The positive regulatory domain I (PRDM1) is a master regulator in the differentiation of mature B lymphocytes to plasma cells. It has 2 isoforms, PRDM1α and PRDM1β, and is regulated by the transcriptional regulator nuclear factor kappa (NF)–κB. PRDM1 protein expression was recently demonstrated in a subset of diffuse large B-cell lymphoma (DLBCL) with aggressive behavior, a type of lymphoma for which rituximab associated with chemotherapy (R-CHOP) is now widely indicated. Using laser microdissection combined with reverse transcription–polymerase chain reaction (RT-PCR) amplification, PRDM1 gene expression was assessed in 82 DLBCL patients. The results showed that both PRDM1α and PRDM1β transcripts were expressed in microdissected lymphoma cells only in the non–germinal center B-cell–like (non-GCB) subtype of DLBCL. PRDM1β gene expression was correlated with short survival time in the non-GCB patients treated with CHOP but not with R-CHOP. In vitro, B-lymphoma cells resistant to chemotherapy expressed PRDM1β. Rituximab suppressed PRDM1β expression, which was concomitant with NF-κB inactivation. The value of PRDM1β expression as a prognostic marker in non-GCB DLBCL might thus be considered. This study confirms the efficiency of rituximab on DLBCL and allows a better understanding of one of its biologic actions.
Introduction
Diffuse large B-cell lymphoma (DLBCL) is the most common type of B-cell lymphoma and represents a heterogeneous group of tumors on morphologic, phenotypic, molecular, and clinical grounds.1 Using gene-expression profiling, DLBCL has been classified as 3 distinct subtypes, which reflect different stages of B-cell differentiation. Germinal center B-cell–like (GCB) DLBCL presumably derives from GC centroblasts and is associated with a good outcome. Activated B-cell–like (ABC) DLBCL has the expression pattern of GC cells undergoing plasmacytic differentiation or of mitogen-activated peripheral B cells and is associated with a poor outcome.2,3 The type 3 group behaves in a manner similar to the ABC group.4 The protein expression patterns of CD10, BCL-6, and IRF4 (also known as MUM1) are alternative means of identifying the GCB and non-GCB groups, the latter corresponding to the microarray-defined ABC and type 3 phenotype.5 Rituximab (Rituxan; IDEC-C2B8), a recombinant chimeric monoclonal antibody against the pan–B-cell marker CD20, has shown promising results in the clinical treatment of patients with DLBCL.6 Addition of rituximab to chemotherapy can modify the adverse prognostic significance of the ABC phenotype.7
To further determine the role of B-cell differentiation on DLBCL development, related genes should be studied. The positive regulatory domain I (PRDM1), belonging to the PRDM gene family of transcriptional repressors, plays a central role in the terminal differentiation of B cells to plasma cells.8,9 PRDM1 is positively regulated by NF-κB. In B cells, PRDM1 gene expression can be induced in M12 and CH12 lymphoma lines. This induction is dependent on NF-κB because Helenalin, an inhibitor specific for NF-κB, inhibits PRDM1 expression.10 PRDM1 was found to be inactivated in the non-GCB subtype of DLBCL and is considered a potential tumor suppressor gene in these lymphomas.11,–13 However, PRDM1 protein expression has been recently reported in a sizable fraction of DLBCL, which displayed more aggressive behavior, with a shorter failure-free survival.14
PRDM1 exists as 2 isoforms, PRDM1α and PRDM1β. Generated from the same gene by alternative transcription, PRDM1β differs from PRDM1α by lacking the amino-terminal 101 amino acids and having a disrupted PR domain. It is functionally impaired, with loss of repressive function on multiple target genes.15 The prognostic significance of PRDM1 isoform expression has not yet been reported in DLBCL. Using laser microdissection combined with reverse transcription–polymerase chain reaction (RT-PCR) amplification, we assessed the respective expression of the 2 PRDM1 isoforms in 82 DLBCL patients and their prognostic value. We further studied the PRDM1 expression in B-lymphoma cell lines and their variations when treated by doxorubicin and/or rituximab.
Patients, materials, and methods
Patients
From January 2001 to June 2006, 82 patients with de novo DLBCL, 49 men and 33 women aged 18 to 84 years (median, 51 years), treated in the Shanghai Institute of Hematology–based patient network with available frozen tumor specimen at diagnosis, were included in this retrospective study. Histologic diagnoses were established according to the World Health Organization classification. The patients were treated with the standard dose of chemotherapy alone (CHOP regimens) or combined with rituximab (R-CHOP regimens). The clinical features of these patients are listed in Table 1. No significant difference in sex, age, Ann Arbor stage, or international prognostic index (IPI) was found between the 2 groups. Informed consent was obtained from all patients, in accordance with regulation of the Shanghai Jiao Tong University School of Medicine Institutional Review Boards.
Clinical characteristics of the DLBCL patients
| Characteristics . | CHOP group, n = 39 . | R-CHOP group, n = 43 . | P . |
|---|---|---|---|
| Sex | |||
| Male | 25 | 24 | |
| Female | 14 | 19 | .445 |
| Age, y | |||
| 60 or younger | 31 | 33 | |
| Older than 60 | 8 | 10 | .764 |
| Ann Arbor stage | |||
| I/II | 13 | 10 | |
| III/IV | 26 | 33 | .310 |
| International prognostic index | |||
| Low risk to intermediate low risk | 20 | 18 | |
| Intermediate high risk to high risk | 19 | 25 | .393 |
| Characteristics . | CHOP group, n = 39 . | R-CHOP group, n = 43 . | P . |
|---|---|---|---|
| Sex | |||
| Male | 25 | 24 | |
| Female | 14 | 19 | .445 |
| Age, y | |||
| 60 or younger | 31 | 33 | |
| Older than 60 | 8 | 10 | .764 |
| Ann Arbor stage | |||
| I/II | 13 | 10 | |
| III/IV | 26 | 33 | .310 |
| International prognostic index | |||
| Low risk to intermediate low risk | 20 | 18 | |
| Intermediate high risk to high risk | 19 | 25 | .393 |
Tissue samples
Tumor samples at time of diagnosis were immediately cut into 2 parts: one part was fixed in formaldehyde and further processed for paraffin embedding and the other was snap-frozen and stored at −80°C.
Cell lines
Cell lines U266, SU-DHL-4, Daudi, and Namalwa were cultured in RPMI-1640 medium supplemented with 10% heat-inactivated fetal bovine serum (Gibco/BRL, Grand Island, NY) in 5% CO2, 95% air-humidified atmosphere at 37°C. Namalwa cells, with features of non-GCB and resistant to doxorubicin,16 were treated with doxorubicin, rituximab, or pyrrolidine dithiocarbamate (PDTC; Sigma, St Louis, MO) that specifically inhibits the NF-κB activation.17 Cell viability was assessed by triplicate count of trypan blue test.
Immunohistochemistry
Immunohistochemical analyses were performed on 5-μm–thick paraffin sections with an indirect immunoperoxidase method, using antibodies against CD10 (Novocastra, Newcastle, United Kingdom; 1:80), Bcl-6 (Dako, Glostrup, Denmark; 1:10), IRF4 (Dako; 1:40), and PRDM1 (one from Novus Biologicals, Littleton, CO; 1:50; and another from the Monoclonal Antibodies Unit, Biotechnology Program, Spanish National Cancer Center, Madrid, Spain; 1:2). The slides were viewed on a Leica CTR MIC microscope (Leica Microsystem, Wetzlar, Germany), photographed by a 3CCD camera (HV-C20AMP, Hitachi Kokusai Electric Inc., Tokyo, Japan) using image-acquisition software Matrox Intellicam Version 2.06 (Matrox Electronic Systems Ltd., Dorval, Quebec, Canada).
Laser microdissection
Seven μm–thick frozen sections of DLBCL were incubated in RNAse-free conditions with anti-CD20 (Dako; 1:50) or IRF4 (Dako; 1:10) antibody for 5 minutes and then in fluorescein-conjugated goat anti–mouse IgG antibody (Jackson ImmunoResearch Laboratories, West Grove, PA; 1:50) for 5 minutes. Laser microdissection of fluorescent cells was immediately performed (Leica Microsystem) for RNA extraction.
Semiquantitative RT-PCR
Total RNA was extracted from whole frozen tissue sections, laser-microdissected lymphoma cells, or cultured cells using Trizol reagent (Invitrogen, Carlsbad, CA). First-strand cDNA was synthesized by Superscript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Semiquantitative PCR was performed using primers listed in Table 2. The housekeeping GAPDH gene was used as control.
Primers used in the semi-quantitative PCR studies
| Gene and position . | Sequences . | Length, bp . |
|---|---|---|
| PRDM1α | ||
| Promoter | 721 | |
| PRDM1α-1F | 5′-TCCTCTGAACTGTGAAACGAC-3′ | |
| PRDM1α-1R | 5′-GTGTCACGGCAGCACTTTGTC-3′ | |
| Promoter | 981 | |
| PRDM1α-2F | 5′-TCTGAGGACTCTTAGGAATTG-3′ | |
| PRDM1α-2R | 5′-TTCCTCTCCTCCCAGAACCCA-3′ | |
| Exon 2/intron 2 junction | 366 | |
| PRDM1α-3F | 5′-AGAAGGAGCCACAGGAACG-3′ | |
| PRDM1α-3R | 5′-AGTAGGGAGATTTGGCACC-3′ | |
| Exon 2–exon 3 | 239 | |
| PRDM1α-4F | 5′-GTTCCTAAGAACGCCAACAGG-3′ | |
| PRDM1α-4R | 5′-GCAAAGTCCCGACAATACCAC-3′ | |
| Exon 2–exon 4 | 354 | |
| PRDM1α-5F | 5′-GGACATGGAGGATGCGGATAT-3′ | |
| PRDM1α-5R | 5′-GTTGCTTTTCTCTTCATTAAAGCCG-3′ | |
| Exon 3 | 463 | |
| PRDM1α-6F | 5′-GCTTGGTTGAGTATTTGCT-3′ | |
| PRDM1α-6R | 5′-GCTGGATACTTATGGGTGA-3′ | |
| Exon 4 | 382 | |
| PRDM1α-7F | 5′-CGGCTGTGTTTATTCTGAG-3′ | |
| PRDM1α-7R | 5′-GAACCGACATTACTGGCAT-3′ | |
| Exon 5 | 489 | |
| PRDM1α-8F | 5′-TTTGGCAGTTTTGCTTCAG-3′ | |
| PRDM1α-8R | 5′-CACAGGGGACACCGTATTC-3′ | |
| Exon 5 | 540 | |
| PRDM1α-9F | 5′-GACCCACGTACATCACTCGC-3′ | |
| PRDM1α-9R | 5′-CGGTAAAGGAGAAGGCACTG-3′ | |
| Exon 5 | 487 | |
| PRDM1α-10F | 5′-GTCTACAGCAATCTCCTCG-3′ | |
| PRDM1α-10R | 5′-CACAAGCATGCACTCACAG-3′ | |
| Exon 5–exon 7 | 471 | |
| PRDM1α-11F | 5′-AGTACGAATGCAACGTTTGCG-3′ | |
| PRDM1α-11R | 5′-TCGATTTCTTCATTGATTCGGGT-3′ | |
| Exon 6 | 292 | |
| PRDM1α-12F | 5′-GGAGCAGAAATGTTAGGTC-3′ | |
| PRDM1α-12R | 5′-GTGTTGGCTTTAACTACGG-3′ | |
| Exon 7 | 413 | |
| PRDM1α-13F | 5′-CCGTTGGCAACTCTTAATC-3′ | |
| PRDM1α-13R | 5′-TGTCATCCTCCACGTCCTC-3′ | |
| Exon 7 | 494 | |
| PRDM1α-14F | 5′-CACAAGAACTACATCCATC-3′ | |
| PRDM1α-14R | 5′-CAGAGCTGGGATTATGTAC-3′ | |
| PRDM1β | ||
| Intron 3–exon 4 | 264 | |
| PRDM1β-F | 5′-TGGTGGGTTAATCGGTTTGAG-3′ | |
| PRDM1β-R | 5′-GGGATGGGCTTAATGGTGTAG-3′ | |
| IRF4 | ||
| Exon 2–exon 3 | 114 | |
| IRF4-F | 5′-AGAACGAGGAGAAGAGCATC-3′ | |
| IRF4-R | 5′-CCTTTAAACAGTGCCCAAG-3′ | |
| GAPDH | ||
| Exon 1–exon 2 | 255 | |
| GAPDH-F | 5′-GAAGGTGAAGGTCGGAGTC-3′ | |
| GAPDH-R | 5′-GAAGATGGTGATGGGATTTC-3′ | |
| Gene and position . | Sequences . | Length, bp . |
|---|---|---|
| PRDM1α | ||
| Promoter | 721 | |
| PRDM1α-1F | 5′-TCCTCTGAACTGTGAAACGAC-3′ | |
| PRDM1α-1R | 5′-GTGTCACGGCAGCACTTTGTC-3′ | |
| Promoter | 981 | |
| PRDM1α-2F | 5′-TCTGAGGACTCTTAGGAATTG-3′ | |
| PRDM1α-2R | 5′-TTCCTCTCCTCCCAGAACCCA-3′ | |
| Exon 2/intron 2 junction | 366 | |
| PRDM1α-3F | 5′-AGAAGGAGCCACAGGAACG-3′ | |
| PRDM1α-3R | 5′-AGTAGGGAGATTTGGCACC-3′ | |
| Exon 2–exon 3 | 239 | |
| PRDM1α-4F | 5′-GTTCCTAAGAACGCCAACAGG-3′ | |
| PRDM1α-4R | 5′-GCAAAGTCCCGACAATACCAC-3′ | |
| Exon 2–exon 4 | 354 | |
| PRDM1α-5F | 5′-GGACATGGAGGATGCGGATAT-3′ | |
| PRDM1α-5R | 5′-GTTGCTTTTCTCTTCATTAAAGCCG-3′ | |
| Exon 3 | 463 | |
| PRDM1α-6F | 5′-GCTTGGTTGAGTATTTGCT-3′ | |
| PRDM1α-6R | 5′-GCTGGATACTTATGGGTGA-3′ | |
| Exon 4 | 382 | |
| PRDM1α-7F | 5′-CGGCTGTGTTTATTCTGAG-3′ | |
| PRDM1α-7R | 5′-GAACCGACATTACTGGCAT-3′ | |
| Exon 5 | 489 | |
| PRDM1α-8F | 5′-TTTGGCAGTTTTGCTTCAG-3′ | |
| PRDM1α-8R | 5′-CACAGGGGACACCGTATTC-3′ | |
| Exon 5 | 540 | |
| PRDM1α-9F | 5′-GACCCACGTACATCACTCGC-3′ | |
| PRDM1α-9R | 5′-CGGTAAAGGAGAAGGCACTG-3′ | |
| Exon 5 | 487 | |
| PRDM1α-10F | 5′-GTCTACAGCAATCTCCTCG-3′ | |
| PRDM1α-10R | 5′-CACAAGCATGCACTCACAG-3′ | |
| Exon 5–exon 7 | 471 | |
| PRDM1α-11F | 5′-AGTACGAATGCAACGTTTGCG-3′ | |
| PRDM1α-11R | 5′-TCGATTTCTTCATTGATTCGGGT-3′ | |
| Exon 6 | 292 | |
| PRDM1α-12F | 5′-GGAGCAGAAATGTTAGGTC-3′ | |
| PRDM1α-12R | 5′-GTGTTGGCTTTAACTACGG-3′ | |
| Exon 7 | 413 | |
| PRDM1α-13F | 5′-CCGTTGGCAACTCTTAATC-3′ | |
| PRDM1α-13R | 5′-TGTCATCCTCCACGTCCTC-3′ | |
| Exon 7 | 494 | |
| PRDM1α-14F | 5′-CACAAGAACTACATCCATC-3′ | |
| PRDM1α-14R | 5′-CAGAGCTGGGATTATGTAC-3′ | |
| PRDM1β | ||
| Intron 3–exon 4 | 264 | |
| PRDM1β-F | 5′-TGGTGGGTTAATCGGTTTGAG-3′ | |
| PRDM1β-R | 5′-GGGATGGGCTTAATGGTGTAG-3′ | |
| IRF4 | ||
| Exon 2–exon 3 | 114 | |
| IRF4-F | 5′-AGAACGAGGAGAAGAGCATC-3′ | |
| IRF4-R | 5′-CCTTTAAACAGTGCCCAAG-3′ | |
| GAPDH | ||
| Exon 1–exon 2 | 255 | |
| GAPDH-F | 5′-GAAGGTGAAGGTCGGAGTC-3′ | |
| GAPDH-R | 5′-GAAGATGGTGATGGGATTTC-3′ | |
Sequence analysis of PRDM1
Genomic DNA was extracted from frozen tissue sections using standard proteinase K digestion and phenol/chloroform procedures. The resultant PCR products were purified on Qiagen columns (Qiagen, Valencia, CA) and sequenced by PRDM1 primers on ABI PRISM 3700 DNA Analyzer using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA).
Western blot
Frozen tissue sections or cultured cells were lysed in 200 μL lysis buffer (0.5 M Tris-HCl, pH 6.8; 2 mM EDTA; 10% glycerol; 2% SDS; and 5% β-mercaptoethanol). Protein extracts (20 μg) were loaded onto 10% polyacrylamide gel containing sodium dodecyl sulfate, subjected to electrophoresis, and transferred to nitrocellulose membranes. Membranes were blocked with 5% nonfat dried milk in Tris-buffered saline (TBS) /0.05% Tween 20 and incubated for 2 hours at room temperature with appropriate primary antibody, followed by treatment with horseradish peroxidase–linked secondary antibody. The immunocomplexes were visualized using chemiluminescence phototope-horseradish peroxidase kit. Detection of β-actin was performed to ensure equivalent protein loading.
Enzyme-linked immunosorbent assay (ELISA) analysis for NF-κB activity
Most actively involved in NF-κB signaling, NF-κB (P65) phosphorylation at serine 276 allows for increased interaction with the transcriptional coactivator p300/CBP to enhance its transcriptional activity.18 Total NF-κB, phosphorylated NF-κB, activated form of NF-κB (nuclear localization sequence of P65), and GAPDH were detected by KC NF-κB Pathway ELISA Kit (KangChen Biotech, Shanghai, China) according to the manufacturer's instructions.
Statistical analysis
Patient characteristics were compared using χ2 analysis and the Fisher exact test. Overall survival (OS) was measured from the time of diagnosis to either death or the end date of August 31, 2006. Event-free survival (EFS) was calculated from the time of diagnosis to the date of progression, death, or the end date. When the end date was not reached, the data were censored at the date of the last follow-up evaluation. Survival functions were estimated using the Kaplan-Meier method and compared using the log-rank test. Multivariate survival analysis was performed using a Cox regression model. P less than .05 was considered to be significant. All statistical analyses were performed on SAS 8.2 software (SAS Institute, Cary, NC).
Results
PRDM1 was predominantly expressed in the non-GCB subtype of DLBCL
As defined by immunostainings on paraffin sections using antibodies against CD10, BCL-6, and IRF4,5 33 (40.2%) of the 82 patients had a GCB phenotype and 49 (59.8%) had a non-GCB phenotype.
PRDM1 protein expression was not significantly different in GCB (13/33, 39.4%) and non-GCB (24/49, 49.0%) DLBCL. However, PRDM1α and PRDM1β transcripts were more frequently observed in the non-GCB subtype (73.5% and 38.8%, respectively) than in the GCB subtype (39.4% and 6.1%, respectively; P = .002 and P < .001; Table 3). While all 13 GCB cases expressed the PRDM1α transcript and protein at the same time, 12 of the 36 non-GCB patients positive for PRDM1α transcript had no PRDM1 protein expression. In turn, PRDM1 protein was detected in the 19 patients expressing PRDM1β transcripts.
PRDM1 transcripts and protein expressions in DLBCL patients
| . | GCB, n=33 . | non-GCB, n=49 . | ||
|---|---|---|---|---|
| CHOP, n=15 . | R-CHOP, n=18 . | CHOP, n=24 . | R-CHOP, n=25 . | |
| Transcripts | ||||
| PRDM1α | 6 | 7 | 20 | 16 |
| PRDM1β | 1 | 1 | 8 | 11 |
| Protein | ||||
| PRDM1 | 6 | 7 | 11 | 13 |
| . | GCB, n=33 . | non-GCB, n=49 . | ||
|---|---|---|---|---|
| CHOP, n=15 . | R-CHOP, n=18 . | CHOP, n=24 . | R-CHOP, n=25 . | |
| Transcripts | ||||
| PRDM1α | 6 | 7 | 20 | 16 |
| PRDM1β | 1 | 1 | 8 | 11 |
| Protein | ||||
| PRDM1 | 6 | 7 | 11 | 13 |
Systematic sequencing of PRDM1 in these 82 patients showed no mutation, including the previously described mutational hot spot (exon 2/intron 2 junction).11
PRDM1 transcripts and protein were expressed in lymphoma cells of non-GCB DLBCL and related to IRF4 expression
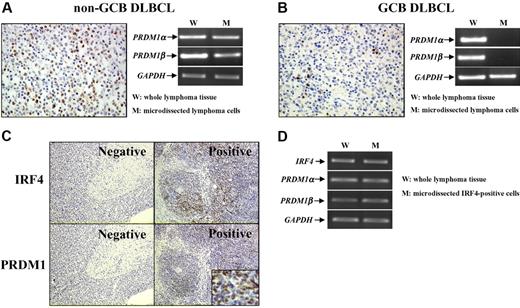
PRDM1 protein was expressed in a larger number of cells in non-GCB DLBCL compared with GCB cases (Figure 1A-B). To confirm the PRDM1 gene expression in lymphoma cells, we isolated CD20+ cells by laser microdissection. Both PRDM1 transcripts were detected in microdissected non-GCB lymphoma cells (Figure 1A). Although expressed in whole lymphoma tissue sections, neither PRDM1α nor PRDM1β messenger RNA (mRNA) was found in the microdissected GCB lymphoma cells at the same level as mRNA for the GAPDH gene (Figure 1B).
PRDM1 was expressed in lymphoma cells of non-GCB DLBCL and related to IRF4 expression. With different protein expression patterns revealed by immunohistochemisty (A-B), PRDM1 transcripts were expressed in microdissected lymphoma cells of DLBCL non-GCB subtype (A) but not of GCB subtype (B) (40×/0.75 NA non-oil objective). PRDM1 expression was related to IRF4 expression (C). IRF4-positive lymphoma cells coexpressed IRF4, PRDM1α, and PRDM1β (D) (10×/0.75 NA non-oil objective; small figure: 150×/0.75 NA non-oil objective).
PRDM1 was expressed in lymphoma cells of non-GCB DLBCL and related to IRF4 expression. With different protein expression patterns revealed by immunohistochemisty (A-B), PRDM1 transcripts were expressed in microdissected lymphoma cells of DLBCL non-GCB subtype (A) but not of GCB subtype (B) (40×/0.75 NA non-oil objective). PRDM1 expression was related to IRF4 expression (C). IRF4-positive lymphoma cells coexpressed IRF4, PRDM1α, and PRDM1β (D) (10×/0.75 NA non-oil objective; small figure: 150×/0.75 NA non-oil objective).
Interestingly, PRDM1 was significantly related to IRF4 expression (Figure 1C): it was found in 17 (65.4%) of 26 IRF4-positive non-GCB DLBCL patients but in only 7 (30.4%) of 23 IRF4-negative cases (P = .015). At the transcriptional level, laser-microdissected IRF4-positive cells expressed mRNA of IRF4, PRDM1α, and PRDM1β (Figure 1D).
PRDM1β expression was independently related to short survival time in non-GCB DLBCL patients treated by CHOP but not in those treated by R-CHOP
Of the 49 non-GCB DLBCL patients, 24 received CHOP and 25 received R-CHOP. In the CHOP group, the 3-year EFS and OS rates (± SE percentage) for patients with PRDM1β gene expression were 14.3% (± 3.2%) and 35.7% (± 19.7%), respectively, significantly shorter than those without PRDM1β gene expression (65.9% [± 14.3%] and 85.7% [± 13.2%], P = .026 and P = .017, respectively). Using multivariate analysis, PRDM1β expression was an independent adverse prognostic factor for EFS and OS (P = .029 and P = .007, respectively). The Cox model selected 4 criteria for survival: age, sex, Ann Arbor stage, and IPI. In the R-CHOP group, however, the decreased survival was no longer observed in patients with PRDM1β expression, both for EFS (P = .213) and OS (P = .358).
No correlation was found between PRDM1α gene expression and survival or between PRDM1 protein expression and survival.
PRDM1β transcript and protein were expressed in lymphoma cells and down-regulated after treatment with rituximab and doxorubicin
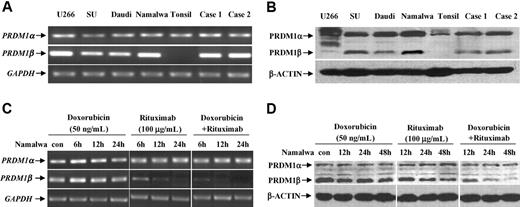
By RT-PCR, PRDM1α transcript was detected in a series of B-lymphoma cell lines, human tonsil, as well as non-GCB DLBCL samples, whereas no PRDM1β transcript was found in human tonsil (Figure 2A). PRDM1α and PRDM1β protein profile revealed by Western blot followed mRNA variations (Figure 2B). PRDM1β protein was identified as a fragment at about 70 kDa in B-lymphoma cell lines and the patient samples, different from an 80-kDa PRDM1β protein found in myeloma cell line U266. No PRDM1β protein was observed in human tonsil.
Rituximab combined with doxorubicin down-regulated PRDM1β expression. B-lymphoma cell lines and primary DLBCL cells expressed PRDM1α and PRDM1β both at transcriptional (A) and protein levels (B). Combined treatment of doxorubicin and rituximab reduced PRDM1β expression (C-D).
Rituximab combined with doxorubicin down-regulated PRDM1β expression. B-lymphoma cell lines and primary DLBCL cells expressed PRDM1α and PRDM1β both at transcriptional (A) and protein levels (B). Combined treatment of doxorubicin and rituximab reduced PRDM1β expression (C-D).
Namalwa cells were treated with different concentrations of doxorubicin (1, 5, 10, 20, and 50 ng/mL), alone or combined with rituximab (100 μg/mL), for 48 hours. Doxorubicin alone did not inhibit cell proliferation at concentrations up to 50 ng/mL, but doxorubicin with rituximab had a significant antiproliferative effect at 20 ng/mL (15.8% ± 0.7% vs 2.1% ± 0.2%, respectively, P > .001) and 50 ng/mL (29.3% ± 1.2% vs 5.2% ± 0.6%, respectively, P < .001).
After 12 and 24 hours of incubation with doxorubicin (50 ng/mL) and rituximab (100 μg/mL), a decreased expression of PRDM1β gene, but not of PRDM1α gene, was found compared with each agent alone (Figure 2C). The PRDM1β protein level followed a similar reduction, observed later at 24 and 48 hours after treatment (Figure 2D).
Rituximab inhibited NF-κB activity in B-lymphoma cells and this could be mimicked by the NF-κB inhibitor PDTC
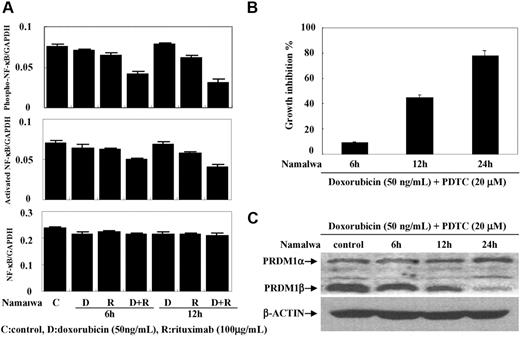
To determine if NF-κB activity was altered, sequential ELISA assays were performed at 6 and 12 hours in Namalwa cells. The level of phosphorylated NF-κB was significantly lower in the cells cotreated with doxorubicin (50 ng/mL) and rituximab (100 μg/mL) compared with each drug alone (Figure 3A). This was concomitant with a reduction of activated NF-κB, without change of total NF-κB.
Rituximab combined with doxorubicin decreased NF-κB activity. Rituximab combined with doxorubicin significantly decreased the phosphorylated and the activated forms of NF-κB (P65) in Namawa cells, without variation of total NF-κB (A). Addition of NF-κB inhibitor PDTC increased the antiproliferative effect of doxorubicin on Namawa cells (B) and induced PRDM1β down-regulation (C).
Rituximab combined with doxorubicin decreased NF-κB activity. Rituximab combined with doxorubicin significantly decreased the phosphorylated and the activated forms of NF-κB (P65) in Namawa cells, without variation of total NF-κB (A). Addition of NF-κB inhibitor PDTC increased the antiproliferative effect of doxorubicin on Namawa cells (B) and induced PRDM1β down-regulation (C).
Discussion
In the present study, we showed that aberrant gene and protein expression of PRDM1 occurred in DLBCL and was particularly relevant to the non-GCB phenotype. Since PRDM1 transcript could be detected on lymphoma cells, as well as associated nonmalignant cells like plasma or reactive T cells,14 laser microdissection allowed us to confirm that lymphoma cells in non-GCB DLBCL expressed both isoforms of PRDM1 gene. Repeated microdissection and semiquantitative RT-PCR, however, did not detect any expression of PRDM1 gene in GCB lymphoma cells. This is in accordance with the new DLBCL classification, which suggests that the GCB subtype of lymphoma derives from GC B centroblasts that do not normally express PRDM1.19
At the protein level, no significant difference in PRDM1 expression was found between the non-GCB and GCB groups. The discrepancy from PRDM1α transcript to protein expression had also been reported in another non-GCB DLBCL series.12 Although no gene mutation was detected, other genetic and epigenetic inactivation as well as defects in protein translation or stability might contribute to the lack of PRDM1 protein expression in non-GCB DLBCL.12
No discrepancy in transcript and protein expressions was found for the isoform PRDM1β notifying it as a specific lesion in non-GCB DLBCL. PRDM1β is highly analogous to the PRDM2 (RIZ) and PRDM3 (MDS1-EVI1), both expressing a truncated protein missing the PR domain. These truncated proteins are expressed in different types of malignant cells and critical for oncogenesis.20,,–23 Our study demonstrated that the PRDM1β was present in both B-lymphoma cell lines and primary DLBCL cells. Statistical analyses showed that PRDM1β expression was an independent adverse prognostic factor for EFS and OS in non-GCB DLBCL. Therefore, PRDM1β may favor lymphoma progression in DLBCL.
PRDM1β coexpressed with IRF4 in 17 of 49 of our non-GCB cases. IRF4 is a lymphoid-specific transcriptional regulator and denotes the final step of GCB cell differentiation toward plasma cells.24,25 Originally identified as the product of a proto-oncogene in multiple myeloma,26 IRF4 is often abnormally expressed in B-cell lymphomas.27 This IFR4-PRDM1β coexpression suggested that PRDM1β happens with IRF4 during later B-cell maturation. It might also interact with IRF4 and contribute to lymphomagenesis.28
An unexpected observation was that PRDM1β expression was significantly correlated with short survival time in patients treated by CHOP but not by R-CHOP. Since it was a retrospective study, patient selection for treatment could possibly influence the results. This must be further verified in a randomized trial. In vitro, rituximab enhances lymphoma cell sensitivity to chemotherapeutic drugs through inhibition of the NF-κB pathway.29 Experimental studies also showed that NF-κB is able to regulate PRDM1.10,30 In our study, rituximab combined with doxorubicin could inhibit NF-κB activity in B lymphoma cells, resulting in reduced PRDM1β expression. To test this hypothesis, we associated PDTC, an inhibitor or NF-κB, with doxorubicin in lymphoma cell cultures. The induced growth inhibition, similar to rituximab treatment, confirmed that NF-κB activity was responsible for PRDM1β down-regulation by rituximab. Rituximab could overcome the adverse prognostic effect of PRDM1β in the non-GCB DLBCL patients. Interestingly, another clinical study also reported the effect of rituximab on chemo-resistant DLCBL with the expression of BCL-2,31 another biomarker regulated by NF-κB.32 Therefore, the action of rituximab in B-lymphoma cells might be linked to interactions of NF-κB with multiple gene products. This further raises the question of the prospective identification of B-cell lymphoma patients for rituximab therapy through a test of NF-κB activity.
In conclusion, abnormal expression of PRDM1, particularly of its isoform PRDM1β, was restricted to lymphoma cells of the non-GCB subtype in DLBCL. Addition of rituximab could down-regulate PRDM1β expression, reversing the negative effect of PRDM1β on chemotherapy-treated patients.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the Chinese National Key Program for Basic Research (973: 2004CB518600), the Chinese National High Tech Program (863: 2006AA02A301 and 863:2006AA02A405), the Key Discipline Program of Shanghai Municipal Education Commission (Y0201), the Shanghai Commission of Science and Technology (44107025 and 05DZ19317), the Shanghai Rising Star Program (05QMX1429), the Scientific Research Foundation for the Returned Overseas Chinese Scholars, the Program de Recherches Avancées (PRA B 03–02 and PRA B 05–02), and the Samuel Waxman Cancer Research Foundation Laboratory.
Authorship
Contribution: W.-L.Z., A.J., S.-J.C., and Z.C. designed research and wrote the paper; Y.-Y.L., C.L., J.-Y.S., and L.W. performed research; J.-F.G. contributed vital new reagents; J.-M.L. and Z.-X.S. collected data; and Y.S. analyzed data.
Y.Y.L., C.L., and J.-Y.S. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Wei-Li Zhao and Sai-Juan Chen, State Key Laboratory of Medical Genomics, Shanghai Institute of Hematology, Shanghai Rui Jin Hospital, Shanghai Jiao Tong University School of Medicine, 197 Rui Jin Er Road, Shanghai 200025, China; e-mail: weili_zhao_sih@yahoo.com and sjchen@stn.sh.cn.