Abstract
Staphylococcus aureus secretes several virulence factors interfering with host-cell functions. Staphylococcal superantigen-like (SSL) proteins are a family of 11 exotoxins with structural homology to superantigens but with generally unknown functions. Recently, we described that chemotaxis inhibitory protein of Staphylococcus aureus (CHIPS31-121), a potent inhibitor of C5a-induced responses, is structurally homologous to the C-terminal domain of SSL5. Here, we identify P-selectin glycoprotein ligand-1 (PSGL-1), involved in the initial rolling of neutrophils along the endothelium, as a target for SSL5. SSL5 specifically bound to Chinese hamster ovary cells stably expressing PSGL-1 (CHO–PSGL-1), which was dependent of sulfation and sialylation. Furthermore, SSL5 bound to PSGL-1/Ig fusion protein immobilized on a biosensor chip. SSL5 affected binding of soluble P-selectin/Fc chimera, the principle ligand of PSGL-1, to CHO–PSGL-1 cells and inhibited adhesion of neutrophils to immobilized P-selectin under static conditions. Under flow conditions SSL5 strongly decreased neutrophil rolling on immobilized P-selectin/Fc and activated human endothelial cells. In conclusion, SSL5 interferes with the interaction between PSGL-1 and P-selectin, suggesting that S aureus uses SSL5 to prevent neutrophil extravasation toward the site of infection. This makes SSL5 a potential lead for the development of new anti-inflammatory compounds for disorders characterized by excessive recruitment of leukocytes.
Introduction
Staphylococcus aureus is a common human pathogen that induces both community-acquired and nosocomial infections. This Gram-positive bacterium is well known for its suppurative diseases such as skin-limited abscesses and boils and more seriously endocarditis, sepsis, and toxic shock syndrome.1,2 Its invasiveness is ascribed to the production of a wide repertoire of cell surface–expressed as well as secreted virulence factors that interfere with host defense.2,3 Superantigens constitute a large portion of the secreted arsenal of staphylococci and modulate immune responses. They trigger nonspecific activation of T lymphocytes by binding to major histocompatibility complex (MHC) class II molecules on antigen-presenting cells outside the antigen-binding cleft and Vβ domains of T-cell receptors (TCRs).4
We have described chemotaxis inhibitory protein of S aureus (CHIPS), an excreted virulence factor of S aureus.5,6 CHIPS is known to inhibit formylated peptide- and complement factor C5a-induced responses in neutrophils through direct binding to the formyl peptide receptor and C5a receptor (C5aR), respectively.7 Thereby, CHIPS inhibits the initial activation and migration of neutrophils to the site of infection; thus, it hampers clearance of S aureus by innate immune cells. Recently, the structure of CHIPS consisting of residues 31 to 121 (CHIPS31-121) was resolved.8 CHIPS31-121 is composed of an α-helix packed onto a 4-stranded antiparallel β-sheet, a domain also present in the C-terminal domain of superantigens. This protein also revealed to be homologous to the C-terminal domain of staphylococcal superantigen-like 5 (SSL5) and SSL7.
SSLs are a family of secreted proteins identified through sequence homology to staphylococcal and streptococcal superantigens.9 Eleven different SSLs exist that are encoded on staphylococcal pathogenicity island 2 in a conserved order. Staphylococci contain 7 to 11 different SSLs, and their homology varies between 36% and 67%. Allelic variants show 85% to 100% homology.10,11 Determination of the crystal structures of SSL512 and SSL713 also revealed their high structural homology to superantigens; the N-terminal oligonucleotide/oligosaccharide-binding fold and the C-terminal β-grasp domain characteristic for superantigens are also observed in SSLs. However, residues important for MHC class II and TCR binding of superantigens are not conserved in SSLs, which may explain their inability to display superantigenic activities.9,10,12 Recently, Langley et al14 described binding of complement component 5 and immunoglobulin A (IgA) by SSL7, suggesting a role for SSLs in staphylococcal defense against host immune responses. SSL7 was subsequently found to bind the Cα2/Cα3 interface of IgA Fc, which is the adhesion site for the FcαRI.15 So far, no other functions have been linked to the SSLs.
Neutrophil recruitment to sites of infection is a multistep process.16 The initial tethering and rolling of neutrophils on the endothelium of vessel walls during inflammation are mediated by P-selectin.17 P-selectin is stored in α-granules of platelets and Weibel-Palade bodies of endothelial cells and is rapidly translocated to the cell surface after stimulation with thrombin and histamine.17-19 P-selectin glycoprotein ligand-1 (PSGL-1) has been identified as the principal ligand for P-selectin.20 PSGL-1 is a disulfide-linked homodimer consisting of 2 glycoprotein chains with a molecular mass of 120 kDa each. It is heavily glycosylated and contains sialylated, fucosylated O-linked glycans, which often terminate with sialyl Lewis x. Further post-translational modifications include up to 3 N-linked glycans and sulfation of at least 1 of the 3 tyrosines at the distal end of PSGL-1.21 PSGL-1 is expressed on most leukocytes, including neutrophils, monocytes, and T lymphocytes, and has been shown to mediate rolling of neutrophils on P-selectin in vitro20,22 and in vivo.23 Several chronic and acute diseases, such as stroke, reperfusion/ischemia, transplant rejection, and rheumatoid arthritis, are caused by an excessive recruitment of leukocytes to the site of tissue damage.24-26 An inhibitor of the interaction between PSGL-1 and P-selectin is therefore an attractive therapeutic with high potential as an anti-inflammatory compound. Here, we demonstrate that SSL5 specifically binds PSGL-1 and functionally inhibits rolling of neutrophils on P-selectin and activated human umbilical vein endothelial cells (HUVECs).
Materials and methods
Antibodies
Phycoerythrin (PE)–conjugated monoclonal antibodies (mAbs) directed against CD4, CD8, CD14, CD16, CD19, CD35, CD47, CD56, CD89, CD114, CDw119, CD132, CD142, and CD162; fluorescein isothiocyanate (FITC)–labeled mAbs directed against CD11a, CD15, CD18, CD46, CD55, CD62L, CD66b, and CDw17; and allophycocyanin (APC)–labeled monoclonal antibodies against CD13, CD14, CD16, and CD45 were purchased from BD Biosciences (San Jose, CA). Monoclonal cyanine 5 (Cy5)–conjugated mAb against CD3 was purchased from Dako (Heverlee, Belgium). Anti–CCR1-PE, –CCR2-PE, –CXCR1-PE, –CXCR2-PE, –TNFRI-FITC, and –TNFRII-FITC were from R&D Systems (Minneapolis, MN). CD10-APC was from Caltag Laboratories (Burlingame, CA). CD63-PE was purchased from Sanquin (Amsterdam, The Netherlands). The monoclonal anti–PSGL-1 antibodies PL1 (function-blocking,22 clone 3E2.25.5) and PL2 (nonblocking,22 clone 5D8.8.12) were obtained from Serotec (Oxford, United Kingdom), and KPL1 (function-blocking27 ) was purchased from BD Biosciences. The mAb against C5aR (clone W17/1) was purchased from Serotec or Hbt (Uden, The Netherlands), the mAb against CD15s was from BD Biosciences, and the mAb against P-selectin (clone WASP12.2; ATCC, Rockville, MD) was purified from hybridoma culture supernatants. Polyclonal goat anti–mouse IgG-FITC was from Dako, and goat anti–human IgG (Fc specific)–FITC was from Sigma-Aldrich (St Louis, MO). Anti–Xpress-FITC was obtained from Invitrogen Life Technologies (Paisley, United Kingdom).
Cloning, expression, and purification of SSL5
For expression of recombinant SSL5, the SSL5 gene (ssl5) of S aureus strain NCTC8325, except for the signal sequence, was cloned into the expression vector pRSETB (Invitrogen Life Technologies) directly downstream of the enterokinase cleavage site. For this purpose, an overhang extension polymerase chain reaction (PCR) was performed. First, we amplified the HIS tag and enterokinase cleavage site from the pRSETB vector using the XbaI recognition sequence and introducing the N-terminal first 29-bp sequence of ssl5 via the reverse primer (5′-GCTCTAGAAATAATTTTGTTTAACTTTAAGAAGGAG-3′, 5′-ACATTTTCATATTTTGCTTTATGTTCACTCTTGTCGTCATCGTCGTACAG-3′, Xba1 recognition site underlined). Second, ssl5 was amplified by PCR (5′-AGTGAACATAAAGCAAAATATG-3′, 5′-CCGGAATTCTTATCTAATGTTGGCTTCTATTTTTTC-3′, EcoR1 recognition site underlined) on chromosomal DNA of S aureus. Finally, a third PCR was performed to anneal the 2 PCR products together using the XbaI and EcoRI primer. All PCR products were amplified using PfuTurbo DNA polymerase (Stratagene, Cedar Creek, TX).
After verification of the correct sequence, the pRSET/SSL5 expression vector was transformed in Rosetta-Gami(DE3)pLysS Escherichia coli according to the manufacturer's protocol (Novagen, Darmstadt, Germany). Expression of HIS-tagged SSL5 (HIS-SSL5) was induced with 1 mM isopropyl-β-D-thiogalactopyranoside (IPTG; Roche, Basel, Switzerland) for 3 hours. HIS-tagged SSL5 was isolated under denaturing conditions on a HiTrap chelating HP column according to the manufacturer's protocol (Amersham Biosciences, Piscataway, NJ). The protein was renatured on the column by gradual exchanging denaturing buffer (8 M urea, 500 mM NaCl, 20 mM Na2HPO4, pH 5.3) for native buffer (500 mM NaCl, 20 mM Na2HPO4, pH 5.3). Bound protein was eluted using 50 mM EDTA (ethylenediaminetetraacetic acid). After dialysis, the HIS tag was removed from SSL5 by cleavage with enterokinase according to the manufacturer's instructions (Invitrogen Life Technologies). Finally, SSL5 was stored in phosphate-buffered saline (PBS), and its purity was examined by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE).
Cells
Informed consent was obtained from all subjects and was provided in accordance with the Declaration of Helsinki. Approval was obtained from the medical ethics committee of the University Medical Center Utrecht (Utrecht, The Netherlands). Leukocytes were isolated by means of the Ficoll-Histopaque gradient method. Venous blood was obtained from healthy volunteers using sodium heparin as anticoagulant (Greiner, Alphen a/d Rijn, The Netherlands). Heparinized blood was diluted with an equal volume of PBS and subsequently layered onto a gradient of Ficoll-Paque PLUS (Amersham Biosciences) and Histopaque-1119 (Sigma-Aldrich). After centrifugation for 20 minutes at 400g, plasma was aspired, and peripheral blood mononuclear cells (PBMCs) were collected from the Ficoll layer and neutrophils from the Histopaque layer. After washing with RPMI 1640 containing 25 mM HEPES (N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid), l-glutamine (BioWhittaker, Walkersville, MD), and 0.05% human serum albumin (HSA; Sanquin) (RPMI/HSA), the neutrophils were subjected to a hypotonic shock with water for 30 seconds to lyse remaining erythrocytes, after which the neutrophils and PBMCs were washed.
Chinese hamster ovary K1 (CHO; ACC 110; DSMZ, Braunschweig, Germany) cells were cultured in Minimum Essential Medium α (Gibco, Paisley, United Kingdom) supplemented with 10% fetal bovine serum (Biochem AG, Laufelfingen, Switzerland) and 10 μg/mL gentamicin (Gibco). The CHO cell line expressing functionally active PSGL-1 by coexpression of core2 β(1,6)-N-acetylglucosaminyltransferase and α(1,3/4)fucosyltransferase has previously been described in Pendu et al28 and was cultured with additional 50 U/mL penicillin (Gibco), 50 μg/mL streptomycin (Gibco), 2 mM glutamine (Gibco), 100 mM methotrexate (MTX; Sigma), and 1 mg/mL G418 (Gibco). In some experiments, 50 mM sodium chlorate (NaClO3; Merck, Dormstadt, Germany) was added to the medium for at least 2 passages.
Human umbilical vein endothelial cells (HUVECs) were isolated as described by Jaffe et al.29 Freshly isolated cells were grown to confluent monolayers in endothelial growth medium-2 (Clonetics, Waltersville, MD).
SSL5 binding to cells
To determine binding of SSL5 to different leukocyte subpopulations, SSL5 was labeled with FITC. Therefore, 1 mg/mL SSL5 was incubated with 100 μg/mL FITC in 0.1 M sodium carbonate buffer (pH 9.6) for 1 hour at 37°C. Using a HiTrap desalting column (Amersham Biosciences), FITC-labeled SSL5 (SSL5-FITC) was separated from unbound FITC. For binding of SSL5-FITC to leukocytes, neutrophils (5 × 106 cells/mL) and PBMCs (1 × 107 cells/mL) were incubated with increasing concentrations of SSL5-FITC in RPMI/HSA for 30 minutes on ice in the presence of PE- or Cy-conjugated, leukocyte-subset–specific antibodies. Leukocyte-subset–specific antibodies include anti-CD3, -CD4, -CD8, -CD14, -CD16, -CD19, and -CD56. After washing, fluorescence was measured on a flow cytometer (FACSCalibur; Becton Dickinson, Franklin Lanes, NJ). In another experiment, PSGL-1–transfected and control CHO cells (5 × 106 cells/mL) were incubated with increasing concentrations of HIS-SSL5 for 30 minutes on ice. Bound HIS-SSL5 was detected with anti–Xpress-FITC. Where indicated, CHO–PSGL-1 cells (2 × 106 cells/mL) were first treated with 0.2 U/mL neuraminidase (from Vibrio cholerae; Sigma) at 37°C for 45 minutes at pH 6.0. Efficacy of the treatment was assessed by staining with the anti–sialyl Lewis x (CD15s) mAb, which was detected with goat anti–mouse IgG-FITC.
Competition for receptor binding
To determine a putative receptor for SSL5, a mixture of neutrophils (5 × 106 cells/mL) and PBMCs (1 × 107 cells/mL) was incubated with 10 μg/mL SSL5 for 15 minutes on ice in the presence of 10% heat-inactivated human pooled serum. Subsequently, FITC-, PE-, and APC-conjugated mAbs directed against a series of cell-surface receptors were added for 30 minutes on ice. After washing, fluorescence was measured using flow cytometry. Neutrophils, monocytes, and lymphocytes were selected by gating. In another experiment, neutrophils (5 × 106 cells/mL) were incubated with increasing concentrations of SSL5 or SSL7 for 30 minutes on ice. After washing, the neutrophils were incubated with 1 μg/mL anti–PSGL-1 mAbs for 30 minutes on ice, washed, and stained with 20 μg/mL goat anti–mouse IgG-FITC. For binding of KPL1 to specific subpopulations of leukocytes in competition with SSL5, a mixture of neutrophils and PBMCs was concurrently stained with PE- and Cy-conjugated, leukocyte-subset–specific antibodies directed against CD3, CD4, CD8, CD14, CD16, CD19, and CD56. In a separate experiment, CHO–PSGL-1 cells were incubated with increasing concentrations of SSL5, SSL7, or anti–PSGL-1 mAbs on ice for 30 minutes, washed, and stained with 1 μg/mL P-selectin/Fc chimera (R&D Systems) for 30 minutes on ice. Bound P-selectin/Fc was detected with 10 μg/mL goat anti–human IgG-FITC.
Surface plasmon resonance (SPR) analysis
SPR analysis was performed using a Biacore 2000 biosensor system (Biacore AB, Uppsala, Sweden). Streptavidin sensorchips were loaded with biotinylated anti-Fcγ F(ab′)2 fragments (Jackson ImmunoResearch Europe, Sirham, United Kingdom) until a density of 2.5 kRU on 2 adjacent channels. The first of these channels (channel 1) was used as a control. Channel 2 was used to capture PSGL-1/Ig fusion protein.28 rPSGL/Ig was passed at a concentration of 0.3 mg/mL in 100 mM NaCl, 0.005% Tween-20, 2.5 mM CaCl2, 25 mM HEPES (pH 7.4) at a flow rate of 5 μL/minute at 25°C to reach a density of 0.25 kRU. Subsequently, both channels were used for the perfusion of SSL5 at a flow rate of 5 μL/minute in the same buffer until equilibrium was reached. Binding to the PSGL-1/Ig–coated channel was corrected for binding to the control channel. Sensorgrams were analyzed using BiaEvaluation software (Biacore) provided by the manufacturer to determine responses at equilibrium (Req). Req was then plotted against protein concentration to calculate apparent affinity as described.30
Adhesion of neutrophils to P-selectin under static conditions
To investigate adhesion of neutrophils to P-selectin, neutrophils were loaded with 4 μM calcein-AM (Molecular Probes, Leiden, The Netherlands) in Hanks buffered salt solution (BioWhittaker) with 0.05% HSA. A 96-well plate (Greiner Bio-One, Frickenhausen, Germany) was coated with 3 μg/mL P-selectin (R&D Systems) for 1 hour at 37°C. After washing with PBS, the plate was blocked with 4% bovine serum albumin (Sigma-Aldrich) for 90 minutes at 37°C. The plate was then washed, and 3 × 105 calcein-labeled neutrophils were added to duplicate wells and allowed to adhere for 15 minutes at room temperature. After washing twice, adherent cells were quantified using a platereader fluorometer (FlexStation; Molecular Devices, Sunnyvale, CA).
Adhesion of neutrophils to P-selectin or HUVECs under flow conditions
To study the effect of SSL5 on the rolling of neutrophils, a flow chamber was used as previously described.31 Briefly, the chamber is a modified form of a transparent parallel-plate perfusion chamber in which a coated coverslip of 18 mm × 18 mm is used as a rolling surface. Glass coverslips were coated with 10 μg/mL P-selectin/Fc for 1 hour at 37°C. After blocking with 1% HSA for 1 hour, the slips were washed with PBS and inserted into the flow chamber. Neutrophils were washed and diluted to 4 × 106 cells/mL in perfusion buffer (20 mM HEPES, 132 mM NaCl, 6 mM KCl, 1 mM MgSO4, 1.2 mM KH2PO4, supplemented with 5 mM glucose, 1 mM CaCl2, and 0.5% HSA) and kept on ice until use. They were put at room temperature for 15 minutes before treatment with a concentration range of SSL5 (0.3-30 μg/mL) or 10 μg/mL anti–PSGL-1 mAb KPL1 for 15 minutes at 37°C. The anti-C5aR antibody W17/1 was used as an isotype control antibody. Neutrophils were then diluted with perfusion buffer to 2 × 106 cells/mL, perfused for 5 minutes through the perfusion chamber containing the P-selectin/Fc–coated coverslip, and subsequently washed for 1 minute with perfusion buffer. The perfusions were performed at 37°C at a shear stress of 1.6 dyn/cm2 by drawing cells through the perfusion chamber with a syringe pump (Harvard Apparatus, South Natick, MA). The perfusion chamber was placed on a microscope stage (DM RXE; Leica, Wetzlar, Germany), which was equipped with a B/W CCD video camera (Sanyo, Osaka, Japan). To the camera a video recorder was connected to record individual perfusions. Recorded images were analyzed by a custom-made program using Optimas 6.1 software (Media Cybernetics Systems, Silver Spring, MD). The number of adherent neutrophils was quantified for at least 40 adjacent high-power fields recorded along the perfusion chamber (total surface at least 1 mm2). Adherent neutrophils appeared as white-centered cells, and experiments in which more than 40% of the cells in control conditions flattened and did not appear as white-centered cells were discarded. In addition, experiments in which adhesion of cells in control conditions was lower than 100 cells/mm2 were also discarded.
In other experiments, neutrophil rolling adhesion on HUVECs was investigated. Therefore, freshly isolated HUVECs were grown to cobblestone confluent monolayers on coverslips precoated with glutaraldehyde-linked gelatin. They were stimulated for 3 minutes with 100 μM histamine (Sigma) to induce surface expression of P-selectin, and neutrophils were immediately perfused at 0.8 dyn/cm.2
Results
SSL5 binds to different leukocyte populations
SSL5 was produced in Rosetta-Gami(DE3)pLysS E coli and isolated with high purity. We used fluorescein-labeled SSL5 (SSL5-FITC) and flow cytometry to determine its binding to different leukocyte populations. Specific cell types were identified through scatter gates and cell type-specific lineage markers. Neutrophils (gated only on scatter), monocytes (CD14+), and natural killer (NK) cells (CD3−, CD16+, and CD56+) stained highly positive for SSL5-FITC, whereas binding of SSL5-FITC to T lymphocytes (CD4+ or CD8+) was lower (Figure 1). Hardly any binding to B lymphocytes (CD19+) was observed.
Binding of SSL5 to leukocytes. Two-color flow cytometry was used to analyze SSL5 binding to different leukocyte subpopulations. (A) Leukocytes were incubated with 10 μg/mL SSL5-FITC for 30 minutes on ice. To differentiate for specific leukocyte subpopulations, monocytes, T lymphocytes, and B lymphocytes were concurrently stained with PE-conjugated antibodies directed against CD14, CD4 or CD8, and CD19, respectively. NK cells were first negatively selected for binding of anti–CD3-Cy and then positively selected for binding anti–CD16-PE and anti–CD56-PE. Neutrophils were selected only by gating. (B) Binding of a concentration range of SSL5-FITC (0.3-10 μg/mL) to the different leukocyte subpopulations. The data represent mean fluorescence of detected SSL5-FITC and are representative of 3 independent experiments.
Binding of SSL5 to leukocytes. Two-color flow cytometry was used to analyze SSL5 binding to different leukocyte subpopulations. (A) Leukocytes were incubated with 10 μg/mL SSL5-FITC for 30 minutes on ice. To differentiate for specific leukocyte subpopulations, monocytes, T lymphocytes, and B lymphocytes were concurrently stained with PE-conjugated antibodies directed against CD14, CD4 or CD8, and CD19, respectively. NK cells were first negatively selected for binding of anti–CD3-Cy and then positively selected for binding anti–CD16-PE and anti–CD56-PE. Neutrophils were selected only by gating. (B) Binding of a concentration range of SSL5-FITC (0.3-10 μg/mL) to the different leukocyte subpopulations. The data represent mean fluorescence of detected SSL5-FITC and are representative of 3 independent experiments.
PSGL-1 is a putative receptor for SSL5
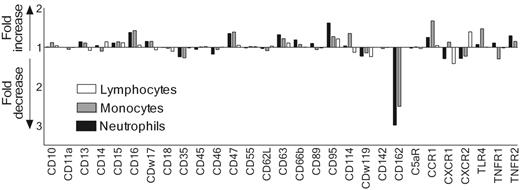
A multiscreening assay for surface-expressed receptors of leukocytes was performed to identify a receptor target for SSL5. For this purpose, we selected a panel of 29 monoclonal antibodies that recognize a variety of leukocyte receptors with distinctive functions, including chemokine, cytokine, and signaling receptors, and receptors involved in adhesion or phagocytosis. SSL5 was screened for its ability to block binding of these antibodies to neutrophils, monocytes, and lymphocytes in the presence of serum. Serum was used to prevent unspecific binding of the antibodies by leukocytes. SSL5 blocked the binding of the anti–PSGL-1 (CD162) mAb KPL1 to neutrophils and monocytes but not to lymphocytes when incubated on ice (Figure 2). Binding of antibodies directed against the other 28 cell-surface receptors was not markedly affected by SSL5. When the experiment was repeated at 37°C, SSL5 again affected only the binding of the anti–PSGL-1 mAb (data not shown). This excludes for an up- or down-regulation of the tested receptors through activation of the cells by SSL5. Thus, PSGL-1 was identified as a putative target of SSL5.
Competition of SSL5 and mAbs directed against surface-expressed receptors in binding to neutrophils, monocytes, and lymphocytes. Neutrophils (▪), monocytes (⊡), and lymphocytes (□) were incubated with 10 μg/mL SSL5 for 15 minutes on ice. The cells were then stained with a panel of FITC-, PE-, or APC-conjugated mAbs directed against cell-surface–expressed receptors in the presence of 10% heat-inactivated serum. Data depict fold increase or decrease of antibody binding to SSL5-treated cells compared with control-treated cells. The data are mean values of 3 independent experiments.
Competition of SSL5 and mAbs directed against surface-expressed receptors in binding to neutrophils, monocytes, and lymphocytes. Neutrophils (▪), monocytes (⊡), and lymphocytes (□) were incubated with 10 μg/mL SSL5 for 15 minutes on ice. The cells were then stained with a panel of FITC-, PE-, or APC-conjugated mAbs directed against cell-surface–expressed receptors in the presence of 10% heat-inactivated serum. Data depict fold increase or decrease of antibody binding to SSL5-treated cells compared with control-treated cells. The data are mean values of 3 independent experiments.
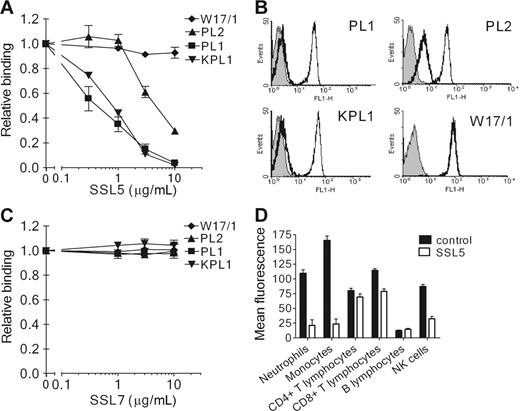
SSL5 competes with antibodies directed against PSGL-1
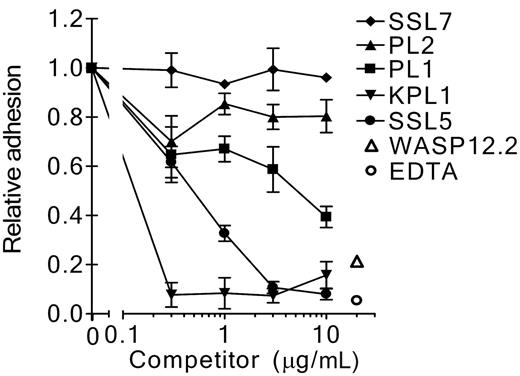
To address the interaction of SSL5 and PSGL-1 further, SSL5 was screened for its ability to block binding of several anti–PSGL-1 mAbs to neutrophils. SSL5 competitively and dose-dependently inhibited binding of all 3 anti–PSGL-1 mAbs tested (Figure 3A-B). Binding of PL1 and KPL1 (two function-blocking antibodies) was strongly inhibited by SSL5. Binding of PL2 (a nonblocking antibody) was also inhibited but to a lower extent; 10-fold more SSL5 was needed to achieve the same inhibition compared with PL1 and KPL1. SSL5 did not affect binding of the isotype control mAb W17/1 directed against C5aR, which is highly expressed on neutrophils. Binding to PSGL-1 was specific for SSL5, as SSL7 did not show competition with anti–PSGL-1 mAbs in this assay (Figure 3C). In addition to neutrophils, competition of SSL5 and KPL1 was examined on other leukocyte subpopulations. SSL5 also inhibited binding of KPL1 to monocytes and NK cells (Figure 3C). Although KPL1 bound well to PSGL-1 on T lymphocytes, SSL5 hardly affected this interaction. B lymphocytes expressed almost no PSGL-1. Thus, competition was observed on those cells that bind SSL5 well.
Competition of SSL5 with anti–PSGL-1 mAbs binding to neutrophils. (A) Neutrophils were incubated with a concentration range of SSL5 (0.3-10 μg/mL) for 30 minutes on ice. After washing, the cells were treated with 1 μg/mL anti–PSGL-1 PL1, PL2, or KPL1, or 1 μg/mL isotype control anti-C5aR W17/1. Bound antibodies were detected with FITC-conjugated goat anti–mouse IgG. Data represent relative binding of PL1 (▪), PL2 (▴), KPL1 (▾), or W17/1 (♦) to cells treated with SSL5 compared with control-treated cells and are mean values ± SEMs of 3 independent experiments. (B) Representative histograms of panel A depict binding of 1 μg/mL PL1, PL2, KPL1, and W17/1 to neutrophils pretreated without (thin continuous line) or with (thick continuous line) 10 μg/mL SSL5. Gray histograms represent cells stained only with the secondary antibody. (C) Experiment as described in Panel A using SSL7 instead of SSL5. Data represent relative binding of PL1 (▪), PL2 (▴), KPL1 (▾), or W17/1 (♦) to cells treated with SSL7 compared with control-treated cells and are mean values ± SEMs of 3 independent experiments. (D) Two-color flow cytometry was used to analyze binding of 1 μg/mL KPL1 to different leukocyte subpopulations in the absence (▪) or presence (□) of 10 μg/mL SSL5. Neutrophils were selected on gating, whereas monocytes, T lymphocytes, and B lymphocytes were stained with anti–CD14-PE, anti–CD4-PE or anti–CD8-PE, and anti–CD19-PE, respectively. NK cells were first negatively selected for anti–CD3-Cy and then positively selected for anti–CD16-PE and anti–CD56-PE. The data represent mean fluorescence of detected KPL1 and are mean values ± SEMs of 3 independent experiments.
Competition of SSL5 with anti–PSGL-1 mAbs binding to neutrophils. (A) Neutrophils were incubated with a concentration range of SSL5 (0.3-10 μg/mL) for 30 minutes on ice. After washing, the cells were treated with 1 μg/mL anti–PSGL-1 PL1, PL2, or KPL1, or 1 μg/mL isotype control anti-C5aR W17/1. Bound antibodies were detected with FITC-conjugated goat anti–mouse IgG. Data represent relative binding of PL1 (▪), PL2 (▴), KPL1 (▾), or W17/1 (♦) to cells treated with SSL5 compared with control-treated cells and are mean values ± SEMs of 3 independent experiments. (B) Representative histograms of panel A depict binding of 1 μg/mL PL1, PL2, KPL1, and W17/1 to neutrophils pretreated without (thin continuous line) or with (thick continuous line) 10 μg/mL SSL5. Gray histograms represent cells stained only with the secondary antibody. (C) Experiment as described in Panel A using SSL7 instead of SSL5. Data represent relative binding of PL1 (▪), PL2 (▴), KPL1 (▾), or W17/1 (♦) to cells treated with SSL7 compared with control-treated cells and are mean values ± SEMs of 3 independent experiments. (D) Two-color flow cytometry was used to analyze binding of 1 μg/mL KPL1 to different leukocyte subpopulations in the absence (▪) or presence (□) of 10 μg/mL SSL5. Neutrophils were selected on gating, whereas monocytes, T lymphocytes, and B lymphocytes were stained with anti–CD14-PE, anti–CD4-PE or anti–CD8-PE, and anti–CD19-PE, respectively. NK cells were first negatively selected for anti–CD3-Cy and then positively selected for anti–CD16-PE and anti–CD56-PE. The data represent mean fluorescence of detected KPL1 and are mean values ± SEMs of 3 independent experiments.
SSL5 directly binds PSGL-1
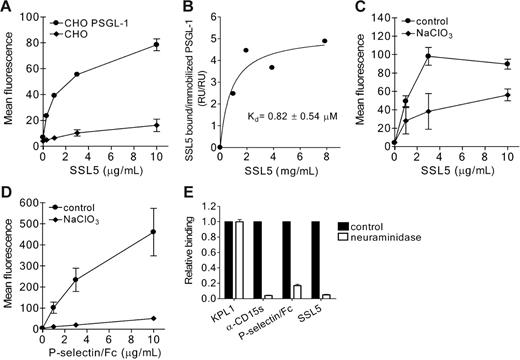
To determine binding of SSL5 to PSGL-1, we used CHO cells expressing functionally active PSGL-1. SSL5 specifically bound to PSGL-1–transfected but not to control CHO cells (Figure 4A). This interaction was strong and dose dependent. Furthermore, direct binding of SSL5 to PSGL-1 at the protein level was determined through SPR analysis. Different concentrations of SSL5 were presented to rPSGL/Ig coated on a SPR surface. SSL5 bound to rPSGL/Ig in a saturable and dose-dependent manner, and the apparent affinity constant (Kd) was calculated to be 0.82 ± 0.54 μM (Figure 4B).
Direct binding of SSL5 to PSGL-1. (A) PSGL-1–transfected (•) and control (♦) CHO cells were incubated with 0.3 to 10 μg/mL HIS-SSL5 for 30 minutes on ice. Bound HIS-SSL5 was detected with FITC-conjugated anti-Xpress. The data represent mean fluorescence of detected HIS-SSL5 and are mean values ± SEMs of 3 independent experiments. (B) Biotinylated F(ab′)2 goat anti–human Fcγ antibodies were immobilized (2.5 kRU) onto 2 adjacent channels of a streptavidin-coated sensor chip. Purified recombinant rPSGL/Ig (0.3 mg/mL) was then applied to the second of these channels to reach a density of 0.25 kRU, whereas channel 1 was used as a reference. The response of SSL5 at equilibrium was determined and plotted against the concentration applied. (C-D) CHO–PSGL-1 cells were cultured in the absence or presence of 50 mM sodium chlorate (NaClO3). Subsequently, cells were tested for binding of HIS-SSL5 (C) and P-selectin/Fc (D). The data represent mean fluorescence of detected HIS-SSL5 and P-selectin/Fc and are mean values ± SEMs of 3 independent experiments. (E) CHO–PSGL-1 cells were treated without (▪) or with (□) 0.2 U/mL neuraminidase for 45 minutes at 37°C. After washing, binding of HIS-SSL5, P-selectin/Fc, KPL1, and anti-CD15s was examined. The data represent relative fluorescence of detected protein and are mean values ± SEMs of 3 independent experiments.
Direct binding of SSL5 to PSGL-1. (A) PSGL-1–transfected (•) and control (♦) CHO cells were incubated with 0.3 to 10 μg/mL HIS-SSL5 for 30 minutes on ice. Bound HIS-SSL5 was detected with FITC-conjugated anti-Xpress. The data represent mean fluorescence of detected HIS-SSL5 and are mean values ± SEMs of 3 independent experiments. (B) Biotinylated F(ab′)2 goat anti–human Fcγ antibodies were immobilized (2.5 kRU) onto 2 adjacent channels of a streptavidin-coated sensor chip. Purified recombinant rPSGL/Ig (0.3 mg/mL) was then applied to the second of these channels to reach a density of 0.25 kRU, whereas channel 1 was used as a reference. The response of SSL5 at equilibrium was determined and plotted against the concentration applied. (C-D) CHO–PSGL-1 cells were cultured in the absence or presence of 50 mM sodium chlorate (NaClO3). Subsequently, cells were tested for binding of HIS-SSL5 (C) and P-selectin/Fc (D). The data represent mean fluorescence of detected HIS-SSL5 and P-selectin/Fc and are mean values ± SEMs of 3 independent experiments. (E) CHO–PSGL-1 cells were treated without (▪) or with (□) 0.2 U/mL neuraminidase for 45 minutes at 37°C. After washing, binding of HIS-SSL5, P-selectin/Fc, KPL1, and anti-CD15s was examined. The data represent relative fluorescence of detected protein and are mean values ± SEMs of 3 independent experiments.
To examine the importance of sulfation of PSGL-1 in SSL5 binding, PSGL-1–transfected CHO cells were cultured in the presence of sodium chlorate, an inhibitor of ATP sulfurylase activity. Binding of SSL5 to CHO–PSGL-1 cells treated with sodium chlorate was partially inhibited compared with untreated cells (Figure 4C), whereas binding of P-selectin/Fc was abolished (Figure 4D). Binding of KPL1 remained unaffected (data not shown), demonstrating that expression of PSGL-1 was not altered by sodium chlorate treatment. PSGL-1–transfected CHO cells were also treated with neuraminidase to investigate the role of sialic acids. Upon treatment, P-selectin/Fc and anti-CD15s binding were abolished, showing effective removal of sialic acids (Figure 4E). SSL5 binding to treated CHO–PSGL-1 cells was also abrogated, suggesting sialic acid residues may be a critical determinant in recognition of PSGL-1 by SSL5. KPL1 binding remained equal compared with untreated cells as binding of this anti–PSGL-1 antibody is not sensitive to glycosylation.
SSL5 inhibits binding of P-selectin to CHO–PSGL-1 cells
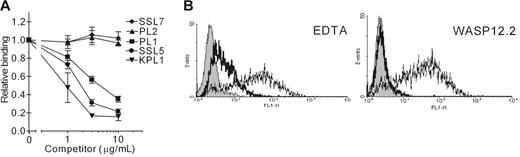
To investigate whether SSL5 interferes with the interaction of PSGL-1 and its ligand P-selectin, we used flow cytometry to measure binding of P-selectin/Fc chimera to CHO–PSGL-1 cells. SSL5 was incubated in a serial dilution with CHO–PSGL-1 cells with a fixed concentration of P-selectin/Fc. SSL5 clearly inhibited binding of P-selectin/Fc (Figure 5A). This effect was dose dependent and comparable to the anti–PSGL-1 mAb KPL1 and PL1. Binding of P-selectin/Fc was not affected by the anti–PSGL-1 mAb PL2 or SSL7. Furthermore, the binding was P-selectin dependent, as it was blocked by EDTA and the function-blocking anti–P-selectin mAb WASP12.2 by 87% and 95%, respectively (Figure 5B).
Competition of SSL5 with P-selectin/Fc binding to CHO–PSGL-1 cells. (A) CHO–PSGL-1 cells were incubated with 0.3 to 10 μg/mL SSL5 (•), SSL7 (♦), or PSGL-1 mAbs PL1 (▪), PL2 (▴), or KPL1 (▾) for 30 minutes on ice. After washing, the cells were treated with 1 μg/mL P-selectin/Fc. Bound P-selectin/Fc was detected with FITC-conjugated goat anti–human IgG. The data represent relative binding of P-selectin/Fc compared with control-treated cells and are mean values ± SEMs of 3 independent experiments. (B) Representative histograms of panel A depict binding of 1 μg/mL P-selectin/Fc to CHO–PSGL-1 cells in the absence (thin continuous line) and presence of 5 mM EDTA or 30 μg/mL anti–P-selectin mAb WASP12.2 (thick continuous line). Gray histograms represent cells stained only with the secondary antibody.
Competition of SSL5 with P-selectin/Fc binding to CHO–PSGL-1 cells. (A) CHO–PSGL-1 cells were incubated with 0.3 to 10 μg/mL SSL5 (•), SSL7 (♦), or PSGL-1 mAbs PL1 (▪), PL2 (▴), or KPL1 (▾) for 30 minutes on ice. After washing, the cells were treated with 1 μg/mL P-selectin/Fc. Bound P-selectin/Fc was detected with FITC-conjugated goat anti–human IgG. The data represent relative binding of P-selectin/Fc compared with control-treated cells and are mean values ± SEMs of 3 independent experiments. (B) Representative histograms of panel A depict binding of 1 μg/mL P-selectin/Fc to CHO–PSGL-1 cells in the absence (thin continuous line) and presence of 5 mM EDTA or 30 μg/mL anti–P-selectin mAb WASP12.2 (thick continuous line). Gray histograms represent cells stained only with the secondary antibody.
SSL5 blocks adhesion of neutrophils to P-selectin under static conditions
Adhesion assays were performed to determine whether SSL5 also inhibits the cell adhesion function of PSGL-1. Under static conditions, binding of neutrophils to a P-selectin–coated surface was analyzed in the presence or absence of SSL5 or anti–PSGL-1 mAbs. In agreement with the results obtained by flow cytometry, SSL5 strongly inhibited binding of neutrophils to P-selectin; maximal inhibition of 90% was achieved at a concentration of 3 μg/mL SSL5 (Figure 6). KPL1 also effectively blocked neutrophil adhesion to P-selectin, whereas PL1 inhibition was weaker, and PL2 and SSL7 had no effect. Adhesion was P-selectin dependent, as EDTA and the anti–P-selectin mAb WASP12.2 abolished neutrophil adhesion (Figure 6).
Effect of SSL5 on adhesion of neutrophils under static conditions. Calcein-labeled neutrophils were incubated with 0.3 to 10 μg/mL SSL5 (•), SSL7 (♦), PSGL-1 mAbs PL1 (▪), PL2 (▴), or KPL1 (▾), or 5 mM EDTA (○) for 10 minutes. Subsequently, the neutrophils were incubated in duplicate wells for 15 minutes in a 96-well microtiterplate to which P-selectin was immobilized. After washing, bound neutrophils were quantified using a microplate reader. Average adherence of untreated cells was 78%. Neutrophil adherence was also examined on the P-selectin surface that was first treated with 30 μg/mL anti–P-selectin mAb WASP12.2 (▵). The data represent relative adhesion of neutrophils compared with untreated cells and are mean values ± SEMs of 3 independent experiments.
Effect of SSL5 on adhesion of neutrophils under static conditions. Calcein-labeled neutrophils were incubated with 0.3 to 10 μg/mL SSL5 (•), SSL7 (♦), PSGL-1 mAbs PL1 (▪), PL2 (▴), or KPL1 (▾), or 5 mM EDTA (○) for 10 minutes. Subsequently, the neutrophils were incubated in duplicate wells for 15 minutes in a 96-well microtiterplate to which P-selectin was immobilized. After washing, bound neutrophils were quantified using a microplate reader. Average adherence of untreated cells was 78%. Neutrophil adherence was also examined on the P-selectin surface that was first treated with 30 μg/mL anti–P-selectin mAb WASP12.2 (▵). The data represent relative adhesion of neutrophils compared with untreated cells and are mean values ± SEMs of 3 independent experiments.
SSL5 inhibits rolling of neutrophils on P-selectin under shear conditions
The effect of SSL5 on rolling of neutrophils on P-selectin was examined under physiologic shear stress by means of a flow chamber. In this flow chamber, neutrophils were perfused over glass coverslips coated with P-selectin/Fc at a shear stress of 1.6 dyn/cm2 in the presence or absence of SSL5. After 5 minutes, the number of accumulated neutrophils per square millimeter was determined. In the absence of SSL5, neutrophils adhered efficiently to P-selectin/Fc (total adhesion was 434 ± 121 cells/mm2), and 95% of the cells was rolling in this situation (data not shown). Treatment of neutrophils with SSL5 strongly and dose dependently inhibited their attachment under flow conditions (Figure 7A). Rolling adhesion was blocked for 72% by 10 μg/mL SSL5. Treatment of neutrophils with KPL1 also abolished rolling adhesion on the P-selectin/Fc surface, whereas isotype control mAb W17/1 directed against the C5aR had no effect (Figure 7B).
Effect of SSL5 on rolling adhesion of neutrophils under shear conditions. (A) Neutrophils were treated with SSL5 (1-30 μg/mL) for 15 minutes at 37°C. Subsequently, the neutrophils were perfused over glass coverslips coated with 10 μg/mL P-selectin/Fc at a shear stress of 1.6 dyn/cm2 during 5 minutes at 37°C. Rolling on the P-selectin/Fc surface was observed in 95% of untreated cells. After washing for 1 minute, accumulated neutrophils were quantified. (B) Effect of 10 μg/mL SSL5 on neutrophil rolling on P-selectin/Fc compared with the effect of 10 μg/mL anti–PSGL-1 KPL1 and the isotype control mAb W17/1 directed against the C5aR. (C) Effect of SSL5 on neutrophil rolling on activated endothelial cells. Freshly isolated HUVECs were cultured on glass coverslips. After stimulation of HUVECs with 100 μM histamine for 3 minutes, neutrophils treated with 10 μg/mL SSL5, SSL7, KPL1, or W17/1 were perfused at 0.8 dyn/cm2 for 5 minutes. After washing for 1 minute, accumulated neutrophils were quantified. The data represent relative accumulation of neutrophils compared with control-treated cells and are mean values ± SEMs of at least 3 independent experiments.
Effect of SSL5 on rolling adhesion of neutrophils under shear conditions. (A) Neutrophils were treated with SSL5 (1-30 μg/mL) for 15 minutes at 37°C. Subsequently, the neutrophils were perfused over glass coverslips coated with 10 μg/mL P-selectin/Fc at a shear stress of 1.6 dyn/cm2 during 5 minutes at 37°C. Rolling on the P-selectin/Fc surface was observed in 95% of untreated cells. After washing for 1 minute, accumulated neutrophils were quantified. (B) Effect of 10 μg/mL SSL5 on neutrophil rolling on P-selectin/Fc compared with the effect of 10 μg/mL anti–PSGL-1 KPL1 and the isotype control mAb W17/1 directed against the C5aR. (C) Effect of SSL5 on neutrophil rolling on activated endothelial cells. Freshly isolated HUVECs were cultured on glass coverslips. After stimulation of HUVECs with 100 μM histamine for 3 minutes, neutrophils treated with 10 μg/mL SSL5, SSL7, KPL1, or W17/1 were perfused at 0.8 dyn/cm2 for 5 minutes. After washing for 1 minute, accumulated neutrophils were quantified. The data represent relative accumulation of neutrophils compared with control-treated cells and are mean values ± SEMs of at least 3 independent experiments.
SSL5 inhibits rolling of neutrophils on HUVECs under shear conditions
To examine whether SSL5 can interfere with neutrophil interaction with endothelial cells, neutrophils were perfused over histamine-activated HUVECs at 0.8 dyn/cm2 in the presence or absence of SSL5. After 5 minutes, the number of accumulated neutrophils per square millimeter was determined. Preincubation of neutrophils with the anti–PSGL-1 mAb KPL1 affected neutrophil accumulation by greater than 90%, demonstrating the involvement of P-selectin and PSGL-1 interactions under these conditions (Figure 7C). Incubation of neutrophils with 10 μg/mL SSL5 inhibited rolling adhesion for 73%. The isotype control mAb W17/1 and SSL7 had no effect. In conclusion, SSL5 specifically inhibited neutrophil rolling to endothelial cells.
Discussion
SSLs have a high structural homology to superantigens but do not show superantigenic properties.9,10,12 Recently, we demonstrated a high structural homology between CHIPS31-121 and the C-terminal β-grasp domain of SSL5 and SSL7.8 The gene encoding for CHIPS is located on a bacteriophage-encoded immune evasion cluster present in greater than 60% of S aureus isolates.6 The SSL genes reside on the staphylococcal pathogenicity island 2 present in all S aureus strains.10,11 Because of the structural homology and high prevalence of CHIPS and SSLs, we postulated that SSLs, as CHIPS, have important immune evasive properties. SSL7 is already described to bind C5 and IgA, properties indicative of an immune-evasive strategy.14 Here, we show that SSL5 binds PSGL-1, the principal ligand for P-selectin, and thereby inhibits rolling adhesion of neutrophils to P-selectin and activated human endothelial cells. These results strongly suggest that S aureus uses the interaction of SSL5 with PSGL-1 to prevent clearance from the host by blocking the initial interaction of neutrophils on the endothelial lining and thus hampering the extravasation of neutrophils from the blood vessels to the infected tissue.
To our knowledge this is the first-described microbial evasion strategy that targets PSGL-1 for immunomodulatory purposes. Only Anaplasma phagocytophilum, an obligate intracellular bacterium that causes human granulocytic anaplasmosis, is known to bind PSGL-1.32,33 However, A phagocytophilum binds PSGL-1 to mediate infection of neutrophils and not to block the function as an evasion strategy.
Arcus et al12 have demonstrated the presence of antibodies directed against SSL5 in several sera of healthy human persons. The presence of anti-SSL5 antibodies in serum might therefore affect the SSL5 activity on PSGL-1. This is however unlikely, as the presence of human pooled serum hardly affected the competition of SSL5 and the anti–PSGL-1 antibody KPL1 on neutrophils and monocytes in the multiscreening assay.
Direct binding of SSL5 to PSGL-1 at the protein level was demonstrated through surface plasmon resonance and exhibited an apparent Kd of 0.82 μM. Similar experiments using neutrophil-derived PSGL-1 and recombinant soluble P-selectin revealed a Kd of 0.3 μM.34 In addition, the Kd of soluble P-selectin and recombinant PSGL-1–Ig was observed to be 0.4 μM.35 Thus, it seems that the affinity of SSL5 and P-selectin for PSGL-1 is in the same order of magnitude.
SSL5 competed well with function-blocking anti–PSGL-1 antibodies, KPL1 and PL1, and to a lower extent with the nonblocking antibody PL2. In addition, SSL5 inhibited binding of P-selectin to neutrophils. These data suggest a binding site for SSL5 at the N-terminal, membrane-distal segment of PSGL-1. Epitopes of the functional blocking antibodies to PSGL-1 span the N-terminal, membrane-distal segment that is involved in P-selectin binding. PL1 binds to the segment of PSGL-1 spanning residues 49 through 62,36 and KPL1 interacts with the tyrosine sulfation consensus motif from residue 46 to 52, but its epitope also extends beyond residue 52.27,37 The nonblocking antibody PL2 recognizes a membrane-proximal epitope of PSGL-1 that is encoded by residues 188 to 235.36 All 3 antibodies recognize PSGL-1 independent of tyrosine sulfation, and they are not influenced by other post-translational modifications as glycosylation.36,37 The interaction of PSGL-1 and its ligand P-selectin is membrane-distal and requires sulfation of at least one tyrosine at residues 46, 48, or 51.38,39 In addition, binding of P-selectin to PSGL-1 requires proper glycosylation.40,41 We found that SSL5 binds strongly to neutrophils, monocytes, and NK cells and hardly to T and B lymphocytes. B lymphocytes express low levels of PSGL-1 and seem not to interact with P-selectin.22,42,43 T lymphocytes express PSGL-1 to a higher extent, but PSGL-1 is hardly glycosylated because of low expression of glycosyltransferases in resting T lymphocytes.42,44 For that reason, P-selectin is unable to bind PSGL-1 on these cells. SSL5 shows the same binding pattern to leukocytes as described for the interaction of P-selectin with PSGL-1, which suggests that SSL5 also requires expression of a functional, properly glycosylated PSGL-1 for binding. Indeed, we have demonstrated the importance of sulfation and sialic acid residues in the binding of SSL5 to PSGL-1.
SSL5 specificity for neutrophils, monocytes, and NK cells and inability to bind lymphocytes show its specificity for cells of the innate immune system. These cells pose the greatest threat for invading staphylococci and targeting the initial step in recruitment of these cells to sites of infection aids in survival of the bacterium in the host.
Here, we report SSL5 as a potent modulator of the interaction between PSGL-1 and P-selectin. SSL5 hinders initial rolling of neutrophils on the endothelial lining, likely preventing extravasation of neutrophils from blood vessels to infected or damaged tissue. Therefore, SSL5 may be a potential lead in treatment of diseases characterized by excessive recruitment of leukocytes.
Authorship
Contribution: J.B. performed research and wrote the article; M.J.J.G.P. performed binding experiments; L.H.U. contributed to analysis of rolling data; P.J.L. performed and analyzed Biacore data; C.V.D. contributed reagents; K.P.M.v.K., J.A.G.v.S., and C.J.C.d.H. participated in designing the research; and all authors checked the final version of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Jovanka Bestebroer, Experimental Microbiology, University Medical Center Utrecht, Heidelberglaan 100, 3584 CX Utrecht, The Netherlands; e-mail: J.Bestebroer@umcutrecht.nl.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Netherlands Genomics Initiative (NGI-050-71-028).



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal