X-linked severe combined immunodeficiency (SCID-X1) results from mutations in the IL2RG gene, which encodes the common gamma chain (γc) of the receptors for interleukin (IL)-2, 4, 7, 9, 15, and 21. Affected infants typically lack T and natural killer (NK) cells as a consequence of loss of signaling via the IL-7 receptor (IL-7R) and the IL-15R, respectively. In some infants, however, autologous NK cells are observed despite failure of T-cell ontogeny. The mechanisms by which mutations in γc differentially impact T- and NK-cell ontogeny remain incompletely understood. We used SCID-X1 patient–derived EBV-transformed B cells to test the hypothesis that the IL-15R–mediated signaling is preferentially retained as γc expression becomes limiting. Signal transduction via the IL-15R was readily detected in control EBV-transformed B cells, and via the IL-7R when modified to express IL-7Rα. Under the same experimental conditions, patient-derived EBV-transformed B cells expressing trace amounts of γc proved incapable of signal transduction via the IL-7R while retaining the capacity for signal transduction via the IL-15R. An equivalent result was obtained in ED-7R cells modified to express varying levels of γc. Collectively, these results confirm that signal transduction via the IL-15R, and hence NK ontogeny, is preferentially retained relative to the IL-7R as γc expression becomes limiting.
Introduction
The X-linked form of severe combined immunodeficiency (SCID-X1) is caused by mutations in the IL2RG gene, which encodes the common γ chain (γc).1 Affected infants typically lack both T and natural killer (NK) cells and have normal or elevated numbers of functionally deficient B cells that are unable to undergo immunoglobulin class switching and antibody production. The common γ chain is an integral component of receptors for interleukin (IL)-2, IL-4, IL-7, IL-9, IL-15, and IL-21 and associates with Janus kinase 3 (Jak3).1,–3 Ligand/receptor binding results in Jak3 activation by transphosphorylation that in turn phosphorylates signal transducers and activators of transcription (STATs), which translocate to the nucleus and modulate gene expression.2,4 Among these γc-dependent receptors, the IL-2R, IL-7R, and IL-15R are recognized to be essential for lymphoid ontogeny and homeostasis,5 and deficient signaling through these receptors as a consequence of γc mutations is thought to account for the classical T−B+NK− SCID-X1 phenotype. A proportion of SCID-X1 infants, however, have atypical phenotypes that remain incompletely understood
Human clinical and laboratory data, complemented with comprehensive studies in knockout mice, have demonstrated the critical role of signal transduction via the IL-7R and IL-15R for T- and NK-cell ontogeny, respectively.6,7 The IL-7R is heterodimeric, consisting of a unique IL-7Rα subunit and γc, while the IL-15R is heterotrimeric, consisting of a unique IL-15Rα subunit, γc and the IL-2Rβ subunit, which is shared with the IL-2R.5,8,9 The unique α subunits confer ligand-binding specificity. Mice and SCID patients with defective expression of IL-7Rα exhibit a T−B+NK+ phenotype, indicating that signal transduction via the IL-7R is required for T-cell development, but is dispensable for NK-cell development.10 Conversely, IL-15Rα knockout mice exhibit a T+B+NK− phenotype, indicating that signal transduction via the IL-15R is essential for NK-cell development in vivo, but is dispensable for T-cell development.11 These conclusions are further supported by the defects in lymphoid ontogeny observed in IL-7 and IL-15 knock-out mice.12,13
A subset of SCID-X1 infants retain the capacity for NK cell ontogeny and exhibit a T−B+NK+ phenotype similar to that observed in patients with defective IL-7Rα expression.10,14 A logical hypothesis to explain this atypical SCID-X1 phenotype is that certain γc mutations result in preferential impairment of IL-7R–mediated signaling. This is supported by the identification of a mutant γc chain (A156V) isolated from an NK+ SCID-X1 infant that, when expressed in the ED-7R T cell line, conferred selective impairment of responses to IL-4 and IL-7 with relative maintenance of responses to IL-2 and IL-15.9 More recently, we have described a novel splice-site mutation in the IL2RG gene in an NK+ SCID-X1 infant15 who was subsequently treated by gene therapy.16 Correctly spliced γc mRNA was reduced to trace levels and γc expression was undetectable on the surface of NK cells and the majority of B cells by fluorescence-activated cell sorter (FACS) analysis. In that report we hypothesized that signal transduction via the IL-15R is preferentially retained relative to the IL-7R as γc expression becomes limiting.
In the current study we formally tested this hypothesis by examining STAT5 phosphorylation in response to γc-dependent cytokine stimulation in healthy control and SCID-X1 patient–derived Epstein Barr Virus (EBV)–transformed B cell lines using immunolabeling and fluorescence microscopy. Common γ chain–dependent signal transduction via both the IL-15R and IL-7R was readily detected in healthy control-derived EBV-transformed B cells, when modified to express IL-7Rα. Under the same experimental conditions, patient-derived EBV-transformed B cells expressing trace amounts of γc, insufficient for detection at the cell surface, proved incapable of signal transduction via the IL-7R while retaining the capacity for signal transduction via the IL-15R. In supporting studies, the same result was obtained in ED-7R cells modified by lentivirus-mediated transduction to express varying levels of γc. Together these results confirm that signal transduction via the IL-15R is preferentially retained relative to the IL-7R as γc expression becomes limiting. Given the requirement for signaling via the IL-7R and IL-15R for T- and NK-cell development, respectively, this finding provides a novel mechanism for selective retention of NK cells in a proportion of SCID-X1 infants, and has important implications for vector design in SCID-X1 gene therapy.
Materials and methods
Approval for the study was received from The Children's Hospital at Westmead Ethics Committee.
Cell lines
The human T-cell lines ED-7R and ED-γc-7R have been described previously.9 Both lines express IL-2Rα, IL-2Rβ, IL-4Rα, IL-7Rα and IL-15Rα. The ED-γc-7R also expresses the common γ chain.9 The healthy donor-derived EBV-transformed B cell line, herein designated EBV-γcwt, was provided by Dr Graham Mann (Millennium Institute, Westmead, NSW, Australia) and the γc-null SCID-X1 patient-derived EBV-transformed B cell line (EBV-DR), herein designated EBV-γcnull, was provided by Professor Marina Cavazzana-Calvo (Hôpital Necker, Paris, France). The EBV-γclow was generated in-house from bone marrow cells from a previously described SCID-X1 patient with the mutation c.468 + 3 A→C15 using EBV provided by Mala Ratnamohan (ICPMR, Westmead, NSW, Australia). The above lines were cultured in RPMI 1640 medium (GIBCO-BRL, Invitrogen Australia, Mt Waverly, VIC, Australia) containing 10% (v/v) fetal bovine serum (FBS) (CSL, Parkville, VIC, Australia) and 2 mM glutamine (GIBCO-BRL). The HEK 293 cell line has been described previously17 and was cultured in Dulbecco modified essential medium (DMEM) (GIBCO-BRL) containing 10% FBS and 2 mM glutamine. All cultures were grown at 37°C in a humidified 5% CO2-air atmosphere.
FACS analysis for the detection of cell-surface markers and DNA ploidy
The cell-surface antibody-labeling methods used have been previously described.18 Flow cytometry was performed on a FACScan cytometer running CellQuest software Version 3.1f (Becton Dickinson, North Ryde, NSW, Australia). Cells were phenotyped with saturating concentrations of murine antibodies to human CD19 (Dako Australia, Botany, NSW, Australia), IL-2Rα (CD25) (BD Biosciences, North Ryde, NSW, Australia), IL-2Rβ (CD122) (Pharmingen, BD Biosciences), IL-7Rα (CD127) (Immunotech, Beckman Coulter, Gladesville, NSW, Australia) conjugated to either fluorescein isothiocyanate (FITC) or phycoerythrin (PE). Biotinylated rat antihuman IL-2Rγ (CD132) (Pharmingen, BD Pharmingen)19 and biotinylated mutant human IL-15/mouse Fc fusion protein (Chimerigen, Bioscientific, Gymea, NSW, Australia) were detected using RPE-conjugated streptavidin (Dako). DNA ploidy in EBV-transformed B cell lines was determined as previously described.20
Western analysis for the detection of γc, Jak3, and STAT5
Cultured cells were pelleted, washed twice with cold PBS, and lysed by resuspension in lysis buffer (10 mM Tris.Cl pH7.5, 150 mM NaCl, 1 mM EDTA, 2 mM Na3VO4, 2 mM Na2MoO4, 1% (v/v) Nonidet P40, 1 × Complete protease inhibitor (Roche Diagnostics, Castle Hill, NSW, Australia), 0.001% (v/w) aprotinin and 1 mM phenylmethylsulphonylfluoride) and gentle agitation at 4°C for 30 minutes. Protein concentrations were determined using a DC Protein Assay kit (Bio-Rad, Hercules, CA). For each sample, 100 μg of protein was separated using SDS-PAGE and transferred to a nitrocellulose membrane. Blots were probed with either biotinylated rat antihuman γc (1:1000), rabbit antihuman Jak3 or rabbit antihuman STAT5 (1:1000, both from Santa Cruz Biotechnology Inc., Santa Cruz, CA). Blots were also probed for actin (1:500, Sigma-Aldrich, St Louis, MO) to ensure equivalent amounts of protein were loaded to each lane. HRP-conjugated goat antirabbit IgG (1:20 000 Bio-Rad) or HRP-streptavidin (1:15 000, PerkinElmer Inc., Waltham, MA) were used as secondary detection reagents and immobilized HRP was detected using enhanced chemiluminescence (Pierce Biotechnology Inc., Rockford, IL).
Cytokine stimulation and detection of STAT5 phosphorylation
Prior to cytokine stimulation cells were washed in RPMI 1640 medium and incubated overnight in RPMI 1640 medium containing 0.02% (w/v) bovine serum albumin (BSA Fraction V; Roche Diagnostics). The following day, cells were incubated with IL-2 (100 pM to 100 nM), IL-7 (100 pM to 1000 nM) or IL-15 (100 pM to 1000 nM) (R& D Systems, Inc, Bioscientific) for 20 minutes at 37°C followed by chilling to 4°C to halt cytokine activation. Cells were then cytospun onto prepared slides and fixed in ice-cold 4% (w/v) paraformaldehyde (pH 7.4) followed by ice cold methanol, each for 30 minutes.21 Cells were then incubated in PBS containing 10% FBS and 0.1% (w/v) sodium azide (Ajax Chemicals, Auburn, NSW, Australia) for at least 1 hour, labeled with rabbit anti-pSTAT5 (Cell Signaling, Genesearch, Arundel, QLD, Australia), and stored overnight at 4°C in a humidified chamber. The following day, cells were stained with goat antirabbit Alexa Fluor 488 antibody (Molecular Probes, Invitrogen Australia), and nuclei counterstained with 4′6′diamidino-2 phenylindole (DAPI) (Sigma-Aldrich). Slides were mounted using 2.5% (w/v) 1,4-Diazabicyclo[2.2.2]octane (DABCO) (Sigma-Aldrich), 50 mM Tris (pH8.0) and 90% (v/v) glycerol and examined using a Leica CLSM confocal microscope (Leica Microsystems AG, Germany) equipped with an argon/krypton laser and Leica objective X40 / NA 1.3. Cells, positive or negative for pSTAT5, were counted either directly from the BX51 Olympus Microscope (Olympus Mt Waverley, Australia) image field using Olympus objectives X20 / NA 0.5, X40 / NA 0.75 and X60 / NA 1.4, or from images captured with a Spot enhanced SP6.O CCD Camera using Spot Software version 4.0.1 (Diagnostic Instruments, SciTech Pty Ltd, VIC, Australia). Images were collated using Adobe Photoshop 6.0 (AdobeSystems Inc., San Jose, CA).
Lentiviral vector construction and production
To obtain the human IL-7Rα cDNA, genomic DNA was extracted from ED-7R cells using the QIAamp DNA Blood Mini Kit (QIAGEN, Doncaster, VIC, Australia). PCR was performed to amplify IL-7Rα cDNA using the following forward and reverse primers, 5′-CCACCATGACAATTCTAGGTACAAC-3′ and 5′-TCACTGGTTTTGGTAGAAGC-3′, respectively. Reaction conditions were 30 cycles of 94°C for 30 seconds, 52°C for 30 seconds, and 72°C for 3 minutes using Pfu Ultra DNA polymerase (Stratagene, Cedar Creek, TX). The purified 1385 bp PCR product was subcloned into pTARGET (Promega, Annandale, NSW, Australia) and the sequence confirmed (GenBank accession number NM_002185). The 1177 bp human elongation factor-1-alpha (EF1α) promoter from pEF/myc/nuc (Invitrogen), the 516 bp human phosphoglycerate kinase (PGK) promoter from pRRLPGK-GFP, the 456 bp Nhe I/Kpn I fragment containing the Molony murine leukemia virus LTR (LTR) promoter from p3′NXL, the 487 bp primary Wiskott-Aldrich syndrome protein (WASP) promoter from pGL-481,22 the 1039 bp Janus kinase 3 (Jak3) -1013 to + 27 promoter fragment,23 and the human γc cDNA (GenBank accession number L19546) were amplified by PCR using Pfu Ultra DNA polymerase (Stratagene) and subcloned into pGEM-T Easy (Promega) for sequencing and to obtain compatible termini for subsequent ligation into lentiviral vector constructs.
Lentiviral vector constructs were derived from pRRLsin18.cPPT.CMV.EGFP.WPRE24 by replacing either the CMV promoter or the EGFP transgene using standard techniques. Vector stocks, pseudotyped with the VSV-G envelope,25 were produced by using a 4-plasmid transfection protocol26 as previously described.27 Vector supernatant was collected 48 and 72 hours after transfection, filtered through a 0.45- μm-pore-size cellulose acetate membrane (Sartorius AG, East Oakleigh, VIC, Australia) and concentrated by using the Vivaspin 20 filtration system with a 100-kDa molecular weight membrane cut-off (Sartorius AG). Lentiviral vector stocks were tested for replication competent virus by HIV-1 p24 ELISA (Perkin-Elmer Life Sciences, Rowville, VIC, Australia) after inoculation of HEK 293 cell cultures with vector supernatant and serial passaging. Transduction titres were determined on HEK 293 cells in the presence of Polybrene (8 μg/mL culture medium; Sigma-Aldrich) by using real-time quantitative PCR and previously published primer and probe sequences.28
Viral transduction
The EBV-transformed B-cell lines, EBV-γcwt and EBV-γclow, were transduced at a multiplicity of infection (MOI) of 25 to produce lines designated EBV-γcwt-IL-7Rα and EBV-γclow-IL-7Rα, respectively. The ED-7R cell line was transduced at an MOI of 10 for EGFP-expressing vectors or 25 for vectors expressing the γc transgene. Transduction was performed in the presence of Polybrene (8 μg/mL of culture medium). Flow cytometry was performed on a FACSCanto cytometer (BD BioSciences) running FACS Diva software (version 5.0.1). Cells expressing EGFP were enumerated and the mean fluorescence intensity (MFI) calculated for each FACS profile.
Results
γc mRNA and γc-dependent cytokine receptor expression in EBV-transformed cell lines
We have previously described a SCID-X1 infant with an atypical T−NK+ phenotype caused by a novel intron 3 splice-site mutation in γc15 . This mutation leads to the production of 2 aberrantly spliced γc mRNA species and trace amounts of correctly spliced message. To facilitate in vitro analysis of the impact of this mutation on γc-dependent signal transduction, patient bone marrow cells harvested prior to treatment with gene therapy16 were transformed with EBV and the resultant cell line designated EBV-γclow. Analysis of γc mRNA species in EBV-γclow cells by RT-PCR confirmed retention of the pattern previously reported in patient peripheral blood mononuclear cells (PBMC) (Fig. 1). As expected, a single γc mRNA species was detected in control EBV-transformed B cells, designated EBV-γcwt, obtained from a healthy donor.
EBV-transformed patient cells exhibit the same aberrant γc mRNA splicing pattern as previously studied patient PBMCs. RT-PCR analysis showed a single 439 bp product representing correctly spliced γc mRNA in healthy control-derived EBV-γcwt cells (lane 4) and 3 products of 439, 499 and 650 bp in patient-derived EBV-γclow cells (lane 6). These represent trace amounts of correctly spliced γc mRNA, a species with aberrant intron 3 splicing and a predominant species with complete failure of intron 3 splicing, respectively. M indicates Molecular Weight Marker VIII (Roche); NTC, no template control; EBV-γcwt, healthy control; EBV-γclow, patient; -, no RT controls.
EBV-transformed patient cells exhibit the same aberrant γc mRNA splicing pattern as previously studied patient PBMCs. RT-PCR analysis showed a single 439 bp product representing correctly spliced γc mRNA in healthy control-derived EBV-γcwt cells (lane 4) and 3 products of 439, 499 and 650 bp in patient-derived EBV-γclow cells (lane 6). These represent trace amounts of correctly spliced γc mRNA, a species with aberrant intron 3 splicing and a predominant species with complete failure of intron 3 splicing, respectively. M indicates Molecular Weight Marker VIII (Roche); NTC, no template control; EBV-γcwt, healthy control; EBV-γclow, patient; -, no RT controls.
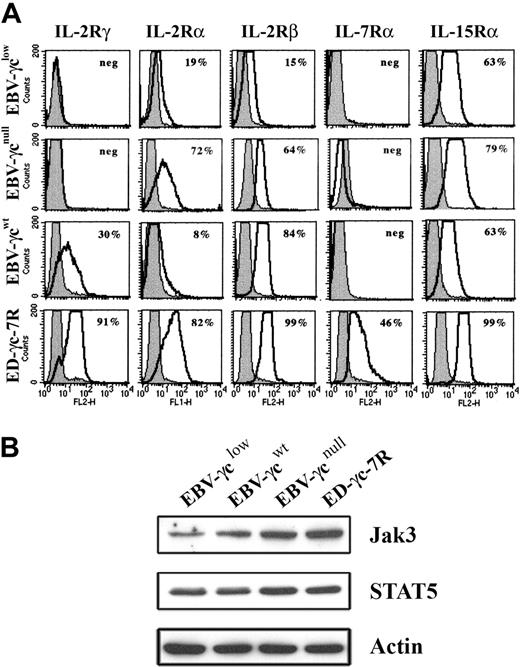
Analysis of EBV-γclow cells by FACS revealed that cell surface γc expression was below the limit of detection (Fig. 2A), consistent with the presence of only trace amounts of correctly spliced γc mRNA. Cell surface γc was similarly undetectable on EBV-transformed B cells, obtained from a SCID-X1 patient with a confirmed null mutation in the γc gene, designated EBV-γcnull, while γc expression was readily detected on healthy donor-derived EBV-γcwt cells. The anti-γc antibody used successfully for FACS had insufficient sensitivity to detect γc by Western analysis (not shown). Further phenotyping of these CD19+ DNA diploid cell lines, for the γc-dependent IL-2-, IL-7- and IL-15-cytokine receptors, demonstrated expression of IL-2Rα, IL-2Rβ and IL-15Rα but not IL-7Rα (Fig. 2A). This latter result is consistent with the known loss of IL-7Rα expression on mature B cells.29,30 Positive control ED-γc-7R cells expressed all receptor subunits examined. In addition, given the intention to evaluate signaling through γc-dependent cytokine receptors, expression of immediate downstream targets in the signal transduction pathway, Jak3 and STAT5, were confirmed in all lines by Western analysis (Fig. 2B).
Expression of cell surface IL-2R, IL-7R, and IL-15R subunits and downstream signaling molecules in EBV-transformed B and ED-γc-7R cell lines. (A) EBV-γclow, EBV-γcnull, EBV-γcwt, and ED-γc-7R cells were labeled with either antibodies to IL-2Rγ, IL-2Rα, IL-2Rβ, IL-7Rα, or IL-15Rα (solid lines) or to the isotype control (gray filled) and subjected to FACS analysis. Percentages shown in each panel represent the proportion of cells expressing detectable levels of the indicated cell surface receptor subunit. (B) Lysates of the above cell lines were analyzed for Jak3, STAT5 or actin using SDS-PAGE and Western blotting. Immobilized proteins were detected using HRP-conjugated secondary reagents and enhanced chemilumiscence.
Expression of cell surface IL-2R, IL-7R, and IL-15R subunits and downstream signaling molecules in EBV-transformed B and ED-γc-7R cell lines. (A) EBV-γclow, EBV-γcnull, EBV-γcwt, and ED-γc-7R cells were labeled with either antibodies to IL-2Rγ, IL-2Rα, IL-2Rβ, IL-7Rα, or IL-15Rα (solid lines) or to the isotype control (gray filled) and subjected to FACS analysis. Percentages shown in each panel represent the proportion of cells expressing detectable levels of the indicated cell surface receptor subunit. (B) Lysates of the above cell lines were analyzed for Jak3, STAT5 or actin using SDS-PAGE and Western blotting. Immobilized proteins were detected using HRP-conjugated secondary reagents and enhanced chemilumiscence.
IL-15 induces STAT5 phosphorylation and nuclear translocation in EBV-γclow cells
Signal transduction by the γc-dependent cytokine receptors IL-7R and IL-15R are essential for T and NK cell ontogeny, respectively.6,7 To explain the atypical T−NK+ phenotype in the SCID-X1 infant described, we hypothesized that limiting levels of γc expression differentially affects signal transduction via the IL-7R and IL-15R receptors with retention of IL-15R-mediated signaling at lower γc expression levels. As a first step toward testing this hypothesis in patient-derived EBV-γclow cells, we investigated STAT5 phosphorylation and nuclear translocation in response to IL-15 stimulation.
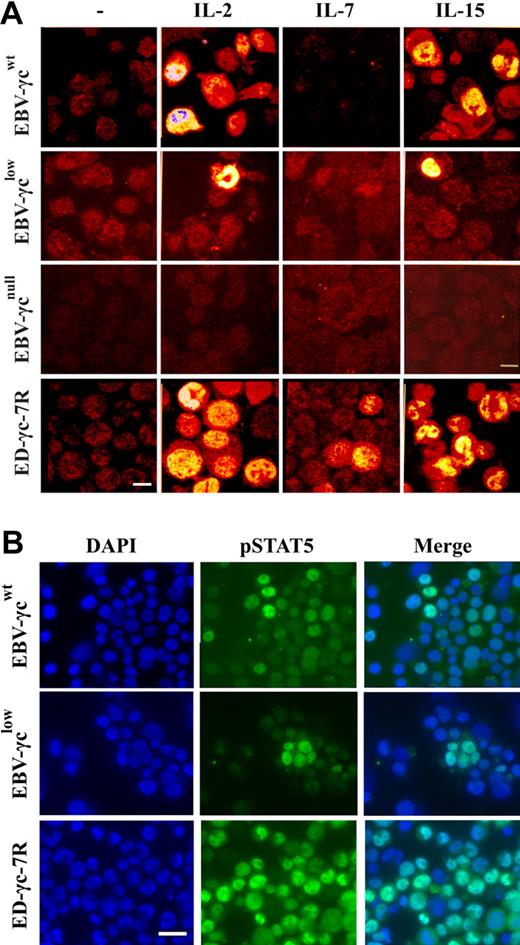
Phosphorylation of STAT5 is commonly measured by Western analysis, however, in order to achieve optimal sensitivity we used an immunofluorescence microscopy method that allows visualization and sub-cellular localization of pSTAT5 in individual cells. Using this assay, STAT5 phosphorylation was observed in a proportion of EBV-γcwt cells following stimulation with saturating concentrations of IL-2 and IL-15 (Fig. 3A). Interestingly, STAT5 phosphorylation was also observed in EBV-γclow cells under the same experimental conditions, albeit in a smaller proportion. This result is consistent with retention of γc-dependent signaling through IL-2R and IL-15R despite undetectable levels of γc at the cell surface (Fig. 2A). In the negative control EBV-γcnull cell line, which completely lacks γc expression, no STAT5 phosphorylation was observed in response to cytokine stimulation confirming the specificity and γc-dependence of the assay (Fig. 3A).
Detection of γc-dependent cytokine signaling by immunocytochemical analysis of STAT5 phosphorylation in EBV-transformed B and ED-γc-7R cell lines. (A) Single xy plane confocal images of EBV-γcwt, EBV-γclow, EBV-γcnull, and ED-γc-7R cell lines labeled with anti-pSTAT5 antibody in the absence (column 1) or presence of IL-2 (5 nM), IL-7 (100 nM), or IL-15 (10 nM or 100 nM) (columns 2, 3, and 4, respectively). Scale bar represents 10 μm. (B) Immunofluorescence images of EBV-γcwt, EBV-γclow, and ED-γc-7R cell lines cells showing nuclear localization of pSTAT5 after IL-15 stimulation (100 nM). Cells labeled with DAPI (column 1), pSTAT5 (column 2), and merged images (column 3). Scale bar represents 20 μm.
Detection of γc-dependent cytokine signaling by immunocytochemical analysis of STAT5 phosphorylation in EBV-transformed B and ED-γc-7R cell lines. (A) Single xy plane confocal images of EBV-γcwt, EBV-γclow, EBV-γcnull, and ED-γc-7R cell lines labeled with anti-pSTAT5 antibody in the absence (column 1) or presence of IL-2 (5 nM), IL-7 (100 nM), or IL-15 (10 nM or 100 nM) (columns 2, 3, and 4, respectively). Scale bar represents 10 μm. (B) Immunofluorescence images of EBV-γcwt, EBV-γclow, and ED-γc-7R cell lines cells showing nuclear localization of pSTAT5 after IL-15 stimulation (100 nM). Cells labeled with DAPI (column 1), pSTAT5 (column 2), and merged images (column 3). Scale bar represents 20 μm.
To better characterize the retention of IL-15R-mediated signaling in patient-derived EBV-γclow cells, the percentage of cells exhibiting STAT5 phosphorylation over a range of IL-15 concentrations was compared with that observed in healthy donor-derived EBV-γcwt cells (Table 1). For both EBV-γcwt and EBV-γclow, STAT5 phosphorylation was detected at concentrations as low as 100 pM and was maximal at 10 nM and above. However, the percentage of cells exhibiting STAT5 phosphorylation was consistently lower in EBV-γclow cells.
Analyses of STAT5 phosphorylation in EBV-transformed cell lines after IL-15 stimulation.
| IL-15 . | NIL . | 100 pM . | 1 nM . | 10 nM . | 100 nM . |
|---|---|---|---|---|---|
| EBV-γcwt | 0 | 1.42 ± 0.85 | 5.40 ± 2.27 | 7.98 ± 3.9 | 8.2 ± 4.1 |
| (n = 5) | (n = 3) | (n = 4) | (n = 4) | (n = 4) | |
| EBV-γclow | 0 | 0.69 ± 0.4 | 0.35 ± 0.17 | 1.5 | 1.46 ± 0.65 |
| (n = 5) | (n = 3) | (n = 4) | (n = 2) | (n = 5) | |
| EBV-γcnull | 0 | 0 | 0 | ||
| (n = 4) | (n = 3) | (n = 3) |
| IL-15 . | NIL . | 100 pM . | 1 nM . | 10 nM . | 100 nM . |
|---|---|---|---|---|---|
| EBV-γcwt | 0 | 1.42 ± 0.85 | 5.40 ± 2.27 | 7.98 ± 3.9 | 8.2 ± 4.1 |
| (n = 5) | (n = 3) | (n = 4) | (n = 4) | (n = 4) | |
| EBV-γclow | 0 | 0.69 ± 0.4 | 0.35 ± 0.17 | 1.5 | 1.46 ± 0.65 |
| (n = 5) | (n = 3) | (n = 4) | (n = 2) | (n = 5) | |
| EBV-γcnull | 0 | 0 | 0 | ||
| (n = 4) | (n = 3) | (n = 3) |
Values are percentage of the sample population exhibiting STAT5 phosphorylation (mean ±SEM). Number of cells counted per sample 1360 ± 194 (mean ± SEM).
As expected, STAT5 phosphorylation was not detected in any of the EBV cell lines following stimulation with a saturating concentration of IL-7 (Fig. 3A), consistent with absence of constitutive cell surface IL-7Rα expression (Fig. 2A). In the positive control ED-γc-7R cell line, STAT5 phosphorylation was observed after stimulation with each of the 3 γc-dependent cytokines tested.
To confirm nuclear translocation of pSTAT5 in EBV-transformed cell lines following IL-15-stimulation, further analyses by nuclear counter-staining with DAPI were performed (Fig. 3B). Characteristic nuclear localization of pSTAT5 was observed in both EBV-γcwt and EBV-γclow, and also in the positive control cell line ED-γc-7R. Collectively these results confirm retention of IL-15R-mediated signaling in EBV-γclow cells consistent with the atypical NK+ phenotype of the SCID-X1 infant from which they were derived.
Patient-derived EBV-γclow-IL-7Rα cells do not respond to IL-7 stimulation
To facilitate analysis of γc-dependent signal transduction via the IL-7R, a lentiviral vector encoding the IL-7Rα cDNA was constructed (Fig. 4A) and used to transduce EBV-γcwt and EBV-γclow cells. Similar levels of IL-7Rα expression were achieved in each cell line (Fig. 4B) with ploidy, CD19, and γc expression levels remaining equivalent to the parental lines (data not shown). The capacity of these cell lines to phosphorylate STAT5 after IL-7 stimulation was then tested over a range of cytokine concentrations (Fig. 4C and Table 2). Following IL-7 stimulation, pSTAT5 was detected in IL-7Rα-expressing EBV-γcwt cells at all concentrations tested down to 100 pM. In contrast, pSTAT5 phosphorylation was not observed in IL-7Rα-expressing EBV-γclow cells following IL-7 stimulation even at cytokine concentrations 10 000 fold higher. These results suggest that the limiting level of γc expression on patient-derived EBV-γclow cells, while sufficient for IL-15R–mediated signaling (and IL-2R–mediated signaling), is insufficient for IL-7R–mediated signaling.
EBV-cell lines genetically modified to express IL-7Rα do not respond to IL-7 signaling when γc is limiting. (A) Lentiviral vector construct encoding the IL-7Rα cDNA used to transduce EBV cell lines. RSV, Rous sarcoma virus hybrid promoter; SD/SA, splice-donor and spice-acceptor sites; ψ, packaging and dimerization signal; GA, fragment of the HIV-1 gag gene; RRE, Rev responsive element; PPT, central polypurine tract; CMV, human cytomegalovirus immediate-early promoter; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) Unmodified (left) and IL-7Rα–modified (right) EBV-transformed B-cell lines were immunolabeled with antibody to IL-7Rα and analyzed by FACS. Percentages shown represent the proportion of cells expressing detectable levels of CD127. (C) EBV-γcwt-IL-7Rα, EBV-γclow-IL-7Rα and ED-γc-7R cells were labeled with antibody to pSTAT5 before and after IL-7 (1 nM) stimulation (columns 1 and 2, respectively). Counterstaining of IL-7–treated cells with DAPI (column 3). Scale bar represents 50 μm.
EBV-cell lines genetically modified to express IL-7Rα do not respond to IL-7 signaling when γc is limiting. (A) Lentiviral vector construct encoding the IL-7Rα cDNA used to transduce EBV cell lines. RSV, Rous sarcoma virus hybrid promoter; SD/SA, splice-donor and spice-acceptor sites; ψ, packaging and dimerization signal; GA, fragment of the HIV-1 gag gene; RRE, Rev responsive element; PPT, central polypurine tract; CMV, human cytomegalovirus immediate-early promoter; WPRE, woodchuck hepatitis virus posttranscriptional regulatory element. (B) Unmodified (left) and IL-7Rα–modified (right) EBV-transformed B-cell lines were immunolabeled with antibody to IL-7Rα and analyzed by FACS. Percentages shown represent the proportion of cells expressing detectable levels of CD127. (C) EBV-γcwt-IL-7Rα, EBV-γclow-IL-7Rα and ED-γc-7R cells were labeled with antibody to pSTAT5 before and after IL-7 (1 nM) stimulation (columns 1 and 2, respectively). Counterstaining of IL-7–treated cells with DAPI (column 3). Scale bar represents 50 μm.
Analyses of STAT5 phosphorylation in EBV-transformed cell lines after IL-7 stimulation.
| IL-7 . | NIL . | 100 pM . | 1 nM . | 10 nM . | 100 nM . | 1000 nM . |
|---|---|---|---|---|---|---|
| EBVγcwt-IL-7Rα | 0 | 1.32 ± 0.76 | 1.45 ± 0.65 | 0.99 ± 0.57 | 1.24 ± 0.72 | 2.9 |
| (n = 3) | (n = 3) | (n = 5) | (n = 3) | (n = 3) | (n = 1) | |
| EBV-γclow-IL-7Rα | 0 | 0 | 0 | 0 | 0 | 0 |
| (n = 4) | (n = 2) | (n = 4) | (n = 3) | (n = 3) | (n = 1) |
| IL-7 . | NIL . | 100 pM . | 1 nM . | 10 nM . | 100 nM . | 1000 nM . |
|---|---|---|---|---|---|---|
| EBVγcwt-IL-7Rα | 0 | 1.32 ± 0.76 | 1.45 ± 0.65 | 0.99 ± 0.57 | 1.24 ± 0.72 | 2.9 |
| (n = 3) | (n = 3) | (n = 5) | (n = 3) | (n = 3) | (n = 1) | |
| EBV-γclow-IL-7Rα | 0 | 0 | 0 | 0 | 0 | 0 |
| (n = 4) | (n = 2) | (n = 4) | (n = 3) | (n = 3) | (n = 1) |
Values are percentage of the sample population exhibiting STAT5 phosphorylation (mean ± SEM). Number of cells counted per sample 2346 ± 398 (mean ± SEM).
Limiting γc expression preferentially impairs IL-7 receptor–mediated signaling
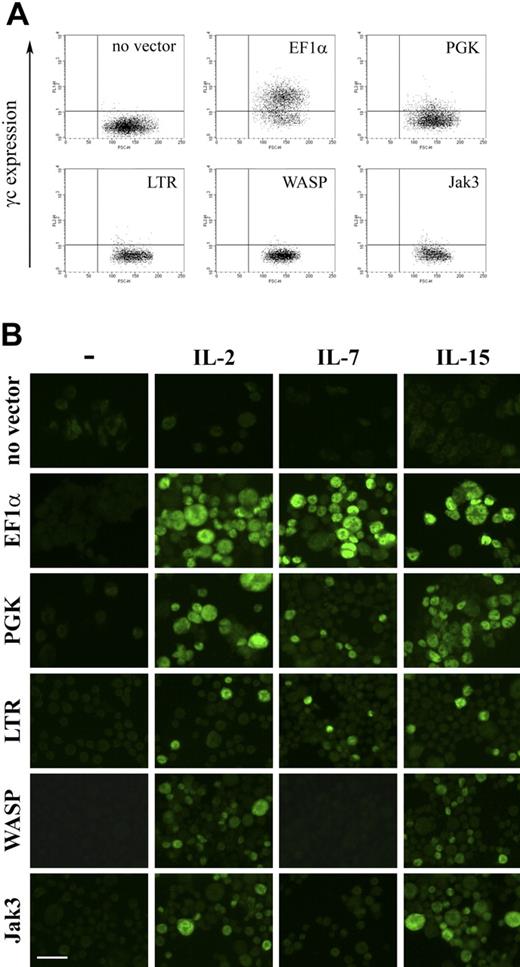
To further substantiate our hypothesis and confirm dependence on γc expression levels, we next sought to examine the effect of titrating γc expression on γc-dependent signal transduction in the γc-null ED-7R cell line. Expression levels of IL-2Rγ, IL-2Rα, IL-2Rβ, IL-7Rα, IL-15Rα, and Jak3 and STAT5 were confirmed by FACS and Western analysis, respectively (not shown), and found to be equivalent to those shown for ED-γc-7R cells (Fig. 2). As an initial step toward the generation of ED-7R cells with varying levels of γc expression, a series of lentiviral vectors encoding either EGFP or γc under the transcriptional control of selected internal heterologous promoters were constructed. To determine the relative strength of each promoter, ED-7R cells were transduced at an MOI of 10 with the EGFP-encoding vector set. This MOI was chosen to ensure that the majority of cells contained single integration events so that the relative strength of each promoter could be accurately assessed. Analysis of EGFP expression by FACS revealed a hierarchy of promoter strengths with the EF1α promoter being the strongest (MFI = 28 276) (Fig. 5). This was 5.0-, 4.7-, 8.8-, and 20.7-fold stronger than the PGK (MFI = 5615), LTR (MFI = 6007), WASP (MFI = 3225), and Jak3 (MFI = 1369) promoters, respectively.
EGFP expression in ED-7R cells following lentiviral vector transduction. ED-7R cells were transduced at an MOI of 10 by vectors encoding EGFP under the transcriptional control of the indicated promoters and analyzed by FACS 7 days later. EF1α indicates human elongation factor-1-alpha promoter; PGK, human phosphoglycerate kinase promoter; LTR, MoMLV LTR promoter/enhancer; WASP, Wiskott-Aldrich syndrome protein promoter; Jak3, Janus kinase 3 promoter.
EGFP expression in ED-7R cells following lentiviral vector transduction. ED-7R cells were transduced at an MOI of 10 by vectors encoding EGFP under the transcriptional control of the indicated promoters and analyzed by FACS 7 days later. EF1α indicates human elongation factor-1-alpha promoter; PGK, human phosphoglycerate kinase promoter; LTR, MoMLV LTR promoter/enhancer; WASP, Wiskott-Aldrich syndrome protein promoter; Jak3, Janus kinase 3 promoter.
The γc-encoding vector set, containing the same promoters, was then used to transduce ED-7R cells and resultant cell surface γc expression determined by FACS. Expression of γc was detectable following expression with vectors containing the EF1α and PGK promoters, but not for vectors containing the LTR, WASP, and Jak3 promoters (Fig. 6A). Analysis of integrated provirus and γc mRNA by quantitative PCR and RT-PCR, respectively, confirmed molecular transduction and γc transgene expression for all vectors (not shown). This result was consistent with γc being expressed on the surface of ED-7R cells transduced with the vectors containing the LTR, WASP, and Jak3 promoters, but at levels below the limit of detection by FACS (Fig. 6A). Each vector was then evaluated for the capacity to reconstitute γc-dependent signaling (Fig. 6B). Following stimulation with the γc-dependent cytokines IL-2, IL-7, and IL-15, STAT5 phosphorylation was detected in ED-7R cells transduced with the EF1α, PGK, and LTR promoter constructs. In ED-7R cells transduced with vectors containing the WASP and Jak3 promoters, however, STAT5 phosphorylation was observed following IL-2 and IL-15 stimulation, but not IL-7. These results are entirely consistent with those obtained using SCID-X1 patient-derived EBV-transformed B cells, and confirm the conclusion that expression of limiting levels of cell surface γc preferentially impairs signaling via the IL-7R.
γc expression and γc-dependent cytokine signaling in ED-7R cells following lentiviral vector transduction. (A) ED-7R cells were transduced at an MOI of 25 by vectors encoding γc under the transcriptional control of the indicated promoters. After 10 days cells were labeled with anti-γc antibody and analyzed by FACS. (B) Immunofluorescence images of the above transduced cell populations labeled with anti-pSTAT5 antibody in the absence (column 1) or presence of IL-2, IL-7, or IL-15 (columns 2, 3 and 4, respectively). Scale bar represents 50 μm.
γc expression and γc-dependent cytokine signaling in ED-7R cells following lentiviral vector transduction. (A) ED-7R cells were transduced at an MOI of 25 by vectors encoding γc under the transcriptional control of the indicated promoters. After 10 days cells were labeled with anti-γc antibody and analyzed by FACS. (B) Immunofluorescence images of the above transduced cell populations labeled with anti-pSTAT5 antibody in the absence (column 1) or presence of IL-2, IL-7, or IL-15 (columns 2, 3 and 4, respectively). Scale bar represents 50 μm.
Discussion
We have previously reported a novel splice-site mutation in the γc-encoding IL2RG gene in a SCID-X1 infant with an atypical NK+ phenotype who was subsequently treated by gene therapy.15,16 This mutation reduced correctly spliced γc mRNA to trace levels such that cell surface γc expression was below the limits of detection by FACS analysis on NK cells and the majority of B cells. Given the known requirement for γc-dependent signaling via the IL-7R and IL-15R for T- and NK-cell ontogeny, respectively, we hypothesized that IL-15R–mediated signaling, and hence NK ontogeny, is preferentially retained as γc expression becomes limiting.
In the current study we first tested this hypothesis using healthy control and SCID-X1 patient-derived EBV-transformed B cells and a highly sensitive and specific assay to measure STAT5 phosphorylation in individual cells in response to γc-dependent cytokine stimulation. Signal transduction via IL-15R was readily detectable at physiologic cytokine concentrations in healthy donor-derived EBV-transformed B cells, and via the IL-7R when modified to express IL-7Rα. Under the same experimental conditions, patient-derived EBV-transformed B cells expressing trace amounts of γc proved incapable of signal transduction via the IL-7R while retaining the capacity for signal transduction via the IL-15R. Moreover, increasing the concentration of IL-7 to in excess of 1000-fold above physiologic levels did not overcome the observed block to IL-7R–mediated signaling. Although not a specific focus of the current study, we also observed retention of IL-2R–mediated signaling in the presence of limiting γc expression. To independently substantiate data obtained using healthy control and SCID-X1 patient-derived EBV-transformed B cells we next confirmed dependence on γc levels by titrating γc expression in the γc-null ED-7R cell line. This was achieved by using γc-encoding lentiviral vectors containing internal promoters with a 20-fold range of transcriptional activities. In this context, free of possible confounding variables present in the EBV-transformed B cell studies, we again observed preferential loss of IL-7R-mediated signaling at low γc expression levels. Indeed, this result was replicated with each of the 2 least transcriptionally active vector constructs. Loss of signaling through the IL-2R and IL-15R was not observed, even at the lowest γc expression levels achieved.
Collectively, these results confirm our hypothesis and reveal a previously unrecognized mechanism by which mutations in the γc-encoding IL-2RG gene can differentially impact T- and NK-cell ontogeny. Another group has previously reported a mutant γc (A156V) isolated from an NK+ SCID-X1 infant, that when expressed in ED-7R cells conferred selective impairment of responses to IL-4 and IL-7 with relative maintenance of responses to IL-2 and IL-15.9 Together these studies show that preferential retention of NK cell ontogeny in SCID-X1 infants can be the consequence of mutations that result in both qualitative and quantitative abnormalities in γc expression. In this latter study the underlying mechanism appeared to be preferential impairment of the ligand-binding affinity of the IL-4R and IL-7R containing the mutant γc. Clearly, a different mechanistic explanation is required to explain our results where the ligand-binding affinity of any γc-dependent cytokine receptors formed would be predicted to be normal.
High-affinity receptors for IL-2 and IL-15 are heterotrimeric, containing unique IL-2Rα and IL-15Rα subunits, a shared IL-2Rβ subunit and γc.31,32 The receptor for IL-7 has substantially lower ligand-binding affinity and is heterodimeric, containing a unique IL-7Rα subunit and γc9. These differences in ligand-binding affinities alone are insufficient to explain our findings as experiments were carried out under conditions where ligand concentrations were in excess and all available γc-dependent receptors would have been saturated. Failure of γc-dependent signal transduction via the IL-7R, as measured by STAT5 phosphorylation, therefore directly implies the presence of insufficient receptor numbers to initiate the intracellular Jak3/STAT5 signal cascade. This being the case, there are 2 possible mechanisms. First, the number of activated IL-2R and IL-15R required to initiate the Jak3/STAT5 intracellular signal cascade may be lower than for the IL-7R. Alternatively, under conditions where the availability of γc is limiting there may be higher numbers of IL-2 and IL-15 receptors, possibly as a consequence of more effective competition for the pathologically low number of γc chains available for receptor assembly. Further dissection of mechanism will be challenging. For example, use of Scatchard analysis, which is commonly used to determine receptor numbers and ligand binding affinities, is confounded by the fact that the IL-15Rα subunit binds IL-15 with extremely high affinity irrespective of whether the subunit is assembled into a functional heterotrimeric receptor.31
In addition to providing insight into the molecular basis of the NK+ SCID-X1 phenotype our data may have implications for the treatment of SCID-X1 by gene therapy. With current gene delivery technologies, successful reconstitution of the T- and NK-cell compartments relies on the selective growth advantage of gene-modified progenitor cells over unmodified counterparts. In the NK+ SCID-X1 infant whose EBV-transformed B cells were investigated in this report, gene therapy resulted in only partial reconstitution of the T-cell compartment and little or no effect on the infant's preexisting but functionally abnormal NK-cell compartment.16 While the relatively low dose of γc+/CD34+ cells used (1.3 × 106/kg) is likely to have been a contributory factor, the infant's partial phenotype may also have exerted an affect. The preexistence of an NK cell compartment before gene therapy, as a consequence of retention of IL-15R–mediated signaling, would have almost certainly resulted in reduced competitive advantage for gene-modified cells undergoing NK–cell ontogeny. Whether there might also have been an impact on reconstitution of the T-cell compartment is more speculative. Given the presence of low-level γc expression, successful signal transduction via the IL-7R in early T-cell ontogeny cannot be ruled out. The functional consequence for IL-7R formation of competition among γc-dependent cytokine receptors for a limiting numbers of γc chains would be determined by the relative levels of expression of subunits for other γc-dependent receptor subunits. As more infants with partial SCID-X1 phenotypes undergo gene therapy the impact of such phenotypes on immunologic reconstitution should become clearer.
Our data have further important implications for researchers involved in the development of safer vectors for SCID-X1 gene therapy. The development of leukemia in 3 of 11 infants treated in the Paris-based SCID-X1 gene therapy trial has been shown to be the consequence of vector insertion in or near oncogene loci, such as LMO2, leading to aberrant activation driven by proximity to strong enhancer elements in the vector LTR promoter.33,34 This has lead to a desire to trial vectors with self-inactivating LTRs and carefully selected internal heterologous promoters with less potent enhancer activity. Our data define a new risk to vector therapeutic efficacy. As promoter enhancer strength is reduced, an increasing proportion of integrated proviral genomes will give rise to limiting levels of γc expression. Based on our results, this phenomenon has the potential to skew reconstitution toward the NK compartment with T-cell reconstitution becoming dependent on increasingly fewer high expressing integration sites. Novel γc-encoding vectors intended for human clinical use should therefore be tested to ensure that they not only minimize transactivation events but also provide adequately robust γc expression to ensure that a high proportion of integration events in progenitor cells express sufficient γc to ensure both T- and NK-cell ontogeny. The assay system used in this report should prove to be a useful tool for such analyses.
Up to the present time, the majority of studies examining the effect of γc mutations on signal transduction have used Western blot analysis and necessitated the biochemical and genetic manipulation of large numbers of cultured cells such as the ED-7R cell line.7,9 In the current study, however, we were faced with the challenge of studying the impact of a novel splice-site mutation that could not be modeled by simple transfection of mutant γc cDNA. This challenge was overcome by directly analyzing patient-derived EBV-transformed B cells. Therefore, anticipating the need to derive data from relatively small cells numbers, we chose to use an intracellular immuno-cytochemical assay system that facilitated analysis of γc-dependent signal transduction in individual cells and required at least an order of magnitude fewer cells than Western blotting.35 Indeed, the relatively low proportion of EBV-transformed B cells from both healthy donor and SCID-X1 patients exhibiting responsiveness to γc-dependent cytokine stimulation precluded the use of more conventional methodologies. In addition to the desired sensitivity, which allowed direct analysis of patient-derived cells, the assay proved to be highly specific with STAT5 phosphorylation events being dependent on the presence of both γc expression and cytokine stimulation.
A tangential, but nevertheless interesting, question arising from the use of immunofluorescence microscopy to detect pSTAT5 in individual cells is the relationship between γc expression levels, the proportion of cells exhibiting STAT5 phosphorylation and the level of phosphorylation per cell. In experiments using both EBV-transformed B cells and ED-7R cells, the proportion of cells exhibiting detectable STAT5 phosphorylation following cytokine stimulation was reduced in cells expressing γc at lower levels. The corresponding level of STAT5 phosphorylation observed in individual cells, however, was not uniformly reduced with some cells remaining capable of high-level STAT5 phosphorylation despite low-level γc expression. This observation suggests that a threshold for triggering STAT5 phosphorylation may exist whereby the number of cell surface receptors increases the probability of the threshold being reached, but exerts relatively less influence on the resultant level of STAT5 phosphorylation.
In summary, using healthy donor and SCID-X1 patient-derived EBV-transformed B cells, lentiviral vector-transduced ED-7R cells, and a sensitive immuno-cytochemical assay to measure STAT5 phosphorylation, we show that limiting γc expression preferentially impairs signal transduction via the IL-7R while signaling via the IL-15 receptor is retained. This result provides a novel explanation for the atypical NK+ SCID-X1 phenotype, and has implications for the treatment of SCID-X1 by gene therapy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Professor Kazuo Sugamura (Tohoku University Graduate School of Medicine, Sendai, Japan) for kindly providing ED-7R and ED-γc-7R cells, Dr Graham Mann (Millennium Institute, Westmead, NSW, Australia) for providing healthy donor-derived EBV-transformed B cells (herein designated EBV-γcwt), Professor Marina Cavazzana-Calvo (Hôpital Necker, Paris, France) for providing γc null SCID-X1 patient-derived EBV-transformed B cells (herein designated EBV-γcnull), Mala Ratnamohan (ICPMR, Westmead, NSW, Australia) for providing the EBV stocks used to generate EBV-γclow cells, Professor Inder Verma (Salk Institute, San Diego, CA) for lentiviral vector reagents, Professor Adrian J Thrasher (Molecular Immunology Unit, Institute of Child Health, London, United Kingdom) for providing the WASP promoter, and Dr John J O'Shea (National Institute of Arthritis and Musculoskeletal and Skin Diseases, Bethesda, MD) for providing the Jak3 promoter. Samantha L Ginn is the recipient of a fellowship honoring the memory of Noel Dowling.
Authorship
Contribution: C.M.S. and S.L.G. contributed equally. All authors contributed to the design of the experimental work and to manuscript preparation. C.M.S., S.L.G., G.J.L., and C.T.D. generated the data reported.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Ian E. Alexander, Gene Therapy Research Unit, The Children's Hospital at Westmead, Locked Bag 4001, Westmead NSW 2145, AUSTRALIA, e-mail: iana@chw.edu.au.