Abstract
CCAAT enhancer-binding protein (CEBP) transcription factors play pivotal roles in proliferation and differentiation, including suppression of myeloid leukemogenesis. Mutations of CEBPA are found in a subset of acute myeloid leukemia (AML) and in some cases of familial AML. Here, using cytogenetics, fluorescence in situ hybridization (FISH), and molecular cloning, we show that 5 CEBP gene family members are targeted by recurrent IGH chromosomal translocations in BCP-ALL. Ten patients with t(8;14)(q11;q32) involved CEBPD on chromosome 8, and 9 patients with t(14;19)(q32;q13) involved CEBPA, while a further patient involved CEBPG, located 71 kb telomeric of CEBPA in chromosome band 19q13; 4 patients with inv(14)(q11q32)/t(14;14)(q11;q32) involved CEBPE and 3 patients with t(14;20)(q32;q13) involved CEBPB. In 16 patients the translocation breakpoints were cloned using long-distance inverse–polymerase chain reaction (LDI-PCR). With the exception of CEBPD breakpoints, which were scattered within a 43-kb region centromeric of CEBPD, translocation breakpoints were clustered immediately 5′ or 3′ of the involved CEBP gene. Except in 1 patient with t(14;14)(q11;q32), the involved CEBP genes retained germ-line sequences. Quantitative reverse transcription (RT)–PCR showed overexpression of the translocated CEBP gene. Our findings implicate the CEBP gene family as novel oncogenes in BCP-ALL, and suggest opposing functions of CEBP dysregulation in myeloid and lymphoid leukemogenesis.
Introduction
CCAAT enhancer-binding proteins (CEBPs) are a family of 6 multifunctional basic leucine zipper (bZIP) transcription factors. They are defined by conserved carboxy-terminal domains consisting of a leucine zipper dimerization and basic DNA-binding domains (Figure 1).1,2 Five family members comprise CEBPA and CEBPG, separated by 71 kb in chromosome band 19q13, CEBPB in chromosome band 20q13, CEBPD in chromosome band 8q11, and CEBPE in chromosome band 14q11. These genes play pivotal roles in proliferation and differentiation. In hematopoiesis, they have been implicated particularly in the control of myeloid differentiation.3 For example, CEBPE is involved in functional maturation and terminal differentiation of myeloid cells; mutations of CEBPE result in neutrophil-specific granule deficiency.4,5
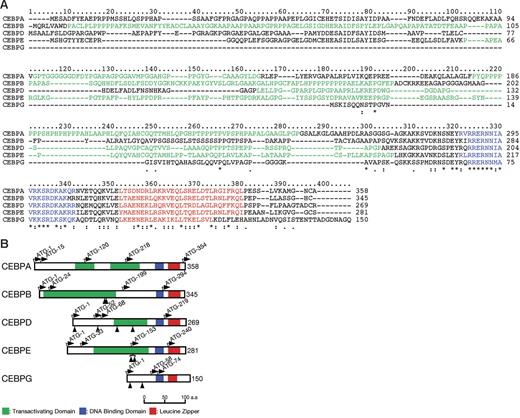
Comparison of CEBP proteins. (A) Multiple amino acid alignment of CEBP proteins. Five members of the CEBP gene family were profiled by the ClustalX program (http://bips.u-strasbg.fr/fr/Documentation/ClustalX). Basic DNA-binding and leucine zipper motifs that compose the bZIP domain are colored blue and red, respectively. A proline- or glycine-rich region suggesting transactivating domains are shown in green. *Positions that have a single, fully conserved residue; a colon indicates strong homology, and a period indicates weak homology, respectively based on the Gonnet Pam250 matrix. (B) Schematic diagram of CEBP structures and mutation sites. Basic DNA-binding motif, leucine zipper motif, and potential transactivating domain are shown in blue, red, and green, respectively. The ATG start codons, including potential initiation codons, are indicated by the arrows with the amino acid numbering. The protein sizes are shown on the right side. Arrowheads indicate the positions of mutations detected in this study.
Comparison of CEBP proteins. (A) Multiple amino acid alignment of CEBP proteins. Five members of the CEBP gene family were profiled by the ClustalX program (http://bips.u-strasbg.fr/fr/Documentation/ClustalX). Basic DNA-binding and leucine zipper motifs that compose the bZIP domain are colored blue and red, respectively. A proline- or glycine-rich region suggesting transactivating domains are shown in green. *Positions that have a single, fully conserved residue; a colon indicates strong homology, and a period indicates weak homology, respectively based on the Gonnet Pam250 matrix. (B) Schematic diagram of CEBP structures and mutation sites. Basic DNA-binding motif, leucine zipper motif, and potential transactivating domain are shown in blue, red, and green, respectively. The ATG start codons, including potential initiation codons, are indicated by the arrows with the amino acid numbering. The protein sizes are shown on the right side. Arrowheads indicate the positions of mutations detected in this study.
Although encoded by genes comprising only 1 or 2 exons, multiple CEBP protein isoforms may nevertheless be produced by the use of different translation initiation sites.6,7 Potential translation initiation sites for the 5 proteins are shown in Figure 1B. CEBPA, for example is typically expressed as 2 major protein isoforms of 42 and 30 kDa that differ significantly in their activities through differences in the amino-terminal region.8 The amino-terminal domains of the CEBP proteins, with the exception of CEBPG, contain transcriptional activation domains. CEBPG lacks any transcriptional activation domain and is therefore thought to represent a naturally occurring dominant-negative form.9 Some isoforms of other CEBP proteins may similarly lack a functional transactivation domain. CEBP proteins form homodimers via the leucine zipper, but may also form complex heterodimers. Under certain circumstances, different family members may replace one another,9,10 but a lack of functional redundancy may also be observed; for example, CEBPE mutant mice express normal levels of other CEBP proteins.11
In terms of malignancy, ectopic expression of CEBPA protein in many experimental situations results in differentiation and loss of proliferation.3,12 For example, CEBPA mutant mice show increased proliferation of myeloid precursors.13 Remarkably, expression of either CEBPA or CEBPB in mouse splenic or bone marrow B cells results in some B cells being reprogrammed to the myeloid lineage.14,15 That CEBPA may function as a tumor suppressor gene in the myeloid lineage in man is supported by the demonstration of mutations of CEBPA in a subset of both sporadic and familial acute myeloid leukemia (AML) with normal karyotypes.3,16,17 These mutations fall either within the amino-terminal region of the protein, resulting in selective translation of a shorter protein product lacking the ability to induce myeloid differentiation, or within the bZIP/DNA-binding domains. CEBPA mutations are frequently biallelic and are associated with a relatively good overall prognosis.18,19 This region of the genome may also exhibit acquired uniparental disomy in AML.20
In view of its potency in reprogramming mature B cells, it is not surprising that CEBPA is subject to strong translational and posttranslational controls. In hematologic malignancies with the translocation t(9;22)(q34;q11), BCR-ABL expression results in down-regulation of CEBPA and CEBPB protein expression at the translational level, through up-regulation of RNA-binding proteins.21 Similar differentiation blocks targeting CEBPA may be achieved in other myeloid malignancies with other chromosomal translocations, either indirectly, through up-regulation of calreticulin, a ubiquitous protein that binds to the GC-rich sequences of CEBPA mRNA and blocks translation, or, in patients with the RUNX1-CBF2T1 fusion, through a direct effect on CEBPA transcription.22,23 Posttranslational mechanisms include phosphorylation of serine 21 via FLT3; phosphorylation of this residue following FLT3 mutation/activation results in loss of the ability of CEBPA to induce granulopoietic differentiation.24
Together, these data highlight the central role of CEBPA in the pathogenesis of various forms of myeloid malignancy. In contrast, the functions of the CEBP gene family in the development of normal lymphocytes and lymphoid malignancies remain largely unknown. Loss of CEBP expression is thought to be concomitant with B-cell differentiation.25 However, some patients with B-cell precursor acute lymphoblastic leukemia (BCP-ALL) have been shown to express CEBPA mRNA in gene expression profiling experiments,26 and similarly, some derived BCP-ALL cell lines may express multiple CEBP mRNAs.27 In mature B-cell malignancies with the translocation t(14;18)(q32;q21), expression of BCL2 from the translocated IGH/BCL2 allele is dependent on CEBPA and CEBPB expression.28 No mutations of any CEBP genes have been reported in lymphoid malignancies.29
Chromosomal translocations involving the immunoglobulin heavy-chain locus (IGH) are a feature of mature B-cell malignancies and result in deregulated expression of the translocated genes due to the proximity of transcriptional enhancers within IGH.30 The partner genes vary according to the disease subtype. Only a small subset of BCP-ALL exhibit IGH translocations. Examples include: t(5;14)(q32;q32), involving IL3; t(1;14)(q21;q32), involving BCL9; t(1;14)(q25;q32), involving LHX4; and t(6;14)(p21;q32), involving ID4. The heterogeneity of these partner genes implicates the involvement of several different pathways in the pathogenesis of this disease.31-34 Interestingly, the t(11;14)(q24;q32) appears to involve directly a microRNA-125b.35 However, all of these translocations have to date only been reported in 1 or 2 sporadic cases.
Recurrent chromosomal translocations t(8;14)(q11;q32) and t(14;19)(q32;q13) involving IGH have been described in BCP-ALL by conventional cytogenetics.36-39 While the involvement of IGH was confirmed by fluorescence in situ hybridization (FISH), the partner genes were unknown. In the t(14;19)(q32;q13) patients the involvement of BCL3, rearranged with IGH in chronic lymphocytic leukemia (CLL) and other mature B-cell malignancies was excluded.38 t(8;14)(q11;q32) has been associated with Down syndrome and the presence of t(9;22)(q34;q11).39 In this study, we demonstrate that these 2 translocations involve CEBPD and CEBPA or CEBPG, while 2 other novel rearrangements, t(14;14)(q11;q32)/inv(14)(q11q32) and t(14;20)(q32;q13), involve CEBPE and CEBPB, respectively. These data suggest an oncogenic role for the CEBP gene family in the pathogenesis of BCP-ALL.
Patients, materials, and methods
Institutional review board approval was obtained for these studies at each of the collaborating centers. Informed consent was obtained in accordance with the Declaration of Helsinki.
Clinical material and derived cell lines
Patients (n = 27) with a diagnosis of BCP-ALL and the chromosomal rearrangements t(14;19)(q32;q13), t(8;14)(q11;q32), t(14;14)(q11;q32)/inv(14)(q11q32), or t(14;20)(q32;p13), were identified from the files of 15 cytogenetic diagnostic centers across Europe and in Australia.
Leukemia cell lines were obtained from the Deutsche Sammlung von Mikroorganismen und Zellkulturen GmbH (Braunschweig, Germany; http://www.dsmz.de), from Dr Linda M. Boxer (Stanford University, Stanford, CA), or from the originating authors; a list of the cell lines used is given in Table S1, available on the Blood website (see the Supplemental Materials link at the top of the online article).
Cytogenetic and FISH analysis
Cytogenetic analysis of diagnostic bone marrow or peripheral blood samples was performed using standard methods. The involvement of IGH was confirmed by FISH using a commercially available dual-color probe (LSI IGH Dual Color, Break Apart Rearrangement Probe; Abbott Diagnostics, Maidenhead, United Kingdom). For mapping the breakpoints of the IGH chromosomal partners, bacterial artificial chromosome (BAC) and P1-derived artificial chromosome (PAC) clones shown in Table 1 were selected. BAC clones (from libraries RPCI11, RPCI13, CTA, CTB, CTC, CTD) and PAC clones (from libraries RPCI1, RPCI3, RPCI4, and RPCI5) positioned along chromosomes 8, 14, 19, and 20 were selected using the Ensembl genome browser (http://www.ensembl.org/Homo_sapiens/index.html) and the Human Genome Browser Gateway (http://genome.ucsc.edu/cgi-bin/hgGateway). Clones were obtained from the The Wellcome Trust Sanger Institute (Hinxton, United Kingdom), the German Resource Centre (RZPD; Berlin, Germany), or Invitrogen (Karlsruhe, Germany). DNA extraction, labeling, and hybridization followed standard procedures. All probes were initially hybridized to normal bone marrow or peripheral blood samples to confirm their location. Visualization of signals was performed on an Axioscope fluorescence microscope (Zeiss, Göttingen, Germany) equipped with appropriate filters and Macprobe (Applied Imaging International, Newcastle, United Kingdom) or ISIS (Metasystems, Altlussheim, Germany) imaging software. For each test, scoring of a minimum of 100 nuclei was performed. Initial breakpoint mapping was carried out by sequential hybridization of selected BAC/PACs to metaphases in patient samples.
BAC and PAC clones used in FISH studies
| Locus, band/clone . | Position in relation to the gene . | Fluorochrome . |
|---|---|---|
| IGH, 14q32 | ||
| Commercial probe | Centromeric | Spectrum Red |
| (Vyis/Abbott Diagnostics) | Telomeric | Spectrum Green |
| CEBPD (1), 8q11 | ||
| RP11-61G18 | Centromeric/Span | Spectrum Green |
| CTD-2655I2 | Telomeric | Spectrum Red |
| CEBPD (2), 8q11 | ||
| RP11-279A10 | Centromeric | Spectrum Green |
| CTD-2655I2 | Telomeric | Spectrum Red |
| CEBPE, 14q11 | ||
| RP11-298I3 | Centromeric | Spectrum Green |
| RP11-124D2 | Telomeric | Spectrum Red |
| CEBPA, 19q13 | ||
| RP11-125M23 | Centromeric | Spectrum Green |
| RP11-54713 | Telomeric | Spectrum Red |
| CEBPG, 19q13 | ||
| CTD-3245C7 | Centromeric | Spectrum Red |
| CTD-3017F12 | Centromeric | Spectrum Green |
| RP11-1101H9 | Telomeric | Spectrum Red |
| CEBPA/G, 19q13 | ||
| RP11-1150B17 | Centromeric | Spectrum Red |
| RP11-298M15 | Centromeric | Spectrum Red |
| CTD-2561O20 | Telomeric | Spectrum Green |
| CTD-2329C7 | Telomeric | Spectrum Green |
| CEBPB, 20q13 | ||
| RP5-1185N5 | Centromeric | Spectrum Green |
| RP4-710H13 | Centromeric | Spectrum Green |
| RP11-290F20 | Telomeric | Spectrum Red |
| RP5-894K16 | Telomeric | Spectrum Red |
| Locus, band/clone . | Position in relation to the gene . | Fluorochrome . |
|---|---|---|
| IGH, 14q32 | ||
| Commercial probe | Centromeric | Spectrum Red |
| (Vyis/Abbott Diagnostics) | Telomeric | Spectrum Green |
| CEBPD (1), 8q11 | ||
| RP11-61G18 | Centromeric/Span | Spectrum Green |
| CTD-2655I2 | Telomeric | Spectrum Red |
| CEBPD (2), 8q11 | ||
| RP11-279A10 | Centromeric | Spectrum Green |
| CTD-2655I2 | Telomeric | Spectrum Red |
| CEBPE, 14q11 | ||
| RP11-298I3 | Centromeric | Spectrum Green |
| RP11-124D2 | Telomeric | Spectrum Red |
| CEBPA, 19q13 | ||
| RP11-125M23 | Centromeric | Spectrum Green |
| RP11-54713 | Telomeric | Spectrum Red |
| CEBPG, 19q13 | ||
| CTD-3245C7 | Centromeric | Spectrum Red |
| CTD-3017F12 | Centromeric | Spectrum Green |
| RP11-1101H9 | Telomeric | Spectrum Red |
| CEBPA/G, 19q13 | ||
| RP11-1150B17 | Centromeric | Spectrum Red |
| RP11-298M15 | Centromeric | Spectrum Red |
| CTD-2561O20 | Telomeric | Spectrum Green |
| CTD-2329C7 | Telomeric | Spectrum Green |
| CEBPB, 20q13 | ||
| RP5-1185N5 | Centromeric | Spectrum Green |
| RP4-710H13 | Centromeric | Spectrum Green |
| RP11-290F20 | Telomeric | Spectrum Red |
| RP5-894K16 | Telomeric | Spectrum Red |
Molecular cloning and analysis of IGH translocation breakpoints
Long-distance inverse–polymerase chain reaction (LDI-PCR) to detect IGHJ translocation breakpoints and/or VDJ rearrangements was carried out as previously described with minor modifications.40,41 Briefly, 100 to 500 ng genomic DNA was digested with restriction enzymes and purified by standard methods. The DNA was diluted to a concentration of 1 μg/mL and incubated at 4°C overnight in the presence of T4 DNA ligase to facilitate intramolecular ligation. The self-ligated circular DNA was used as a template for nested PCR.
PCR primers were modified as follows: IGH/201, 5′-TTCACCCACTCCGACAGTTCTCTTTCCAGCCAATA-3′; IGH/202, 5′-TCAGGAAACCCCACAGGCAGTAGCAGAAAACAAAG-3′; Eμ/203, 5′-CAGATTCTGTTCCGAATCACCGATGCGGCGTCAGC-3′; and Eμ/204, 5′-GCCCCAGCCCTTGTTAATGGACTTGGAGGAATGAT-3′.
The position and orientation of the primers are illustrated in Figure 2. PCR cycling variables as well as the contents of the reaction mixture for LDI-PCR DNA targets were previously described in detail.40 Aliquots of the PCR products were analyzed by agarose gel electrophoresis. Non–germ-line products were purified by gel extraction (Qiagen, Hilden, Germany). The PCR products in some cases were cloned into pGEM-T Easy (Promega, Madison, WI). Nucleotide sequencing of PCR products or cloned DNA was performed with BigDye Terminator Cycle Sequencing Kit (Applied Biosystems, Foster City, CA), and the sequencing reactions were resolved on an ABI 377 automated sequencer (Applied Biosystems). Sequences of the regions of interest were analyzed via the University of California Santa Cruz Genome Bioinformatics database using BLAT.43
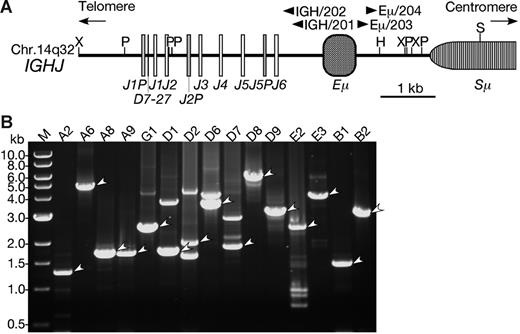
LDI-PCR of the IGH locus in BCP-ALL with CEBP/IGH translocations. (A) Restriction enzyme map of the IGHJ region. The map was constructed based on the GenBank database (accession number NG_001019).42 Boxes represent diverse and junctional region of the IGH (D, J, respectively), IGH-specific enhancer region (Eμ), and switch region (Sμ). Arrowheads are the positions of primers for LDI-PCR. Restriction sites are as follows: H indicates HindIII; P, PstI; S, SacI; and X, XbaI. (B) Ethidium bromide–stained gel electrophoresis of LDI-PCR showing CEBP/IGH fusion genes. The LDI-PCR products representing fusions are indicated by arrowheads. An aliquot of 2 to 10 μL was loaded in each lane and electrophoresed through a 0.7% agarose gel. A DNA ladder (1 kb) was used as a molecular-weight marker.
LDI-PCR of the IGH locus in BCP-ALL with CEBP/IGH translocations. (A) Restriction enzyme map of the IGHJ region. The map was constructed based on the GenBank database (accession number NG_001019).42 Boxes represent diverse and junctional region of the IGH (D, J, respectively), IGH-specific enhancer region (Eμ), and switch region (Sμ). Arrowheads are the positions of primers for LDI-PCR. Restriction sites are as follows: H indicates HindIII; P, PstI; S, SacI; and X, XbaI. (B) Ethidium bromide–stained gel electrophoresis of LDI-PCR showing CEBP/IGH fusion genes. The LDI-PCR products representing fusions are indicated by arrowheads. An aliquot of 2 to 10 μL was loaded in each lane and electrophoresed through a 0.7% agarose gel. A DNA ladder (1 kb) was used as a molecular-weight marker.
CEBP and oncogene mutational analysis
CEBPA, CEBPB, CEBPD, CEBPE, and CEBPG were amplified using long-distance PCR. The oligonucleotide primers were as follows: CEBPA forward, 5′-GCGGGTCCGGGACAGGCCTKGTTCTGGCTTTGAAA-3′; CEBPA reverse, 5′-CCTTCGGGCCTCGCAGGGGTAGGGTGTAGCCACAT-3′; CEBPB forward, 5′-ACCTGGGGAGGAGGTGGGAGTTTACGGGAGGAAGG-3′; CEBPB reverse, 5′-CCAGCTGCAACACCCCCACCCAACCACCAAAACCT-3′; CEBPD forward, 5′-CGCGCTGCGGCCAAGTCCTGGTTTTGATTTCACTC-3′; CEBPD reverse, 5′-CATGACAGGCCATGGTTAACTACATCAGATACACG-3′; CEBPE forward, 5′-TAAGGCTTACATCTCTCCCTCTGGGGTGTGTCCTG-3′; CEBPE reverse, 5′-GCAGATGAGGAAACTGAGGCACAGAAAGACATAAT-3′; CEBPG forward, 5′-ATTCACTTCATCTCACCCTAACAACAAAGCACAGC-3′; and CEBPG reverse, 5′-GGTGATTAAGCGGGAACCCATTTATTCAGTTATTC-3′.
PCR cycling variables as well as the contents of the reaction mixture were as described,44 although the annealing and extension times were reduced to 6 minutes. PCR products were sequenced directly and data from the coding regions of the CEBP genes were compared with the wild-type sequences.
Exons 3 and 13 of PTPN11, exons 14, 15, and 20 of FLT3, and exons 1 and 2 of NRAS and KRAS2 genes were amplified as previously reported.45,46 Exons 5 to 8 of TP53 were amplified as described.47 PCR products were sequenced directly and analyzed using BLAST (http://www.ncbi.nlm.nih.gov/blast).
Quantitative RT-PCR
The methods used were SYBR green in an initial experiment on patient E1, and Qiagen QuantiTect or Applied Biosystems TaqMan gene expression assays (MGB probes) for all other clinical samples and cell lines. Real-time PCR was performed on the ABI Prism 7000 or 7500 Sequence Detection System. Total RNA (1 μg) was DNase treated (Qiagen) and converted to cDNA by reverse transcription (Invitrogen Superscript III kit or Promega M-MLV RT kit) according to the manufacturers' instructions. cDNA (1-5 ng) was used as the template for the PCR reaction; SYBR Green PCR Master Mix and TaqMan PCR Master Mix (Applied Biosytems) were used according to the manufacturer's instructions. The housekeeping control gene was TBP (TATA box–binding protein) for the SYBR green RT-PCR and B2M for the MGB probes. The primers were: CEBPE sense, 5′-GCGTTCTCAAGGCCCCTT-3′; CEBPE antisense, 5′-GGGAGGGCGCCTTCAG-3′; TBP sense, 5′-CACGAACCACGGCACTGAT-3′; and TBP antisense, 5′-TGGAAAACCCAACTTCTGTACAACT-3′.
The PCR reaction was 40 cycles (50°C for 2 minutes followed by a step of denaturation 95°C for 10 minutes and 40 cycles of denaturation at 95°C for 15 seconds, with annealing and extension at 59°C for 1 minute). Primers were validated and optimized with the HL-60 cell line.
cDNA was assayed using the QuantiTect multiplex PCR kit according to the manufacturer's instructions (Qiagen). Primers and probes were designed using the Qiagen online primer design software: CEBPA sense, 5′-GGATAACCTTGTGCCTTG-3′; CEBPA antisense, 5′-CTCCCCTCCTTCTCTCAT-3′; CEBPA probe, 5′-TATTTGGAGGTTTCCTG-3′; CEBPB sense, 5′-GCGACGAGTACAAGATCC-3′; CEBPB antisense, 5′-AGCTGCTTGAACAAGTTCC-3′; CEBPB probe, 5′-AGAAGAAGGTGGAGCA-3′; CEBPD sense, 5′-CCATGTACGACGACGAGA-3′; CEBPD antisense, 5′-GCCTTGTGATTGCTGTTGAAGA-3′; CEBPD probe, 5′-GCTGTGCCACGACGAG-3′; CEBPE sense, 5′-CCGAGGCAGCTACAAT-3′; CEBPE antisense, 5′-CCAAAGGGGCCTTGAGA-3′; CEBPE probe, 5′-CAGACAGCCATGCACC-3′; CEBPG sense, 5′-CAAAAAGAGTTCGCCCAT-3′; CEBPG antisense, 5′-TGCAGTGTGTCTTGTGCTTTC-3′; and CEBPG probe, 5′-GAGAGAGGAACAACATGG-3′.
The target probes were labeled with FAM fluorescent dye. For control purposes the QuantiTect endogenous control assay targeting B2M was used and labeled with YY fluorescent dye. The reactions were duplexed and amplified for 40 cycles. Data evaluation was carried out using the ABI Prism 7000 sequence detection system. Each sample was run in triplicate for the quantification of the CEBPE gene in patient E1 as compared with TBP. For the quantification of CEBP genes in the other cell lines and clinical samples, B2M was used as the control.
Western blotting
Nuclear pellets were isolated by differential centrifugation from whole-cell homogenates, prepared using a modified digitonin (0.04%) permeablization/homogenization technique.48 The final nuclear pellet was harvested at 20 000g for 5 minutes and then solubilized in Laemelli SDS-PAGE sample buffer. To detect the low-abundant CEBPA proteins present in BCP-ALL cell lines, nuclear samples (1 mg/well) were electrophoresed on 11% large-format gels (16 × 18 cm) using standard SDS-denaturing conditions. Proteins were transferred onto nitrocellulose overnight (Hybond C extra; GE Healthcare, Chalfont, United Kingdom) and probed with goat polyclonal antibodies raised to either C-terminal (C-18) or N-terminal (N-19) peptides of CEBPA (Santa Cruz Biotechnology, Santa Cruz, CA). Antibody binding was detected using rabbit antigoat secondary antibody conjugated to horseradish peroxidase (DAKO, Glostrup, Denmark) and by chemiluminescence detected by ECL (GE Healthcare).
Results
Patient characteristics
The clinical and laboratory characteristics of the 27 patients with BCP-ALL studied are shown in Tables 2 and 3. The rearrangements involving 14q32 and the partner chromosomes were identified by cytogenetic analysis: t(14;19)(q32;q13) (n = 10); t(8;14)(q11;q32) (n = 10); t(14;14)(q11;q32) or inv(14)(q11q32) (n = 4); and t(14;20)(q32;q13) (n = 3) (Figure 3A). In 7 patients, the IGH translocation was the sole detectable acquired chromosomal abnormality. Two patients (D2 and D6) with t(8;14)(q11;q32) and 1 patient (E2) with inv(14)(q11q32) had a constitutional gain of chromosome 21 consistent with Down syndrome. The translocation t(9;22)(q34;q11) was present in 2 patients (G1 and D1). In patient G1, the t(14;19)(q32;q13) was shown by cytogenetics and FISH to be secondary to t(9;22)(q34;q11). All patients had a diagnosis of BCP-ALL with a median age of 15 years (range, 3-49 years). The 4 patients younger than 10 years had t(8;14)(q11;q32). The median white blood cell (WBC) count was 6 × 109/L (range, 1-140 × 109/L); in only 5 patients was the count greater than 50 × 109/L, which was not associated with a particular translocation. The prognostic significance of the various translocations, and whether all are associated with a similar prognosis, remains unclear. Of the 23 patients where data were available, 5 died (including the patient G1 with t(9;22)(q34;q11) as the primary change) within 12 months of diagnosis, while 7 remain alive and off treatment more than 3 years after diagnosis (Table 2).
Molecular cytogenetic and CEBP mutational analysis of cases with CEBP/IGH translocations
| Patient no. . | Age, y/sex . | WBC, × 109/L . | OS, mo . | Karyotype* . | IGH FISH (%)† . | CEBP FISH (%)† . | Breakpoint‡ . | CEBP expression/mutation (%) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEBPA patients (CEBPA chr19:38, 485,160-38,482,776‡) | ||||||||||||||||
| A1 | 10/M | 1 | 111+ | 46,XY,t(14;19)(q32;q13)[8]§ | 1R1G1F (93) | 1R1G1F (56) | ||||||||||
| A2 | 11/F | 6 | 62+ | 46,XX,t(14;19)(q32;q13)[4]/46,idem,add(20)(p13)[3]§ | 1R1G1F (87) | 1R1G1F (59) | 3′ UTR of CEBPA chr19:38,483,001 | CEBPA (170) | ||||||||
| A3 | 28/F | 7 | 6 | 46,XX,t(14;19)(q32;q13),add(15)(q26)[5]/46,idem,del(X)(q2?)[2]§ | 1R1G1F (76) | 1R1G1F (72) | ||||||||||
| A4 | 19/F | 4 | 51+ | 46,XX,t(14;19)(q32;q13)[9]§§ | 0R1G2F (60) | 1R1G1F (72) | Not cloned (breakpoint in IGHV region) | CEBPA (2790) | ||||||||
| A5 | 44/F | 5 | 20+ | 57,XX,+X,+X,+4,+6,+10,+14,t(14;19)(q32;q13), ins(15)(q15),+17,+18,+21,+21,+mar[3]/57,idem,−17,+22[2] | 1R1G1F (82) | 1R1G1F (80) | CEBPG mutations Q7Stop/P36S | |||||||||
| A6 | 12/F | 71 | 20+ | 46,XX,dup(3)(q21q2?7),t(14;19)(q32;q13)[6]/46,XX,add(5)(q35),t(14;19) (q32;q13)[3] | 1R1G1F (97) | 1R1G1F (92) | 31 kb centromeric of CEBPA chr19:38,451,468 | CEBPA (40) | ||||||||
| A7 | 40/M | 5 | 12 | 46,XY,t(14;19)(q32;q13)[12]/46,idem,idic(8)(p11)[6] | 1R1G1F (76) | 1R1G1F (66) | CEBPA (60) | |||||||||
| A8 | 32/F | 17 | 84+ | 47,XX,+2,t(14;19)(q32;q13)[20] | 1R1G1F (77) | 1R1G1F (87) | 3′ UTR of CEBPA chr19:38,482,997 | |||||||||
| A9 | 15/F | 1 | 1 | 46,XX,der(9)t(9;?)(p24;?),t(14;19)(q32;q13)[20] | No material | No material | 3′ UTR of CEBPA chr19:38,482,996 | CEBPA (20) | ||||||||
| CEBPG patient (CEBPG chr19:38,556,448-38, 565,431) | ||||||||||||||||
| G1 | 38/F | 94 | < 1 | 46,XX,t(9;22)(q34;q11)[9]/46,idem,i(7)(p10)[2]/47,idem,+8,t(14;19)(q32;q13)[8]/48,idem,+6,+8,t(8;9)(q?;q11), t(14;19)(q32;q13),+der(22)t(9;22)∥ | 1R1G1F (70) | 1R1G1F (100)¶ | 52 kb centromeric of CEBPG chr19:38,504,319 | |||||||||
| CEBPD patients (CEBPD chr8:48,240,783-48, 242,619) | ||||||||||||||||
| D1 | 3/F | 19 | 138+ | 46,XX,t(8;14)(q11.2;q32),t(9;22)(q34;q11)[9]# | No material | No material | 37 kb centromeric of CEBPD chr8:48,775,440 | |||||||||
| D2 | 5/M | 3 | 41+ | 47,XY,t(8;14)(q11;q32),+21c[5] | 1R1G1F (37) | 1R1G1F (54) | 39 kb centromeric of CEBPD chr8:48,773,480 | |||||||||
| D3 | 9/M | 6 | NK | 46,XY,t(8;14)(q11;q32),del(18)(p11.2p11.3), der(19)t(8;19)(?;p13)[9]# | 1R1G1F (72) | 1R1G1F (72) | ||||||||||
| D4 | 15/F | 5 | 32 | 46,XX,t(8;14)(q11;q32)[2] | 1R1G1F (77) | 1R1G1F (82) | ||||||||||
| D5 | 8/M | 48 | NK | 47,XY,+4,t(8;14)(q11;q32)[7] | 1R1G1F (95) | 1R1G1F (85) | ||||||||||
| D6 | 15/F | 5 | 29+ | 47,XX,t(8;14)(q11;q32),+21c[29] | 1R1G1F (82) | 1R1G1F (97) | 36 kb centromeric of CEBPD chr8:48,775,934 | |||||||||
| D7 | 13/M | 8 | 120+ | 47,XY,t(8;14)(q11;q32),+21[11] | 1R1G1F (95) | 1R1G1F (80) | 42 kb centromeric of CEBPD chr8:48,769,988 | |||||||||
| D8 | 49/F | N/A | 1+ | 46,X,t(X;4)(q26;p16),t(8;14)(q11;q32)[10] | 1R1G1F (91) | 1R1G1F (96) | 18 kb centromeric of CEBPD chr8:48,794,358 | |||||||||
| D9 | 24/F | 140 | 6 | 46,XX,t(8;14)(q11.2;q32)[29]** | No material | No material | 61 kb centromeric of CEBPD chr8:48,751,074 | CEBPD (826) | ||||||||
| D10 | 20/M | 2 | 1+ | 45,X,−Y,t(2;14;8)(p11;q32;q11)[9] | 1R1G1F (75) 1Gk(2p11) | 1R1G1F (90) | CEBPD (2818) 1R1G1F (91) | |||||||||
| CEBPE patients (CEBPE chr14:22, 658,314-22,656,354) | ||||||||||||||||
| E1 | 45/M | 1 | 48+ | 45,XY,dup(5)(q14q21),−7,t(14;14)(q11;q32)[17] | 2R1G0F (72) | 1R1G1F (89) | 1.1 kb centromeric of CEBPE chr14:22,655,292 | CEBPE (50) | ||||||||
| E2 | 15/M | 39 | NK | 47, XY,inv(14)(q11q32),+21c | 1R1G1F (94) | 1R1G1F (92) | 0.8 kb telomeric of CEBPE chr14:22,659,084 | |||||||||
| E3 | 25/M | N/A | NK | 46,XY,inv(9)(p21q11),inv(14)(q11q32)[3]/6,idem,del(12)(p11p12)[3] | 1R1G1F (30) | 1R1G1F (58) | 0.8 kb telomeric of CEBPE chr14:22,659,097 | CEBPE (10) CEBPE mutation L155M | ||||||||
| E4 | 45/M | 24 | 19+ | 46,XY,der(2)t(1;2)(q1?;q3?7),inv(14)(q11q32),inc[3] | 1R1G1F (53) | 1R1G1F (57) | ||||||||||
| CEBPB patients (CEBPB chr20:48, 240,783-48,242,619) | ||||||||||||||||
| B1 | 15/M | 3 | 87+ | 46,XY,inv(9)(p1?3q3?2),t(14;20)(q32;q13)[14] | 1R1G1F (83) | 1R1G1F (88) | 1.3 kb centromeric of CEBPB chr20:48,239,531 | CEBPB (1490) | ||||||||
| B2 | 13/F | 103 | 42+ | 47,XX,t(14;20)(q32;q13),add(21)(p1?),+add(21)(p1?)[9] | 1R1G1F (77) | 1R1G1F (85) | 1.3 kb centromeric of CEBPB chr20:48,239,523 | CEBPB (31) | ||||||||
| B3 | 35/F | 75 | 7+ | 46,XX,i(7)(q10)[47].ish t(14;20)(q32;q13)(IGH cen+, IGHV−;IGH cen−,IGHV+)[3] | 1R1G1F (85) | 1R1G1F (72) | ||||||||||
| Patient no. . | Age, y/sex . | WBC, × 109/L . | OS, mo . | Karyotype* . | IGH FISH (%)† . | CEBP FISH (%)† . | Breakpoint‡ . | CEBP expression/mutation (%) . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| CEBPA patients (CEBPA chr19:38, 485,160-38,482,776‡) | ||||||||||||||||
| A1 | 10/M | 1 | 111+ | 46,XY,t(14;19)(q32;q13)[8]§ | 1R1G1F (93) | 1R1G1F (56) | ||||||||||
| A2 | 11/F | 6 | 62+ | 46,XX,t(14;19)(q32;q13)[4]/46,idem,add(20)(p13)[3]§ | 1R1G1F (87) | 1R1G1F (59) | 3′ UTR of CEBPA chr19:38,483,001 | CEBPA (170) | ||||||||
| A3 | 28/F | 7 | 6 | 46,XX,t(14;19)(q32;q13),add(15)(q26)[5]/46,idem,del(X)(q2?)[2]§ | 1R1G1F (76) | 1R1G1F (72) | ||||||||||
| A4 | 19/F | 4 | 51+ | 46,XX,t(14;19)(q32;q13)[9]§§ | 0R1G2F (60) | 1R1G1F (72) | Not cloned (breakpoint in IGHV region) | CEBPA (2790) | ||||||||
| A5 | 44/F | 5 | 20+ | 57,XX,+X,+X,+4,+6,+10,+14,t(14;19)(q32;q13), ins(15)(q15),+17,+18,+21,+21,+mar[3]/57,idem,−17,+22[2] | 1R1G1F (82) | 1R1G1F (80) | CEBPG mutations Q7Stop/P36S | |||||||||
| A6 | 12/F | 71 | 20+ | 46,XX,dup(3)(q21q2?7),t(14;19)(q32;q13)[6]/46,XX,add(5)(q35),t(14;19) (q32;q13)[3] | 1R1G1F (97) | 1R1G1F (92) | 31 kb centromeric of CEBPA chr19:38,451,468 | CEBPA (40) | ||||||||
| A7 | 40/M | 5 | 12 | 46,XY,t(14;19)(q32;q13)[12]/46,idem,idic(8)(p11)[6] | 1R1G1F (76) | 1R1G1F (66) | CEBPA (60) | |||||||||
| A8 | 32/F | 17 | 84+ | 47,XX,+2,t(14;19)(q32;q13)[20] | 1R1G1F (77) | 1R1G1F (87) | 3′ UTR of CEBPA chr19:38,482,997 | |||||||||
| A9 | 15/F | 1 | 1 | 46,XX,der(9)t(9;?)(p24;?),t(14;19)(q32;q13)[20] | No material | No material | 3′ UTR of CEBPA chr19:38,482,996 | CEBPA (20) | ||||||||
| CEBPG patient (CEBPG chr19:38,556,448-38, 565,431) | ||||||||||||||||
| G1 | 38/F | 94 | < 1 | 46,XX,t(9;22)(q34;q11)[9]/46,idem,i(7)(p10)[2]/47,idem,+8,t(14;19)(q32;q13)[8]/48,idem,+6,+8,t(8;9)(q?;q11), t(14;19)(q32;q13),+der(22)t(9;22)∥ | 1R1G1F (70) | 1R1G1F (100)¶ | 52 kb centromeric of CEBPG chr19:38,504,319 | |||||||||
| CEBPD patients (CEBPD chr8:48,240,783-48, 242,619) | ||||||||||||||||
| D1 | 3/F | 19 | 138+ | 46,XX,t(8;14)(q11.2;q32),t(9;22)(q34;q11)[9]# | No material | No material | 37 kb centromeric of CEBPD chr8:48,775,440 | |||||||||
| D2 | 5/M | 3 | 41+ | 47,XY,t(8;14)(q11;q32),+21c[5] | 1R1G1F (37) | 1R1G1F (54) | 39 kb centromeric of CEBPD chr8:48,773,480 | |||||||||
| D3 | 9/M | 6 | NK | 46,XY,t(8;14)(q11;q32),del(18)(p11.2p11.3), der(19)t(8;19)(?;p13)[9]# | 1R1G1F (72) | 1R1G1F (72) | ||||||||||
| D4 | 15/F | 5 | 32 | 46,XX,t(8;14)(q11;q32)[2] | 1R1G1F (77) | 1R1G1F (82) | ||||||||||
| D5 | 8/M | 48 | NK | 47,XY,+4,t(8;14)(q11;q32)[7] | 1R1G1F (95) | 1R1G1F (85) | ||||||||||
| D6 | 15/F | 5 | 29+ | 47,XX,t(8;14)(q11;q32),+21c[29] | 1R1G1F (82) | 1R1G1F (97) | 36 kb centromeric of CEBPD chr8:48,775,934 | |||||||||
| D7 | 13/M | 8 | 120+ | 47,XY,t(8;14)(q11;q32),+21[11] | 1R1G1F (95) | 1R1G1F (80) | 42 kb centromeric of CEBPD chr8:48,769,988 | |||||||||
| D8 | 49/F | N/A | 1+ | 46,X,t(X;4)(q26;p16),t(8;14)(q11;q32)[10] | 1R1G1F (91) | 1R1G1F (96) | 18 kb centromeric of CEBPD chr8:48,794,358 | |||||||||
| D9 | 24/F | 140 | 6 | 46,XX,t(8;14)(q11.2;q32)[29]** | No material | No material | 61 kb centromeric of CEBPD chr8:48,751,074 | CEBPD (826) | ||||||||
| D10 | 20/M | 2 | 1+ | 45,X,−Y,t(2;14;8)(p11;q32;q11)[9] | 1R1G1F (75) 1Gk(2p11) | 1R1G1F (90) | CEBPD (2818) 1R1G1F (91) | |||||||||
| CEBPE patients (CEBPE chr14:22, 658,314-22,656,354) | ||||||||||||||||
| E1 | 45/M | 1 | 48+ | 45,XY,dup(5)(q14q21),−7,t(14;14)(q11;q32)[17] | 2R1G0F (72) | 1R1G1F (89) | 1.1 kb centromeric of CEBPE chr14:22,655,292 | CEBPE (50) | ||||||||
| E2 | 15/M | 39 | NK | 47, XY,inv(14)(q11q32),+21c | 1R1G1F (94) | 1R1G1F (92) | 0.8 kb telomeric of CEBPE chr14:22,659,084 | |||||||||
| E3 | 25/M | N/A | NK | 46,XY,inv(9)(p21q11),inv(14)(q11q32)[3]/6,idem,del(12)(p11p12)[3] | 1R1G1F (30) | 1R1G1F (58) | 0.8 kb telomeric of CEBPE chr14:22,659,097 | CEBPE (10) CEBPE mutation L155M | ||||||||
| E4 | 45/M | 24 | 19+ | 46,XY,der(2)t(1;2)(q1?;q3?7),inv(14)(q11q32),inc[3] | 1R1G1F (53) | 1R1G1F (57) | ||||||||||
| CEBPB patients (CEBPB chr20:48, 240,783-48,242,619) | ||||||||||||||||
| B1 | 15/M | 3 | 87+ | 46,XY,inv(9)(p1?3q3?2),t(14;20)(q32;q13)[14] | 1R1G1F (83) | 1R1G1F (88) | 1.3 kb centromeric of CEBPB chr20:48,239,531 | CEBPB (1490) | ||||||||
| B2 | 13/F | 103 | 42+ | 47,XX,t(14;20)(q32;q13),add(21)(p1?),+add(21)(p1?)[9] | 1R1G1F (77) | 1R1G1F (85) | 1.3 kb centromeric of CEBPB chr20:48,239,523 | CEBPB (31) | ||||||||
| B3 | 35/F | 75 | 7+ | 46,XX,i(7)(q10)[47].ish t(14;20)(q32;q13)(IGH cen+, IGHV−;IGH cen−,IGHV+)[3] | 1R1G1F (85) | 1R1G1F (72) | ||||||||||
Deceased patients are indicated by italics.
R indicates red signal; G, green signal; F, fusion signal; and NK, not known.
Karyotypes written according to ISCN (2005).49 The normal clone has been omitted from abnormal karyotypes.
Only predominant aberrant signal pattern is shown.
BLAT search results using May 2004 freeze. Chromosome is given first, followed by position.
Previously reported.36
previously reported.50
In metaphase nuclei, FISH and molecular cloning discriminated CEBPG from CEBPA in this case. In patient B3, no 14q32 translocation was seen by cytogenetics but FISH detected an IGH breakpoint. Verification of t(14;20)(q32;q13) was obtained by further FISH experiments in metaphase and interphase preparations. CEBP expression was determined by QRT-PCR and is expressed relative to levels of expression seen in the myeloid cell line HL-60.
Previously reported.37
**Previously reported.38
Analysis of immunophenotype, VDJ assembly, and RAS mutation in CEBP/IGH translocation cases
| Patient no. . | Immunophenotype* . | VDJ assembly . | RAS mutation . | |||
|---|---|---|---|---|---|---|
| Positive . | Negative . | VDJ status . | Homology of VH, % . | NRAS . | KRAS2 . | |
| A1 | CD10, CD19, CD3, HLA-DR | CD13, CD14, CD33 | — | — | — | — |
| A2 | CD10, CD19, CD2, CD7, TdT, HLA-DR | CD33, CD13, CD20 | Only germline detected | WT | WT | |
| A3 | CD19, CD34, TdT, HLA-DR | CD10, CD14, slgM, clgM | — | — | — | — |
| A4 | CD10, CD19, CD22, TdT, IgM, HLA-DR | CD13, CD33, CD34 | — | — | — | — |
| A5 | — | — | V3-23D2-21J6, D5-12J6 | 100.0, — | WT | Codon 13 |
| A6 | CD10, CD19, HLA-DR | CD2, CD7, slgM | V2-5D2-15J6 | 99.8 | WT | WT |
| A7 | CD45, CD19, CD22, CD24, HLA-DR, CD34, TdT, cCD79a | CD2, CD3, CD4, CD5, CD7, CD8, CD20, CD10, CD14, CD13, CD15, CD33, CD41, CD65, CD117 | — | — | — | — |
| A8 | CD10, CD19, TdT, HLA-DR | CD3, CD5, CD7, CD13, CD33, CD34 | D6-6J6 | — | Codon 12 | WT |
| A9 | CD19, CD10, CD22, CD24, HLA-DR, CD34, TdT, CD79a | MPO, CD13, CD14, CD15, CD33, CD65 clgM, cCD3, CD7 | V5-51D3-22J4 | 100.0 | WT | WT |
| G1 | HLA-DR, CD34, CD10, CD19, CD22, CD2 | CD20, slgM, clgM, slgk, slgl, CD7, CD5, CD1a, CD4, CD8, CD3, cCD3, CD56, CD13, CD33, CD14, CD15, CD65w, CD36, CD42, CD61 | V3-43D†J1 | 98.2 | WT | WT |
| D1 | — | — | V4-34D3-16J5 | 100.0 | WT | WT |
| D2 | CD10, CD19, HLA-DR, CD22, CD34, CD45, TdT | CD2, CD7, CD13, CD33 | V7-4.1D3-9J6 | 100.0 | WT | WT |
| D4 | CD10, CD19, CD22, CD79a, clgM, CD2, CD7, HLA-DR, TdT, CD34 | CD3, CD13, CD14, CD15, CD33, CD117 | — | — | — | — |
| D6 | CD19, CD34, CD10, CD22, HLA-DR, TdT, cCD79a, clgM | CD45, CD20, slgk, slgl | V4-39D3-22J6 | 99.0 | WT | WT |
| D7 | CD10, CD19, CD24, TdT, HLA-DR | CD11c, CD3, CD4, CD7, CD8, CD33 | D6-19J4 | — | WT | WT |
| D8 | CD10, CD19, CD34, HLA-DR, CD13, CD33 | CD2, CD5, CD7, CD8, CD14, CD15, CD36, CD56, CD61, CD64 | D2-2J6 | — | WT | WT |
| D9 | CD10 | — | — | — | WT | WT |
| D10 | CD19, CD10, HLA-DR, CD135, TdT (96%), CD79a (25%), CD13 (53%), CD235a (52%), CD90 (48%) | — | — | — | — | — |
| E1 | CD10, CD19, CD34, CD38 | CD5, CD7, CD13, CD33, CD117 | — | — | — | — |
| E2 | CD19, CD10, TdT, cCD3 | V7-4.1D6-19J6, V4-4D6-19J6 | 100.0, 100.0 | WT | WT | |
| E3 | CD19, CD24, Tdt, CD10 | CD5, CD7, CD2, sCD3, CD13, CD33 | V1-46D3-22J3, V3-21D3-16J4 | 100.0, 100.0 | Codon 61 | WT |
| B1 | CD19, CD20, CD22, CD45, CD10, CD34, TdT | CD2, CD3, CD5, CD4, CD8, CD23, CD37, CD7, CD13, CD33, CD14, CD11b, CD11c, CD61, clgM | V3-02,1PD3-3J6 | 99.0 | Codon 13 | WT |
| B2 | CD10, CD19, CD22, HLA-DR, CD45, CD34 | CD20, CD5, CD3, CD7, CD33, CD13, CD41a, CD14 | V3-23D3-22J4 | 100.0 | WT | WT |
| B3 | CD19, CD34, CD10, HLA-DR, TdT | CD3, CD4, CD7, CD13, CD14, CD33 | — | — | — | — |
| Patient no. . | Immunophenotype* . | VDJ assembly . | RAS mutation . | |||
|---|---|---|---|---|---|---|
| Positive . | Negative . | VDJ status . | Homology of VH, % . | NRAS . | KRAS2 . | |
| A1 | CD10, CD19, CD3, HLA-DR | CD13, CD14, CD33 | — | — | — | — |
| A2 | CD10, CD19, CD2, CD7, TdT, HLA-DR | CD33, CD13, CD20 | Only germline detected | WT | WT | |
| A3 | CD19, CD34, TdT, HLA-DR | CD10, CD14, slgM, clgM | — | — | — | — |
| A4 | CD10, CD19, CD22, TdT, IgM, HLA-DR | CD13, CD33, CD34 | — | — | — | — |
| A5 | — | — | V3-23D2-21J6, D5-12J6 | 100.0, — | WT | Codon 13 |
| A6 | CD10, CD19, HLA-DR | CD2, CD7, slgM | V2-5D2-15J6 | 99.8 | WT | WT |
| A7 | CD45, CD19, CD22, CD24, HLA-DR, CD34, TdT, cCD79a | CD2, CD3, CD4, CD5, CD7, CD8, CD20, CD10, CD14, CD13, CD15, CD33, CD41, CD65, CD117 | — | — | — | — |
| A8 | CD10, CD19, TdT, HLA-DR | CD3, CD5, CD7, CD13, CD33, CD34 | D6-6J6 | — | Codon 12 | WT |
| A9 | CD19, CD10, CD22, CD24, HLA-DR, CD34, TdT, CD79a | MPO, CD13, CD14, CD15, CD33, CD65 clgM, cCD3, CD7 | V5-51D3-22J4 | 100.0 | WT | WT |
| G1 | HLA-DR, CD34, CD10, CD19, CD22, CD2 | CD20, slgM, clgM, slgk, slgl, CD7, CD5, CD1a, CD4, CD8, CD3, cCD3, CD56, CD13, CD33, CD14, CD15, CD65w, CD36, CD42, CD61 | V3-43D†J1 | 98.2 | WT | WT |
| D1 | — | — | V4-34D3-16J5 | 100.0 | WT | WT |
| D2 | CD10, CD19, HLA-DR, CD22, CD34, CD45, TdT | CD2, CD7, CD13, CD33 | V7-4.1D3-9J6 | 100.0 | WT | WT |
| D4 | CD10, CD19, CD22, CD79a, clgM, CD2, CD7, HLA-DR, TdT, CD34 | CD3, CD13, CD14, CD15, CD33, CD117 | — | — | — | — |
| D6 | CD19, CD34, CD10, CD22, HLA-DR, TdT, cCD79a, clgM | CD45, CD20, slgk, slgl | V4-39D3-22J6 | 99.0 | WT | WT |
| D7 | CD10, CD19, CD24, TdT, HLA-DR | CD11c, CD3, CD4, CD7, CD8, CD33 | D6-19J4 | — | WT | WT |
| D8 | CD10, CD19, CD34, HLA-DR, CD13, CD33 | CD2, CD5, CD7, CD8, CD14, CD15, CD36, CD56, CD61, CD64 | D2-2J6 | — | WT | WT |
| D9 | CD10 | — | — | — | WT | WT |
| D10 | CD19, CD10, HLA-DR, CD135, TdT (96%), CD79a (25%), CD13 (53%), CD235a (52%), CD90 (48%) | — | — | — | — | — |
| E1 | CD10, CD19, CD34, CD38 | CD5, CD7, CD13, CD33, CD117 | — | — | — | — |
| E2 | CD19, CD10, TdT, cCD3 | V7-4.1D6-19J6, V4-4D6-19J6 | 100.0, 100.0 | WT | WT | |
| E3 | CD19, CD24, Tdt, CD10 | CD5, CD7, CD2, sCD3, CD13, CD33 | V1-46D3-22J3, V3-21D3-16J4 | 100.0, 100.0 | Codon 61 | WT |
| B1 | CD19, CD20, CD22, CD45, CD10, CD34, TdT | CD2, CD3, CD5, CD4, CD8, CD23, CD37, CD7, CD13, CD33, CD14, CD11b, CD11c, CD61, clgM | V3-02,1PD3-3J6 | 99.0 | Codon 13 | WT |
| B2 | CD10, CD19, CD22, HLA-DR, CD45, CD34 | CD20, CD5, CD3, CD7, CD33, CD13, CD41a, CD14 | V3-23D3-22J4 | 100.0 | WT | WT |
| B3 | CD19, CD34, CD10, HLA-DR, TdT | CD3, CD4, CD7, CD13, CD14, CD33 | — | — | — | — |
WT indicates wild type; and —, data not available or not done, due to lack of suitable clinical material.
Immunophenotypic data were obtained at diagnosis using variable antibody panels as indicated. Patient D8 coexpressed CD13 and CD33; interestingly, this patient only exhibited DJ rather than VDJ rearrangement. Detailed immunophenotyping was performed in patient D10; all blasts were CD19, CD10, and TdT positive. Coexpression of CD13 and CD235a was seen in about 50% of cells, but these did not express either myeloperoxidase or CD33. Neither immunophenotypic nor mutational data were available from patients D3, D5, and E4.
D segment was not identified by BLAST research.
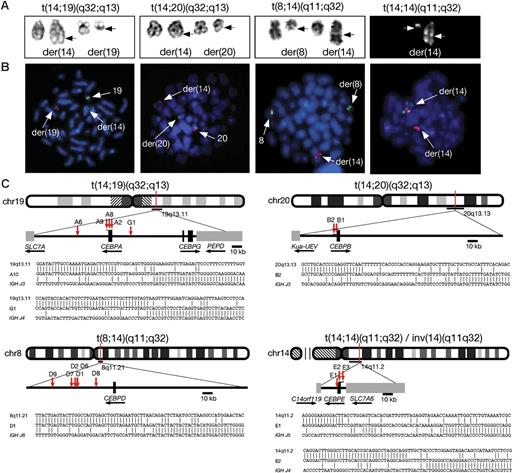
Molecular cytogenetic analysis of patients with CEBP/IGH translocations. (A) Representative partial karyotypes of the 4 IGH translocation from left to right: G-banded, t(14;19)(q32;q13), t(14;20)(q32;q13), t(8;14)(q11;q32); and R-banded, t(14;14)(q11;q32). In the G-banded images the normal chromosomes are shown on the left and the abnormal (der) chromosomes are shown on the right (breakpoints arrowed). There are no normal chromosomes 14 in the R-banded image as both are involved in the translocation. (B) Equivalent FISH images of DAPI-stained metaphases hybridized with specific probes (from left to right) for CEBPA, CEBPB, CEBPD, and CEBPE (probe details are provided in Table 1). A normal red/green fusion signal is seen on the normal chromosomes at 19q13, 20q13, and 8q11. In t(14;14) the normal fusion is seen at 14q11 on the larger der(14) chromosome. A splitting of 1 fusion signal between the derivative chromosomes is shown, with the centromeric signal remaining on the derivative partner chromosome, while the telomeric signal has translocated to the IGH locus in 14q32 on the der(14) (arrows). (C) Idiograms to show the location of breakpoints cloned by LDI-PCR from the IGHJ6 segment. Representative breakpoint sequences with identity to IGHJ and the corresponding CEBP locus are shown.
Molecular cytogenetic analysis of patients with CEBP/IGH translocations. (A) Representative partial karyotypes of the 4 IGH translocation from left to right: G-banded, t(14;19)(q32;q13), t(14;20)(q32;q13), t(8;14)(q11;q32); and R-banded, t(14;14)(q11;q32). In the G-banded images the normal chromosomes are shown on the left and the abnormal (der) chromosomes are shown on the right (breakpoints arrowed). There are no normal chromosomes 14 in the R-banded image as both are involved in the translocation. (B) Equivalent FISH images of DAPI-stained metaphases hybridized with specific probes (from left to right) for CEBPA, CEBPB, CEBPD, and CEBPE (probe details are provided in Table 1). A normal red/green fusion signal is seen on the normal chromosomes at 19q13, 20q13, and 8q11. In t(14;14) the normal fusion is seen at 14q11 on the larger der(14) chromosome. A splitting of 1 fusion signal between the derivative chromosomes is shown, with the centromeric signal remaining on the derivative partner chromosome, while the telomeric signal has translocated to the IGH locus in 14q32 on the der(14) (arrows). (C) Idiograms to show the location of breakpoints cloned by LDI-PCR from the IGHJ6 segment. Representative breakpoint sequences with identity to IGHJ and the corresponding CEBP locus are shown.
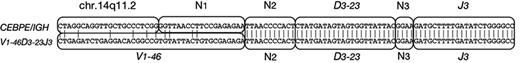
Immunophenotypic results are given in Table 3. The panels of monoclonal antibodies varied from left to left. However, there was no consistent expression of myeloid differentiation antigens, which were only coexpressed by 2 patients (D8 and D10). Intrachromosomal IGH (V)-D-J rearrangements were analyzed using LDI-PCR. The results were consistent with the diagnosis of BCP-ALL as shown in Table 3. All but 1 patient showed clonal immunoglobulin VDJ or DJ recombination, with unmutated VH gene segment usage. Of 11 VDJ rearrangements, 9 were out of frame, indicating that the CEBP/IGH translocations occurred in B-cell precursors destined to undergo apoptosis.51 Interestingly, 1 patient (E3; Figure 4) exhibited an identical DJ rearrangement in both a VDJ rearrangement and in the allele translocated to CEBPE. This translocation may therefore have arisen due to an error in VDJ replacement.52 These data indicate that the CEBPE translocation in this case occurred after an unidentified initiating oncogenic event.
Sequence analysis of patient E3. One of 2 VDJ rearrangements in patient E3 comprised V1-46, D3-23, and J3, whereas the breakpoint region of the CEBPE/IGH fusion is a nucleotide sequence from chr.14q11.2, and the same DJ including some N segments (N2 and N3). N represents nucleotides of unknown origin.
Sequence analysis of patient E3. One of 2 VDJ rearrangements in patient E3 comprised V1-46, D3-23, and J3, whereas the breakpoint region of the CEBPE/IGH fusion is a nucleotide sequence from chr.14q11.2, and the same DJ including some N segments (N2 and N3). N represents nucleotides of unknown origin.
FISH analysis and molecular cloning
FISH using flanking probes confirmed the involvement of IGH in 24 patients with available fixed-cell suspensions. Sequential FISH was used to map the chromosomal breakpoints and indicated the partner genes to be 1 of the members of the CEBP gene family in the same 24 patients: CEBPA in t(14;19)(q32;q13), CEBPD in t(8;14)(q11;q32), CEBPE in t(14;14)(q11;q32)/inv(14)(q11q32), and CEBPB in t(14;20)(q32;q13) (Figure 3B). Breakpoint cloning by LDI-PCR with primers to IGHJ6 was successful in 16 patients of 17 with high-molecular-weight DNA available, including the 3 patients in which FISH was not possible, and confirmed that the CEBP genes were the targets (Figure 3C; Table 2). Among the patients with t(14;19)(q32;q13), FISH and molecular cloning discriminated between translocations involving CEBPA (n = 9) and CEBPG (n = 1). As CEBPG is located only 71 kb distal of CEBPA, it was detected using the same FISH probes as the other 9 patients with t(14;19)(q32;q13). Further FISH probes telomeric of CEBPA, specific for CEBPG (Table 1) and hybridized to metaphases, verified the LDI-PCR result in this patient (data not shown).
Analysis of the breakpoint junctions showed that for CEBPA, CEBPB, and CEBPE the breakpoints were mostly located either within the 3′ untranslated region (UTR) or immediately adjacent to the gene. With the exception of the CEBPD translocations, breakpoints were tightly clustered. In 3 of the 4 patients with t(14;19)(q32;q13), the breakpoints fell within 6 nucleotides of each other within the 3′ UTR of CEBPA. One CEBPA breakpoint fell about 20 kb centromeric of CEBPA. CEBPD breakpoints were scattered over a region 18 to 61 kb centromeric of the 3′ end of CEBPD. CEBPE was involved in both rearrangements involving (14q11;) breakpoints of the inversion, inv(14)(q11q32), were located immediately 5′ of the gene, while in the translocation, t(14;14)(q11;q32), it was located 1000 bp of the 3′ UTR (Figure 3C).
Quantitative RT-PCR and Western blotting
Quantitative RT-PCR was used to assess CEBP gene expression in 11 of the patients with CEBP/IGH translocations, comprising more than 1 patient from each translocation group. Comparison was made with derived BCP-ALL cell lines (none of which exhibited a CEBP/IGH translocation) and the myeloid cell line, HL-60, which expresses CEBPA and CEBPE constitutively. Detection of CEBP transcripts is technically difficult since CEBPA, CEBPB, and CEBPD are intronless, with the consequence that genomic DNA contamination has to be eliminated to obtain meaningful results. Overall, the quality of RNA obtained from several clinical samples was poor, which particularly affected reproducibility of the CEBPD quantifications. Of the 18 BCP-ALL cell lines, 16 showed very low or undetectable levels of expression of CEBPA, CEBPB, CEBPD, and CEBPE, with low expression of CEBPG in all cases (Table S1). In contrast, all patient samples showed high-level CEBP expression of the translocated gene, comparable or in some instances higher than that seen in HL-60 (Table 2). No material was available to assess the expression level of CEBPG in the 1 patient involving this gene (G1). It is unlikely that CEBP expression resulted from contamination by normal myeloid cells, since the samples were from BCP-ALL at diagnosis in which most of the bone marrow was replaced by leukemic blasts.
No suitable material was available from our clinical cases to determine CEBP protein expression. Western blot of derived BCP-ALL cell lines confirmed expression of CEBPA protein in some but not all patients with CEBPA mRNA expression. Three BCP-ALL cell lines (LK63, REH, and NALM-27) expressed relatively low levels of CEBPA. Interestingly, the LK63 BCP-ALL cell line53 exhibited preferential expression of the 30-kDa CEBPA isoform (data not shown). The NALM-27 cell line expressed high levels of CEBPA mRNA27 but failed to express any detectable CEBPA protein (data not shown). This cell line exhibits t(9;22)(q34;q11) and thus expresses the BCR-ABL kinase that specifically suppresses CEBPA expression.22
CEBP mutational analysis
Given the tumor-suppressive function of CEBPA in AML, we sought mutations of the CEBP genes in the patients with CEBP/IGH translocations (Table 2). With the exception of 1 patient with inv(14)(q11q32) that showed a missense mutation of CEBPE, none of the patients with CEBP/IGH translocations showed CEBP mutations of the involved CEBP gene. Specifically, no mutations in the 5′ region of CEBPA comparable with those seen in AML were detected. However, mutations of all CEBP genes, with the exception of CEBPA, were seen at low frequency in 1 patient (A5) and in 4 cell lines. The one BCP-ALL cell line (REH) that expressed CEBPB also showed 2 missense mutations within one CEBPB allele (Table S1). No patients with CEBP/IGH translocations exhibited PTPN11 or FLT3 mutations, although 4 of 16 patients tested showed NRAS or KRAS2 mutations; no patients had TP53 mutations (Table 2).
Discussion
In this study we define a new subgroup of BCP-ALL with IGH translocations involving 5 members of the CEBP gene family. It was difficult to assess the precise frequency of CEBP/IGH translocations in BCP-ALL from these highly selected patients. However, 2% of childhood ALL (15 of 671 patients screened prospectively by FISH) and 8% of adult ALL (12 of 153 patients) showed rearrangements of the IGH locus (C.J.H. and A. V. Moorman, unpublished observations, July 2006; Leukaemia Research UK Cancer Cytogenetics Group Karyotype Database in Acute Leukaemia54 ). Patients with CEBP/IGH translocations collectively comprised approximately 1% of BCP-ALL. CEBP/IGH translocations occurred predominantly in older children and adults, with no consistent expression of myeloid differentiation antigens. Their possible prognostic significance remains to be determined from larger, prospective studies. A parallel study from the Groupe Francophone de Cytogénétique Hématologique (GFCH) has recently reported the involvement of CEBPA in the t(14;19)(q32;q13) in BCP-ALL.50 However, to our knowledge, this is the first report of multiple members of a single gene family being involved with the same locus in chromosomal translocations within one disease.
Our data indicate that deregulated expression of unmutated CEBP genes can occur in B-cell precursors and contribute to malignant transformation. CEBPA has been previously implicated as a tumor-suppressor gene in AML by the demonstration of CEBPA mutations and by the down-regulation of CEBPA mRNA or protein as a consequence of several leukemic fusion transcripts, including BCR-ABL, CBFB-MYH11, and RUNX1-CBF2T1.22,23,55 Although in our study we were unable to evaluate CEBP protein expression due to lack of suitable clinical material, it was shown for CEBPA by GFCH;50 there is no reason to assume that protein expression of the other CEBP genes should be different.
How deregulated CEBP expression might transform B-cell precursors remains unclear. If there is a mechanism common to the 5 translocations, it is likely to be mediated by the conserved carboxy-terminal leucine zipper and DNA-binding domains. The levels of CEBP mRNA expression in patients with translocations were high, and often comparable to those seen in myelomonocytic cell lines. CEBP expression in uncommitted hemopoietic stem cells usually leads to up-regulation of PU.1 and down-regulation of PAX5, with consequent suppression of B-cell differentiation and commitment to the myeloid lineage. This program must therefore be subverted in some B-cell precursors, since the patients studied here were “typical” BCP-ALL with no overt immunophenotypic evidence for commitment to the myeloid lineage.
Possible mechanisms include 1 or more of the following: (1) translational control: selective expression of shorter CEBP protein isoforms such as the 30-kDa isoform of CEBPA that lacks the amino-terminal transcriptional activation domain and thus the ability to induce myeloid proliferation. (2) posttranslational modification: both FLT3 and RAS/phosphorylation modify CEBP functions.24,56 However, no consistent mutation of NRAS, KRAS2, FLT3, or PTPN11 were seen in the patients studied here. (3) CEBP translocation as a secondary event, following “blocking” of the myeloid differentiation program. However, the nature of possible antecedent events remains obscure. Interestingly, the lack of transforming ability of overexpression of CEBPA alone in normal B-cell precursors has recently been shown in a mouse transgenic H2K-CEBPA-Eμ model.57
We currently lack in vitro models that recapitulate the cases studied here. Whatever the mechanism(s), our findings implicate the CEBP gene family as novel oncogenes in the pathogenesis of BCP-ALL, and suggest opposing functions of CEBP dysregulation in myeloid and lymphoid leukemogenesis. Similar cell-type specific effects have recently been reported for the KLF gene that may act as either a dominant oncogene or a tumor-suppressor gene depending on cellular context.58
Authorship
Contribution: T.A., T.B., L.J.R., K.-j.S., A.M., R.W., E.L.K., D.G.B., K.C., L.H., S.G., J.I.M.-S., H.W., and J.C.S. performed the research reported here and analyzed data; M.G.A., M.B., M.J.C., T.D., O.A.H., A.H., H.K., M.L., D.M.L., S.M., F.N.-K., I.R.-W., C.S., S.S., P.T., and M.J.W. provided clinical material and reagents; and C.J.H., R.S., and M.J.S.D. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
T.A., T.B., and L.J.R. contributed equally to this manuscript. C.J.H., R.S., and M.J.S.D. share senior authorship.
Correspondence: Martin J. S. Dyer, MRC Toxicology Unit/Leicester University, Hodgkin Bldg, Rm 402, Lancaster Road, Leicester, United Kingdom LE1 9HN; e-mail:mjsd1@le.ac.uk.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
The expert technical assistance of the cytogenetic, molecular cytogenetic, and scientific staff of all the involved groups is gratefully acknowledged. We thank Dr Linda M. Boxer (Stanford University) for kindly providing some of the cell lines used in these studies.
This work was supported by grants from Leukaemia Research, the Medical Research Council, the Bud Flanagan Leukaemia Fund, Deutsche Krebshilfe, Wilhelm Sander-Stiftung (grant no. 2001.074.2), and the Biotechnology and Biological Sciences Research Council. L.J.R. is supported by a Leukaemia Research Gordon Piller Studentship.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal