and IFN-inducible chemokines. This might be relevant to early life immunity and also to allogeneic stem cell transplantation because responsiveness of donor antigen-presenting cells to LPS represents an important risk factor for acute graft-versus-host disease.43 Indeed, the incidence and severity of this complication was shown to be reduced after cord blood transplantation.44
Authorship
Contribution: E.A. and V.A. performed key experiments and designed the research. M.N. performed the research. J.-F.L. performed microarray experiments, and J.-L.R. performed the statistical analysis of these data. D.D.W. and F.W. designed the research and analyzed data. M.G. and S.G. designed the research, analyzed the data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
E.A. and V.A. contributed equally to this study.
Correspondence: Michel Goldman, Institute for Medical Immunology, 8 rue Adrienne Bolland, B-6041 Charleroi-Gosselies, Belgium; e-mail: mgoldman@ulb.ac.be.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
We thank the staff from the obstetric departments of CHU Tivoli-La Louvière, CHU-Charleroi and Clinique Notre Dame, Charleroi for their kind support. We also thank Thierry Coche, Cindy Castado and Swann Gaulis for helpful discussion on the microarray results.
This work was supported by the Fonds National de la Recherche Scientifique (FNRS; Belgium) and an Interuniversity Attraction Pole of the Belgian Federal Science Policy. The Institute for Medical Immunology is sponsored by the government of the Walloon Region and GlaxoSmithKline Biologicals. S.G. is a postdoctoral researcher of the FNRS.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal