To study the role of the JAK2-V617F mutation in leukemic transformation, we examined 27 patients with myeloproliferative disorders (MPDs) who transformed to acute myeloid leukemia (AML). At MPD diagnosis, JAK2-V617F was detectable in 17 of 27 patients. Surprisingly, only 5 of 17 patients developed JAK2-V617F–positive AML, whereas 9 of 17 patients transformed to JAK2-V617F–negative AML. Microsatellite analysis in a female patient showed that mitotic recombination was not responsible for the transition from JAK2-V617F–positive MPD to JAK2-V617F–negative AML, and clonality determined by the MPP1 polymorphism demonstrated that the granulocytes and leukemic blasts inactivated the same parental X chromosome. In a second patient positive for JAK2-V617F at transformation, but with JAK2-V617F–negative leukemic blasts, we found del(11q) in all cells examined, suggesting a common clonal origin of MPD and AML. We conclude that JAK2-V617F–positive MPD frequently yields JAK2-V617F–negative AML, and transformation of a common JAK2-V617F–negative ancestor represents a possible mechanism.
Introduction
Myeloproliferative disorders (MPDs) are a heterogeneous group of diseases characterized by increased hematopoiesis leading to elevated numbers of nonlymphoid cells and/or platelets in the peripheral blood. An acquired somatic mutation in the JAK2 gene resulting in a valine-to-phenylalanine substitution at position 617 (JAK2-V617F) is frequently found in patients with MPDs.1,,–4 Using a sensitive allele-specific polymerase chain reaction (AS-PCR) assay, the JAK2-V617F mutation is detectable in more than 90% of patients with polycythemia vera (PV), more than 50% of essential thrombocythemia (ET), and more than 50% of idiopathic myelofibrosis (IMF).2,5,6 Transformation to acute myeloid leukemia (AML) is a known complication of MPDs.7,–9 The frequency of JAK2-V617F in patients with de novo AML was around 1%.10,,,,–15 In contrast, the JAK2-V617F mutation in AML secondary to MPD was in the range of 50%.12,14,16 The question of whether the presence of the JAK2-V617F mutation favors leukemic transformation is currently unanswered. Because of the small number of patients with secondary AML, prospective studies are difficult to perform and the data are at present inconclusive.17 Here, we compared the JAK2-V617F status before and at leukemic transformation in 27 MPD patients and performed molecular studies on purified cell populations
Materials and methods
Patients and samples
We performed a retrospective study on 27 patients with AML secondary to MPD (8 PV, 12 ET, 7 IMF) from whom samples at MPD and AML diagnosis were available. Informed consent from all patients alive was obtained in accordance with the Declaration of Helsinki. The study was conducted according to the guidelines of the local ethics committees at centers in France (Nantes, Dijon, Bordeaux) and Switzerland (Basel). The diagnosis of MPD was made according to the criteria of the World Health Organization (patients from Basel) or of the Polycythemia Vera Study Group (Nantes, Dijon, Bordeaux).18,–20
Cell separations, RNA and DNA isolation
Bone marrow or peripheral blood smears were scraped and DNA was isolated using the QIAmp DNA Blood Mini kit (Qiagen, Hilden, Germany). Leukemic blasts were isolated by fluorescence-activated cell sorting (FACS) using side scatter and anti-CD34 antibodies or by laser capture microdissection of bone marrow films stained with May-Grünwald/Giemsa. Cells of the granulocytic lineage and T-cells were sorted by FACS using anti-CD15 and anti-CD3 antibodies, respectively. Purity was greater than 95%. Preparations of DNA and RNA and cDNA syntheses were performed as described.6,21,22
Molecular analyses
The methods for detecting loss of heterozygosity on chromosome 9p (9pLOH) and for determining the copy number of chromosome 9p were described earlier.4 A quantitative PCR assay for JAK2-V617F was performed with 2 different methods,6,22 and analysis of clonality by X chromosome inactivation in female patients heterozygous for a single nucleotide polymorphism in the MPP1 gene was assessed as described.22
Statistical analysis
To compare continuous variables among the groups we used the Mann-Whitney U test.
Results and discussion
We determined the JAK2-V617F mutation status by AS-PCR in 27 MPD patients (8 PV, 12 ET, and 7 IMF) from whom we obtained paired DNA samples at MPD diagnosis and leukemic transformation. At MPD diagnosis, the JAK2-V617F mutation was present in 17 of 27 patients (63%) (Table 1). Surprisingly, in 7 of these 17 patients, the JAK2 mutation was undetectable in unfractionated peripheral blood or bone marrow samples at transformation to AML. Our AS-PCR assays have a detection limit of less than 1%T.6,22 In 2 additional patients, we found that leukemic blast cells purified by FACS (patient DI-35) or laser capture microdissection (patient NA-48) were negative for JAK2-V617F, although the JAK2 mutation was detectable in unfractionated AML samples (Table 1). Thus, 9 of 17 patients (53%) with JAK2-V617F–positive MPD displayed JAK2-V617F–negative leukemic blasts at diagnosis of AML. Purified granulocytes or CD15-positive myeloid cells obtained in 6 of these 9 patients were also negative for the JAK2 mutation, suggesting that the AML clone exerted a suppressive effect on the JAK2-V617F–positive MPD clone (Table 1). In 5 patients (29%), we found JAK2-V617F both at MPD diagnosis and in leukemic blasts at transformation. In these 5 patients, the allelic ratio of JAK2-V617F in blasts was more than 50%T, suggesting that at least a proportion of blasts were homozygous for JAK2-V617F. In the remaining 3 of 17 patients (18%) with JAK2-V617F–positive MPD, the mutational status of the AML blasts could not be determined with certainty because of a lack of purified cells.
Summary of patient characteristics and molecular analyses in AML secondary to MPD
| UPN . | Sex . | MPD . | AML . | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dx . | Age, y . | Nantes, %T . | Basel, %T . | Source . | Cytoreductive treatment . | Patient characteristics . | Unfractionated cell samples . | Purified cells . | |||||||||||||||||
| Granulocytic lineage . | Purified leukemic blasts . | T-cells . | |||||||||||||||||||||||
| FAB . | Age, y . | Delay, y . | Molecular and/or P cytogenetic abnormalities . | %Blasts . | source . | Nantes, %T . | Basel, %T . | source . | %T . | Source . | Method . | %T . | Source . | Method . | %T . | source . | Method . | ||||||||
| V617F-positive MPD/ V617F-negative AML | |||||||||||||||||||||||||
| NA-02 | F | PV | 70 | 79* | 94† | GRA/BM-CD15+ | Pipo | M2 | 72 | 2 | normal | 10 | BM smear | 0 | 0 | BMMC | 0 | PB-GRA | Ficoll | 0 | PB-CD34+ | FACS | 0 | PB-CD3+ | FACS |
| NA-32 | F | PV | 70 | 42 | 52 | PB GRA | Pipo | M1 | 72 | 2 | nd | 70 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| p128 | M | PV | 75 | nd | 30 | PB GRA | HU | M4 | 80 | 5 | Flt-3-ITD | 58 | PB smear | nd | 0 | PBMC | 0 | PB-CD15+ | MACS | 0 | PB-CD34+ | MACS | na | na | na |
| NA-01 | M | PV | 64 | 7 | 12 | BM smear | Pipo | M0 | 72 | 8 | nd | 91 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-03 | M | ET | 89 | 38 | 24 | PB GRA | HU | na | 91 | 2 | −Y | 42 | PB smear | 0 | 0 | PBMC | 0 | PB-GRA | Ficoll | 0 | PB-CD34+ | FACS | 0 | PB-CD3+ | FACS |
| BO-04 | F | ET | 52 | 21 | nd | PB GRA | None | M1 | 54 | 2 | t(10;16) | 93 | PB smear | 0 | 0 | PBMC | 0 | PB-GRA | Ficoll | na | na | na | na | na | na |
| DI-05 | M | ET | 69 | 12 | nd | BM smear | HU | M6 | 71 | 2 | complex, del(5q), +7, −Y | 52 | BM smear | 0 | 0 | BMMC | 0 | BM-CD15+ | FACS | 0 | BM-CD34+ | FACS | na | na | na |
| DI-35 | F | ET | 68 | 30 | 53 | BM smear | HU | M4 | 72 | 4 | complex, del(5q) | 45 | BM smear | 4 | 2 | BMMC | 0 | BM-CD15+ | FACS | 0 | PB/BM-CD34+ | FACS | 0 | PB/BM-CD3+ | FACS |
| NA-48 | M | IMF | 57 | 31 | 56 | BM smear | HU/Thal | M2 | 59 | 2 | complex, del(11q23) | 35 | BM smear | nd | 51 | BM smear | na | na | na | 0 | LC-blasts | LC | na | na | na |
| V617F-positive MPD/ V617F-positive AML | |||||||||||||||||||||||||
| NA-59 | M | PV | 64 | 46 | nd | PB GRA | HU/Pipo | na | 67 | 3 | nd | 15 | PB smear | 75 | nd | PBMC | 90 | PB-GRA | Ficoll | 85 | PB-CD34+ | FACS | 10 | PB-CD3+ | FACS |
| p185 | M | PV | 65 | nd | 30 | BM smear | HU | M2 | 83 | 18 | +8 | 75 | BM smear | nd | 69 | PB-CD34+ | 65 | PB-CD15+ | FACS | 69 | PB-CD34+ | FACS | na | na | na |
| NA-57 | F | ET | 47 | 82 | 89 | PB GRA‡ | HU | na | 60 | 13 | complex, del(20q), del(5q) | 15 | BM smear | 71 | 80 | PBMC | 78 | PB-GRA | Ficoll | 100 | PB-CD34+ | FACS | 0 | PB-CD3+ | FACS |
| DI-58 | F | ET | 64 | 26 | 34 | BM smear | na | M2 | 78 | 14 | complex, del(20q), −5, −7, +8 | 30 | BM smear | 41 | 66 | BMMC | 46 | PB-GRA | Ficoll | 56 | PB-CD34+ | FACS | 1 | PB-CD3+ | FACS |
| DI-60 | M | IMF | 71 | 75 | nd | BM smear | None | na | 74 | 3 | nd | 73 | PB smear | 80 | nd | PBMC | na | na | na | na | na | na | na | na | na |
| V617F-positive MPD/V617F-uninformative AML | |||||||||||||||||||||||||
| NA-30 | M | PV | 61 | 44 | nd | BM smear | 32P/Pipo | M2 | 68 | 7 | nd | 18 | BM smear | 43 | 67 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-33 | F | ET | 63 | 16 | nd | BM smear | HU/Pipo | M1 | 74 | 11 | nd | 91 | BM smear | 15 | nd | BM smear | na | na | na | na | na | na | na | na | na |
| NA-52 | M | IMF | 64 | 44 | 40 | BM smear | HU | M7 | 69 | 5 | nd | 24 | BM smear | nd | 59 | BM smear | na | na | na | na | na | na | na | na | na |
| V617F-negative MPD/V617F-negative AML | |||||||||||||||||||||||||
| NA-07 | M | PV | 53 | 0 | nd | BM smear | Pipo | M6 | 64 | 11 | nd | 75 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| NA-10 | M | ET | 67 | 0 | 0 | BM smear | HU | M4 | 74 | 7 | nd | 65 | PB smear | nd | 0 | PB smear | na | na | na | na | na | na | na | na | na |
| NA-11 | F | ET | 57 | 0 | 0 | BM smear | HU/32P/IFN/Ana | M2 | 73 | 16 | nd | 46 | PB smear | nd | 0 | PB smear | na | na | na | na | na | na | na | na | na |
| DI-12 | M | ET | 47 | 0 | nd | BM smear | HU | M7 | 49 | 2 | complex | 34 | BM smear | 0 | nd | BM smear | na | na | na | na | na | na | na | na | na |
| DI-13 | F | ET | 81 | 0 | 0 | BM smear | HU | M7 | 83 | 2 | complex, del(5q),−7 | 36 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-14 | F | ET | 75 | 0 | 0 | BM smear | HU | M2 | 77 | 2 | nd | 43 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| NA-08 | F | IMF | 62 | 0 | 0 | BM smear | HU/VP-16 | na | 69 | 7 | nd | 48 | PB smear | nd | 0 | PB smear | na | na | na | na | na | na | na | na | na |
| NA-09 | F | IMF | 62 | 0 | 0 | BM smear | Pipo/IFN | bi | 73 | 11 | nd | 38 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-20 | M | IMF | 65 | 0 | nd | BM smear | na | M7 | 67 | 2 | nd | 14 | BM smear | 0 | 0 | BMMC | na | na | na | na | na | na | na | na | na |
| p202 | M | IMF | 75 | nd | 0 | BM smear | HU | na | 80 | 5 | nd | 95 | PB smear | nd | 0 | PBMC | na | na | na | na | na | na | na | na | na |
| UPN . | Sex . | MPD . | AML . | ||||||||||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Dx . | Age, y . | Nantes, %T . | Basel, %T . | Source . | Cytoreductive treatment . | Patient characteristics . | Unfractionated cell samples . | Purified cells . | |||||||||||||||||
| Granulocytic lineage . | Purified leukemic blasts . | T-cells . | |||||||||||||||||||||||
| FAB . | Age, y . | Delay, y . | Molecular and/or P cytogenetic abnormalities . | %Blasts . | source . | Nantes, %T . | Basel, %T . | source . | %T . | Source . | Method . | %T . | Source . | Method . | %T . | source . | Method . | ||||||||
| V617F-positive MPD/ V617F-negative AML | |||||||||||||||||||||||||
| NA-02 | F | PV | 70 | 79* | 94† | GRA/BM-CD15+ | Pipo | M2 | 72 | 2 | normal | 10 | BM smear | 0 | 0 | BMMC | 0 | PB-GRA | Ficoll | 0 | PB-CD34+ | FACS | 0 | PB-CD3+ | FACS |
| NA-32 | F | PV | 70 | 42 | 52 | PB GRA | Pipo | M1 | 72 | 2 | nd | 70 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| p128 | M | PV | 75 | nd | 30 | PB GRA | HU | M4 | 80 | 5 | Flt-3-ITD | 58 | PB smear | nd | 0 | PBMC | 0 | PB-CD15+ | MACS | 0 | PB-CD34+ | MACS | na | na | na |
| NA-01 | M | PV | 64 | 7 | 12 | BM smear | Pipo | M0 | 72 | 8 | nd | 91 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-03 | M | ET | 89 | 38 | 24 | PB GRA | HU | na | 91 | 2 | −Y | 42 | PB smear | 0 | 0 | PBMC | 0 | PB-GRA | Ficoll | 0 | PB-CD34+ | FACS | 0 | PB-CD3+ | FACS |
| BO-04 | F | ET | 52 | 21 | nd | PB GRA | None | M1 | 54 | 2 | t(10;16) | 93 | PB smear | 0 | 0 | PBMC | 0 | PB-GRA | Ficoll | na | na | na | na | na | na |
| DI-05 | M | ET | 69 | 12 | nd | BM smear | HU | M6 | 71 | 2 | complex, del(5q), +7, −Y | 52 | BM smear | 0 | 0 | BMMC | 0 | BM-CD15+ | FACS | 0 | BM-CD34+ | FACS | na | na | na |
| DI-35 | F | ET | 68 | 30 | 53 | BM smear | HU | M4 | 72 | 4 | complex, del(5q) | 45 | BM smear | 4 | 2 | BMMC | 0 | BM-CD15+ | FACS | 0 | PB/BM-CD34+ | FACS | 0 | PB/BM-CD3+ | FACS |
| NA-48 | M | IMF | 57 | 31 | 56 | BM smear | HU/Thal | M2 | 59 | 2 | complex, del(11q23) | 35 | BM smear | nd | 51 | BM smear | na | na | na | 0 | LC-blasts | LC | na | na | na |
| V617F-positive MPD/ V617F-positive AML | |||||||||||||||||||||||||
| NA-59 | M | PV | 64 | 46 | nd | PB GRA | HU/Pipo | na | 67 | 3 | nd | 15 | PB smear | 75 | nd | PBMC | 90 | PB-GRA | Ficoll | 85 | PB-CD34+ | FACS | 10 | PB-CD3+ | FACS |
| p185 | M | PV | 65 | nd | 30 | BM smear | HU | M2 | 83 | 18 | +8 | 75 | BM smear | nd | 69 | PB-CD34+ | 65 | PB-CD15+ | FACS | 69 | PB-CD34+ | FACS | na | na | na |
| NA-57 | F | ET | 47 | 82 | 89 | PB GRA‡ | HU | na | 60 | 13 | complex, del(20q), del(5q) | 15 | BM smear | 71 | 80 | PBMC | 78 | PB-GRA | Ficoll | 100 | PB-CD34+ | FACS | 0 | PB-CD3+ | FACS |
| DI-58 | F | ET | 64 | 26 | 34 | BM smear | na | M2 | 78 | 14 | complex, del(20q), −5, −7, +8 | 30 | BM smear | 41 | 66 | BMMC | 46 | PB-GRA | Ficoll | 56 | PB-CD34+ | FACS | 1 | PB-CD3+ | FACS |
| DI-60 | M | IMF | 71 | 75 | nd | BM smear | None | na | 74 | 3 | nd | 73 | PB smear | 80 | nd | PBMC | na | na | na | na | na | na | na | na | na |
| V617F-positive MPD/V617F-uninformative AML | |||||||||||||||||||||||||
| NA-30 | M | PV | 61 | 44 | nd | BM smear | 32P/Pipo | M2 | 68 | 7 | nd | 18 | BM smear | 43 | 67 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-33 | F | ET | 63 | 16 | nd | BM smear | HU/Pipo | M1 | 74 | 11 | nd | 91 | BM smear | 15 | nd | BM smear | na | na | na | na | na | na | na | na | na |
| NA-52 | M | IMF | 64 | 44 | 40 | BM smear | HU | M7 | 69 | 5 | nd | 24 | BM smear | nd | 59 | BM smear | na | na | na | na | na | na | na | na | na |
| V617F-negative MPD/V617F-negative AML | |||||||||||||||||||||||||
| NA-07 | M | PV | 53 | 0 | nd | BM smear | Pipo | M6 | 64 | 11 | nd | 75 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| NA-10 | M | ET | 67 | 0 | 0 | BM smear | HU | M4 | 74 | 7 | nd | 65 | PB smear | nd | 0 | PB smear | na | na | na | na | na | na | na | na | na |
| NA-11 | F | ET | 57 | 0 | 0 | BM smear | HU/32P/IFN/Ana | M2 | 73 | 16 | nd | 46 | PB smear | nd | 0 | PB smear | na | na | na | na | na | na | na | na | na |
| DI-12 | M | ET | 47 | 0 | nd | BM smear | HU | M7 | 49 | 2 | complex | 34 | BM smear | 0 | nd | BM smear | na | na | na | na | na | na | na | na | na |
| DI-13 | F | ET | 81 | 0 | 0 | BM smear | HU | M7 | 83 | 2 | complex, del(5q),−7 | 36 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-14 | F | ET | 75 | 0 | 0 | BM smear | HU | M2 | 77 | 2 | nd | 43 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| NA-08 | F | IMF | 62 | 0 | 0 | BM smear | HU/VP-16 | na | 69 | 7 | nd | 48 | PB smear | nd | 0 | PB smear | na | na | na | na | na | na | na | na | na |
| NA-09 | F | IMF | 62 | 0 | 0 | BM smear | Pipo/IFN | bi | 73 | 11 | nd | 38 | BM smear | 0 | 0 | BM smear | na | na | na | na | na | na | na | na | na |
| DI-20 | M | IMF | 65 | 0 | nd | BM smear | na | M7 | 67 | 2 | nd | 14 | BM smear | 0 | 0 | BMMC | na | na | na | na | na | na | na | na | na |
| p202 | M | IMF | 75 | nd | 0 | BM smear | HU | na | 80 | 5 | nd | 95 | PB smear | nd | 0 | PBMC | na | na | na | na | na | na | na | na | na |
UPN indicates unique patient number; M, male; F, female; Dx, diagnosis; nd, not done; na, not available; %T, percentage of chromosomes 9 carrying JAK2-V617F determined by AS-PCR; Nantes, AS-PCR was done in Nantes; Basel, AS-PCR was done in Basel; PB, peripheral blood; BM, bone marrow; GRA, granulocytes; PBMC, peripheral blood mononuclear cells; BMMC, bone marrow mononuclear cells; Pipo, pipobroman; HU, hydroxyurea; Thal, thalidomide; 32P, phosphorus 32; VP-16, etoposide; IFN, interferon-α; Ana, anagrelide; none, no cytoreductive treatment; FAB, AML diagnosis according to FAB classification; bi, biphenotypic leukemia; delay, interval between MPD and AML diagnosis in years; %Blasts, percentage of leukemic blasts on smear; LC-blasts, leukemic blasts isolated by laser capture microdissection.
%T determined in PB GRA.
%T determined in BM CD15+ cells.
Sample was taken 9 months before transformation to AML (not at MPD diagnosis), but no signs of AML were present in PB at this time.
Patients positive for JAK2-V617F at MPD diagnosis who transformed to JAK2-V617F–negative AML had a markedly shorter interval between diagnosis of MPD and leukemic transformation than those who transformed to JAK2-V617F–positive AML (3 ± 2 versus 10 ± 7 years; n = 14; P = .013) (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Information on treatment during the MPD phase was available in 25 of 27 patients (93%). Two patients transformed to AML without cytoreductive treatment, whereas the remaining patients received hydroxyurea (15 patients), pipobroman (6 patients), or other cytoreductive drugs (Table 1).
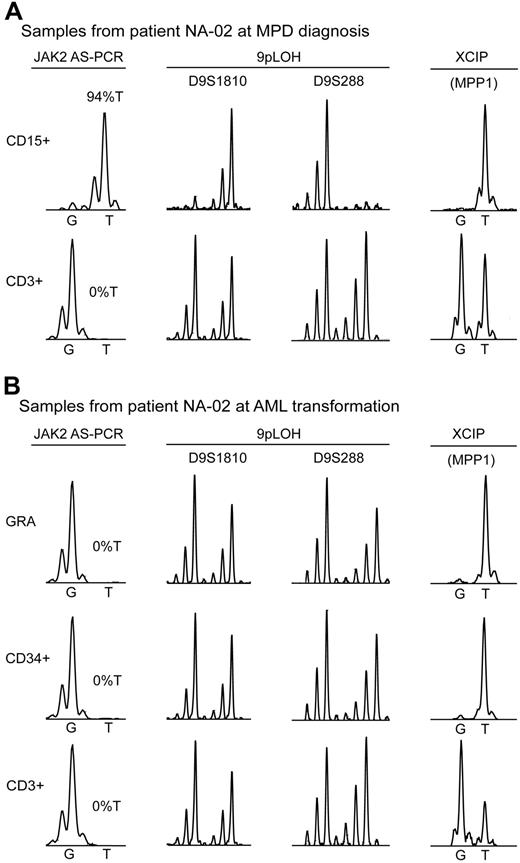
To address the question whether the MPD and AML clones in these patients arose de novo from different progenitor or stem cells, or represent subclones derived from a common ancestor, we performed molecular analyses in 2 cases. In one female patient with PV (NA-02), we compared clonality by 9pLOH and X chromosomal inactivation pattern (XCIP) in purified cell populations at both stages of the disease (Figure 1). At diagnosis of MPD, this patient had 94%T and displayed 9pLOH (Figure 1A). Deletion of chromosome 9p was excluded by determining the copy number using quantitative PCR (Figure S1). A phase resembling neutrophilic leukemia was observed in this patient 3 months before transformation to AML (white blood cells [WBCs], 159 × 109/L, with 75% neutrophils, 6% metamyelocytes, and 14% myelocytes, but no blasts). At this stage, the neutrophils were already negative for JAK2-V617F (not shown). At transformation to AML, the purified granulocytes and leukemic blast cells isolated by FACS were negative for JAK2-V617F and displayed no 9pLOH (Figure 1B). Thus, the AML clone did not arise from a MPD progenitor heterozygous for JAK2-V617F that lost the mutated JAK2 allele during mitotic recombination. A similar conclusion was reached in another MPD patient who transformed to AML.23 Using the MPP1 expression assay to determine clonality by XCIP,24,25 we found that the granulocytes at MPD diagnosis were clonal (Figure 1A). At diagnosis of AML, peripheral blood granulocytes and leukemic blast cells purified from peripheral blood by FACS were clonal and expressed MPP1 from the same parental X chromosome as the granulocytes at MPD diagnosis (Figure 1B). This result is compatible with a common origin of the MPD and AML clone, but does not rule out that 2 clones arose independently, because the AML blasts could fortuitously have inactivated the same X chromosome as the MPD clone. In a second patient (NA-48) with initially JAK2-V617F–positive MPD, we found at the time of leukemic transformation JAK2-V617F to be absent in isolated blast cells, but present in unfractionated bone marrow cells with an allelic ratio of 51%T (Table 1; Figure S2). Del(11q) was found in 20 of 20 metaphases by cytogenetic analysis and in 99 of 100 cells by interphase fluorescence in situ hybridization (FISH). These data suggest that the JAK2-V617F–negative blasts and the JAK2-V617F–positive bone marrow cells carry the same deletion, implying that they share a common clonal origin. Interestingly, the same region on chromosome 11 was previously identified in a genome-wide screening for LOH.26
Analysis of clonal markers in patient NA-02. (A) Bone marrow samples at MPD diagnosis. The presence of the JAK2-V617F mutation was determined by AS-PCR using DNA from purified cells: CD15+, myeloid cells isolated by FACS; CD3+, T cells isolated by FACS. %T indicates the allelic ratio between the mutant and wild-type JAK2 allele. 9pLOH was determined with the microsatellite markers D9S1810 and D9S288 using DNA from purified cells as for JAK2. XCIP was determined by AS-PCR for a G/T polymorphism in the MPP1 mRNA. The relative expression of the 2 MPP1 alleles was determined by comparing the G and T peak intensities obtained by the allele-specific reverse transcription-PCR assay. (B) Peripheral blood samples at AML transformation. GRA, Granulocytes isolated by Ficoll density centrifugation; CD34+, leukemic blasts isolated by FACS; CD3+, T cells isolated by FACS. The assays were performed as in (A).
Analysis of clonal markers in patient NA-02. (A) Bone marrow samples at MPD diagnosis. The presence of the JAK2-V617F mutation was determined by AS-PCR using DNA from purified cells: CD15+, myeloid cells isolated by FACS; CD3+, T cells isolated by FACS. %T indicates the allelic ratio between the mutant and wild-type JAK2 allele. 9pLOH was determined with the microsatellite markers D9S1810 and D9S288 using DNA from purified cells as for JAK2. XCIP was determined by AS-PCR for a G/T polymorphism in the MPP1 mRNA. The relative expression of the 2 MPP1 alleles was determined by comparing the G and T peak intensities obtained by the allele-specific reverse transcription-PCR assay. (B) Peripheral blood samples at AML transformation. GRA, Granulocytes isolated by Ficoll density centrifugation; CD34+, leukemic blasts isolated by FACS; CD3+, T cells isolated by FACS. The assays were performed as in (A).
In summary, the unexpected finding of JAK2-V617F–negative leukemia in patients with previously JAK2-V617F–positive MPD could be explained by 2 models: first, MPD and AML represent 2 independent clones that arose de novo from different progenitor or stem cells, or second, MPD and AML are 2 subclones derived from a common ancestor. Our data on patient NA-02 are compatible with both models, whereas the results from patient NA-48 favor a common clonal origin of MPD and AML and suggest that an additional clonal event may precede the acquisition of JAK2-V617F in the pathogenesis of MPD.
The online version of this manuscript contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Verena Jäggin (Basel, Switzerland) for FACS sorting; Danielle Pineau (Nantes, France) for excellent technical assistance; Francine Mugneret (Dijon, France) for cytogenetic and FISH analyses; Céline Schaeffer (Dijon) for collecting patients' data; Jean-Luc Harousseau (Nantes) for allowing access to the patients' archived files; Denis Caillot (Dijon) and Bruno Salles (Châlon-sur-Saône, France) for providing samples and patient data; and Alois Gratwohl (Basel), Jürg Schwaller (Basel), Ralph Tiedt (Basel), and Robert Kralovics (Vienna, Austria) for comments on the manuscript. A. Theocharides was a fellow of the Postgraduate Course in Experimental Biology and Medicine funded by the Freiwillige Akademische Gesellschaft Basel and the Novartis Foundation.
This work was supported by grants 310000-108006/1 from the Swiss National Science Foundation and the Swiss Cancer League (OCS-01742-08-2005) (R.C.S.) and from the Program Hospitalier de Recherche Clinique of the Région Bourgogne (F.G.), and from the Grand Ouest (S.H.) and Aquitaine-Charente (E.L.) Committees of the Ligue Nationale contre le Cancer.
Authorship
Contribution: A. Theocharides performed research, analyzed data, and wrote the paper; M.B. performed research and analyzed data; F.G., R.G., and E.L. provided essential reagents and analyzed data; S.-S.T. performed research; P.T. performed cytogenetics and FISH studies; A. Tichelli analyzed data; S.H. designed and performed research and analyzed data; and R.C.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Radek C. Skoda, Department of Research, Experimental Hematology, University Hospital Basel, Hebelstrasse 20, 4031 Basel, Switzerland; e-mail: radek.skoda@unibas.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal