Abstract
Two distinct dendritic cell (DC) subsets, conventional DCs (cDCs) and plasmacytoid DCs (pDCs), have been shown to develop via either the myeloid or the lymphoid pathway in murine hematopoiesis. Lineage-specific phenotypes or functions of “myeloid” and “lymphoid” DCs, however, still remain elusive. Furthermore, such analysis has been particularly difficult in humans, due to lack of an assay system appropriate for the analysis of human stem and progenitor cell differentiation. Here, using a highly efficient xenotransplantation model, we extensively analyze the origin and the molecular signature of human DCs. Purified human common myeloid progenitors (CMPs) and common lymphoid progenitors (CLPs) were intravenously transplanted into nonobese diabetic–severe combined immunodeficiency (NOD-scid)/IL2rγnull newborn mice. CMPs and CLPs displayed significant expansion in the xenogeneic host, and human cDC and pDC progeny were isolatable. Strikingly, each human DC subset possessed indistinguishable expression patterns of surface phenotype and gene transcripts regardless of their CMP or CLP origin, even at the genome-wide level. Thus, cDC and pDC normally develop after cells have committed to the myeloid or the lymphoid lineage in human hematopoiesis, while their transcriptional signatures are well preserved irrespective of their lineage origin. We propose that human DCs use unique and flexible developmental programs that cannot be categorized into the conventional myeloid or lymphoid pathway.
Introduction
Dendritic cells (DCs) play a crucial role in maintaining the immune system.1,2 They are particularly efficient in presenting antigenic peptides in the context of major histocompatibility complex (MHC) to T cells. Two DC subsets, conventional dendritic cells (cDCs) and plasmacytoid dendritic cells (pDCs), have been identified in murine and human hematopoietic tissues.3-7 cDCs are known to initiate primary T-cell responses, thereby orchestrating and potentiating the acquired immunity against foreign antigens. pDCs were originally reported as a cell type that is morphologically similar to plasma cells in the T-cell zones of human lymphoid organs.8 In contrast to cDCs, pDCs do not stimulate naive T cells into cycle but can be converted into cDC-like cells upon activation and function as efficient antigen-presenting cells (APCs).9,10 Of note, pDCs but not cDCs are capable of secreting a large amount of type-I interferons (IFNs) in response to unmethylated CpG oligonucleotides and viral and self-nucleic acids.5,11-14 In the context of the TH1/TH2 balance, human pDCs play an essential role in inducing differentiation of TH2, whereas cDCs preferentially induce a TH1 response.15 Each DC subset displays different groups of Toll-like receptors (TLRs) to effectively sense a wide array of invading pathogens.10
Developmental pathways for these human DC subsets, however, still remain enigmatic. cDCs have been shown to develop from monocytes in the presence of IL-4 and GM-CSF and are sometimes called “myeloid” DCs. On the other hand, pDCs are often referred to as “lymphoid” DCs, mainly because they possess several critical “lymphoid” markers such as the rearrangement of immunoglobulin heavy chain (IgH) D-J16,17 and pre-T-cell receptor α (pTα) transcripts.18,19 This simple classification proved to be incorrect, given that in murine hematopoiesis, cDCs and pDCs can be generated from either myeloid- or lymphoid-committed bone marrow progenitor populations in vivo and in vitro.17,20-22 Therefore, mature pDCs appear to ectopically activate some genetic programs primarily associated with early lymphocyte development regardless of their lineage origin. In human hematopoiesis, transplantation of hCD34+ human bone marrow or cord blood cells into adult NOD.Cg-Prkdcscid (NOD-scid [nonobese diabetic–severe combined immunodeficiency]) or newborn Rag2−/−γc−/− mice resulted in the generation of DCs in vivo.23,24 The hCD34+ fraction is a heterogeneous population containing hematopoietic stem cells (HSCs), common myeloid progenitors (CMPs), common lymphoid progenitors (CLPs), and a variety of monopotent progenitors. A recent study has shown that purified human CMPs and CLPs can give rise to cDCs and pDCs, at least when cultured in the presence of murine stromal cells and human cytokines.25 Several critical questions, however, remain to be addressed. It is still unclear whether myeloid and lymphoid pathways contribute to the formation of human cDC or pDC pools in vivo; and if they do, it is unclear whether such DC subsets of myeloid or lymphoid origin are intrinsically different, thereby enabling them to exert distinct functions or display distinct phenotypes. One of the obstacles to address these questions has been the lack of an assay system efficient enough to evaluate lineage potential of committed progenitors in vivo.
We have recently established a highly efficient xenogeneic transplantation system by using newborns of NOD-scid/IL2rγnull mice.26 This xenotransplantation system can recapitulate hematopoietic development from the most primitive hCD34+hCD38− long-term human HSC fraction.26 In the present study, by adoptive transfer of prospectively purified human hematopoietic progenitors into NOD-scid/IL2rγnull newborns, we found that both cDCs and pDCs are efficiently produced from the human CMPs and CLPs in vivo and that each DC subset of CMP or CLP origin shared identical gene-expression profiles. Thus, the common human DC developmental program could be activated even after the cells are committed into the myeloid or the lymphoid lineage, resulting in the formation of single human cDC and pDC pools in vivo. These data collectively suggest that the DC developmental program is operated independently of myeloid or lymphoid ones, at least in human hematopoiesis.
Materials and methods
Mice
NOD.Cg-PrkdcscidIL2rgtmlWjl/Sz (NOD/SCID/IL2rγnull) and NOD/LtSz-Prkdcscid/B2mnull (NOD/SCID/β2mnull) mice were developed at the Jackson Laboratory (Bar Harbor, ME). The NOD/SCID/IL2rγnull strain was established by backcrossing a complete null mutation at the γc locus onto the NOD.Cg-Prkdcscid strain.27 All experiments were performed according to the guidelines of the Institutional Animal Committee of Kyushu University.
Purification of human HSCs, CMPs, and CLPs
Cord blood was harvested, after written informed consent was obtained in accordance with the Declaration of Helsinki, following the protocol approved by the institutional review board of RIKEN. Lin+ cells were depleted from mononuclear cells by using mouse anti-hCD3, anti-hCD4, anti-hCD8, anti-hCD11b, anti-hCD19, anti-hCD20, anti-hCD56, and anti–human glycophorin A (hGPA) monoclonal antibodies (BD Immunocytometry, San Jose, CA). In purification of CMPs, anti-hCD7 and anti-hCD10 antibodies were also added for depleting Lin+ cells. Samples were enriched for hCD34+ cells by using anti-hCD34 microbeads (Miltenyi Biotec, Auburn, CA). These cells were further stained with anti-hCD34 and anti-hCD38 antibodies (BD Immunocytometry). Lin−hCD34+hCD38− HSCs, Lin−hCD34+hCD38+hIL-3RαlohCD45RA− CMPs, and Lin−hCD34+hCD38−hCD7+ CLPs were purified by using a FACSAria (Becton Dickinson, San Jose, CA). Purity of each progenitor was higher than 97%. Sublethally irradiated (100 cGy) NOD/SCID/IL2rγnull newborns received 104 cells of each population via a facial vein. The human cDC and pDC progeny was analyzed at 4 to 5 weeks after transplantation. Reconstitution capacity of each stem/progenitor fraction was determined in 4 independent experiments.
Analysis of human cell engraftment
At 4 to 5 weeks after transplantation, samples of peripheral blood, bone marrow, spleen, and thymus were harvested from recipient mice. Cells from each tissue were isolated using collagenase (Worthington Biochemical, Lakewood, NJ) and DNAase I (Sigma-Aldrich, St Louis, MO) to ensure release of tissue-associated DCs.28,29 The development of human cDCs and pDCs was analyzed by the percentages of hCD45+HLA-DR+ hIL-3RαlohCD11c+ and hCD45+HLA-DR+ hIL-3RαhihCD11c− populations. For lymphoid development, hCD45+hCD19+ B cells, hCD45+hCD56+ natural killer (NK) cells, and hCD45+hCD3+ T cells were analyzed. For myeloid development, hGPA+ erythroid cells, hCD41a+ mature platelets, and HLA-DR−hCD33+ granulocytes were analyzed.
Cytospin specimens were prepared using the imaging media MGK-S (Matsunami Glass, Kishiwada, Japan). Micrographs for cDCs and pDCs were obtained with a BX51 microscope (Olympus, Tokyo, Japan; oil objective lens 100×/1.35 NA), using a charge-coupled device camera (ProgRes C3; JENOPTIK, Jena, Germany). Image-acquisition software was ProgRes PlugIn for PCI version 5.0 (JENOPTIK), and image-processing software was Adobe Photoshop 7.0 (Adobe, San Jose, CA).
Quantitative real-time PCR
Total RNA was extracted from sorted human cDCs and pDCs using Isogen reagent (Nippon Gene, Tokyo, Japan) and treated with DNase I (Invitrogen, Carlsbad, CA) to remove contaminating genomic DNA. First-strand cDNA was synthesized using a QuantiTect reverse transcription kit (Qiagen, Hilden, Germany). Quantitative real-time polymerase chain reaction (PCR) was carried out in the ABI Prism 7700 Sequence Detector (Applied Biosystems, Foster City, CA). Reactions were performed in triplicate wells in 96-well plates. TaqMan target mixes for IFNA2, IFNB1, FLT3, ID2, PTCRA, NOTCH1, SPII, SPIB, TLR2, TLR4, TLR7, TLR9, and IL3RA were purchased from Applied Biosystems. Human β-actin mRNA was separately amplified in the same plate to be used as an internal control for variances in the amount of cDNA in PCR. Collected data were analyzed with the Sequence Detector software v.1.2 (Applied Biosystems). Relative expression levels were represented as a fold change of mRNA levels in pDCs relative to those in cDCs.
Microarray analysis
Complementary RNA (cRNA) with biotin16-UTP (Roche Diagnostics, Basel, Switzerland) for hybridization with Illumina Sentrix arrays was prepared using the Illumina TotalPrep RNA amplification kit (Ambion, Austin, TX). Briefly, reverse transcription to synthesize first-strand cDNA was primed with the T7 oligo (dT) primer to synthesize cDNA containing a T7 promoter sequence. Single-stranded cDNA was then incubated with DNA polymerase and RNase H to simultaneously degrade the RNA and synthesize second-strand cDNA. After purification, double-stranded cDNA was subject to in vitro transcription to synthesize multiple copies of biotinylated cRNA. Amplified cRNA was purified and verified by a spectrophotometer and an agarose-gel electrophoresis. Subsequently, cRNA (1.5 μg) was added to hybridization mix (HybE1 and formamide), applied onto Illumina BeadChip 6 × 2 (Illumina, San Diego, CA), and incubated at 55°C for 20 hours. After hybridization, the BeadChip was washed with E1BC solution and 100% ethanol, blocked with Block E1 buffer, and immobilized with Cy3-streptavidin (GE Healthcare, Slough, United Kingdom). After washing, the hybridized BeadChip was scanned using an Illumina BeadArray reader, which converts fluorescent signal intensity to a numeric value.
Microarray data analysis
An Illumina beads station was used as the first tool for data analysis. This software (Illumina Bead Studio software v.1.5.0.34) allows for calculation of the signal intensity and determination of whether each probe set is present. The signal intensity was then normalized both per chip and per gene using GeneSpring software v.7.2 (Agilent Technologies, Palo Alto, CA). Three separate experiments were implemented to enhance the reliability of the gene-expression data. The Pearson correlation was calculated using Microsoft Excel software v.11.3.3 (Microsoft, Redmond, WA). The genes were grouped according to their functional characteristics after analysis through the Gene Ontology database.30 K-means clustering was performed to identify expression patterns in both cDCs and pDCs, and the identified clusters were further evaluated with filtering procedures. The TIGR Mev software v.3.1 (The Institute for Genomic Research, Rockville, MD), which exhibits color-coding changes in gene expression, was used to make a heat map of expression data to compare expression patterns of specific genes. The differentially expressed genes between cDCs and pDCs were defined as a greater than 3-fold change in the mean gene expression between 2 groups and converted to “expression ratio” by dividing each value by the mean signal value of that gene in the control group.
Results
Human HSCs can generate cDCs and pDCs in NOD-scid/IL2rγnull mice
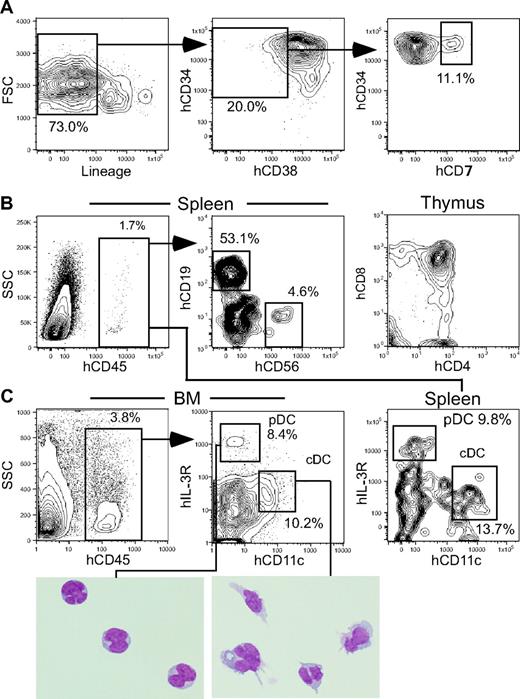
We first tested whether human HSCs can develop cDCs and pDCs together with other blood cells in our xenogeneic transplantation system.26 Sublethally irradiated NOD-scid/IL2rγnull newborns26 received 104 intravenously injected cells of the Lin−hCD34+hCD38− cord blood fraction (Figure 1A), which is enriched for human long-term HSCs.31 Six months after transplantation, the chimerism of human cells represented by the percentage of hCD45+ cells was consistently greater than 50% in recipients' bone marrow. In addition to myeloerythroid and lymphoid cells, 2 distinct human dendritic cell subsets, hCD11c+hIL-3Rαlo cDCs and hCD11c−hIL-3Rαhi pDCs,5,12 were detected in recipients' bone marrow, spleen, and blood (Figure 1B). The bone marrow of NOD-scid/IL2rγnull recipients contained hCD34+hCD38− human HSCs and committed progenitors such as Lin−hCD34+hCD38+hIL-3RαlohCD45RA− CMPs32 and Lin−hCD34+hCD38−hCD7+ CLPs,33 recapitulating the stepwise human hematopoietic development (Figure 1C). HSC-derived cDCs and pDCs were phenotypically isolatable by fluorescence-activated cell sorting (FACS; Figure 1D). Purified pDCs but not cDCs had basal levels of IFNα2 and β1 mRNA, and treatment with unmethylated CpG oligonucleotides induced significant up-regulation of IFN mRNAs only in pDCs (Figure 1E), suggesting that the xenotransplantation system supports the functional maturation of at least the pDC subset.
Human cord blood Lin−hCD34+hCD38− HSCs give rise to DC subsets as well as other progenitors. (A) Purification of Lin−hCD34+hCD38− HSCs from the CD34+ cell–enriched cord blood. (B) cDC and pDC progeny in the bone marrow, spleen, and peripheral blood of NOD-scid/IL2rγnull recipients of human cord blood HSCs. (C) Human HSCs gave rise to myeloid and lymphoid progenitors in the recipients' bone marrow, recapitulating normal human hematopoietic development. (D) The phenotype of purified HSC-derived cDCs and pDCs by FACS. (E) Changes in quantity of IFN mRNA in purified cDCs and pDCs before and after CpG stimulation. Representative data of 2 independent experiments are shown. FSC indicates forward scatter; and BM, bone marrow. Error bars represent the SEM of triplicate cultures.
Human cord blood Lin−hCD34+hCD38− HSCs give rise to DC subsets as well as other progenitors. (A) Purification of Lin−hCD34+hCD38− HSCs from the CD34+ cell–enriched cord blood. (B) cDC and pDC progeny in the bone marrow, spleen, and peripheral blood of NOD-scid/IL2rγnull recipients of human cord blood HSCs. (C) Human HSCs gave rise to myeloid and lymphoid progenitors in the recipients' bone marrow, recapitulating normal human hematopoietic development. (D) The phenotype of purified HSC-derived cDCs and pDCs by FACS. (E) Changes in quantity of IFN mRNA in purified cDCs and pDCs before and after CpG stimulation. Representative data of 2 independent experiments are shown. FSC indicates forward scatter; and BM, bone marrow. Error bars represent the SEM of triplicate cultures.
Human myeloid and lymphoid progenitors can give rise to both cDCs and pDCs
We wished to evaluate whether myeloid or lymphoid pathways can contribute to DC pool formation in vivo. CMPs and CLPs were purified from 4 normal human cord blood samples, and 104 cells of each population were transplanted into sublethally irradiated NOD-scid/IL2rγnull newborns. Because the expansion potential of hematolymphoid progenitors is limited,34 percentages of donor chimerism in these mice were relatively low compared with those in human HSC recipients (Figure 1). DC progeny was detectable at 4 weeks after transplantation. At this time, reconstitution of donor-derived progenitors was mainly seen in the bone marrow and the spleen but not in the lymph nodes.
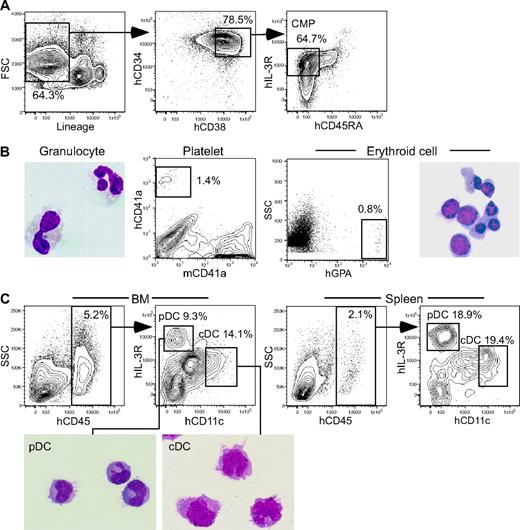
In all CMP recipients, percentages of hCD45+ cells were approximately 5% in the bone marrow and 2% in the spleen. Recipients of CMPs (Figure 2A) were reconstituted with hGPA+ erythroid cells, hCD41a+ platelets, and HLA-DR−CD33+ granulocytes (Figure 2B). In the bone marrow, both HLA-DR+hCD11c+hIL-3Rαlo cDC and HLA-DR+hCD11c−hIL-3Rαhi pDC populations were clearly seen (Figure 2C), which accounted for 13.1% and 11.1% of hCD45+ cells in average of 4 independent experiments, respectively. In the spleen, cDCs and pDCs constituted 18% and 13% of hCD45+ cells, respectively.
Human CMPs give rise to cDCs and pDCs in NOD-scid/IL2rγnull recipients.(A) Purification of human cord blood CMPs based on the expression of lineage antigens hCD34, hCD38, hIL-3Rα, and hCD45RA. (B) Myeloid progeny in the bone marrow 4 weeks after CMP xenotransplantation is shown. Mature hCD13+ granulocytes, hCD41a+ platelets, and hGPA+ erythroid cells were seen in the bone marrow. (C) CMP-derived cDCs and pDCs developed in both the bone marrow and the spleen of CMP recipients. Morphology of purified cDC and pDC progeny is also shown (May-Grünwald-Giemsa staining). See “Analysis of human cell engraftment” for complete image acquisition information. BM indicates bone marrow.
Human CMPs give rise to cDCs and pDCs in NOD-scid/IL2rγnull recipients.(A) Purification of human cord blood CMPs based on the expression of lineage antigens hCD34, hCD38, hIL-3Rα, and hCD45RA. (B) Myeloid progeny in the bone marrow 4 weeks after CMP xenotransplantation is shown. Mature hCD13+ granulocytes, hCD41a+ platelets, and hGPA+ erythroid cells were seen in the bone marrow. (C) CMP-derived cDCs and pDCs developed in both the bone marrow and the spleen of CMP recipients. Morphology of purified cDC and pDC progeny is also shown (May-Grünwald-Giemsa staining). See “Analysis of human cell engraftment” for complete image acquisition information. BM indicates bone marrow.
On the other hand, in all CLP recipients, percentages of hCD45+ cells were approximately 4% in the bone marrow and approximately 2% in the spleen at 4 weeks after transplantation. Human CLPs (Figure 3A) differentiated into B and NK cells in the spleen, whereas T cells were found almost exclusively in the thymus at this time point (Figure 3B). In the bone marrow, cDCs and pDCs constituted 10.7% and 11.2% of hCD45+ cells in average of 4 independent experiments, respectively (Figure 3C). In the spleen, cDCs and pDCs accounted for 13.8% and 9.0% of hCD45+ cells, respectively. Measurable levels of DCs were not found in the thymus of recipients of CMPs or CLPs.
Human CLPs give rise to cDCs and pDCs in NOD-scid/IL2rγnull recipients. (A) Purification of human cord blood CLPs based on the expression of lineage antigens hCD34, hCD38, and hCD7. (B) The analysis of recipients 4 weeks after CLP xenotransplantation. B- and NK-cell progeny and T-cell progeny were found in the spleen and the thymus, respectively. (C) hCD45+ human CLP progeny included cDCs and pDCs in both the bone marrow and the spleen. Morphology of purified cDC and pDC progeny is also shown (May-Grünwald-Giemsa staining). The pDC images presented in 3 separate boxes were taken from a single slide. See “Analysis of human cell engraftment” for complete image acquisition information. BM indicates bone marrow.
Human CLPs give rise to cDCs and pDCs in NOD-scid/IL2rγnull recipients. (A) Purification of human cord blood CLPs based on the expression of lineage antigens hCD34, hCD38, and hCD7. (B) The analysis of recipients 4 weeks after CLP xenotransplantation. B- and NK-cell progeny and T-cell progeny were found in the spleen and the thymus, respectively. (C) hCD45+ human CLP progeny included cDCs and pDCs in both the bone marrow and the spleen. Morphology of purified cDC and pDC progeny is also shown (May-Grünwald-Giemsa staining). The pDC images presented in 3 separate boxes were taken from a single slide. See “Analysis of human cell engraftment” for complete image acquisition information. BM indicates bone marrow.
Each DC subset displays similar expression profiles of representative surface antigens and genes, regardless of their lineage origin
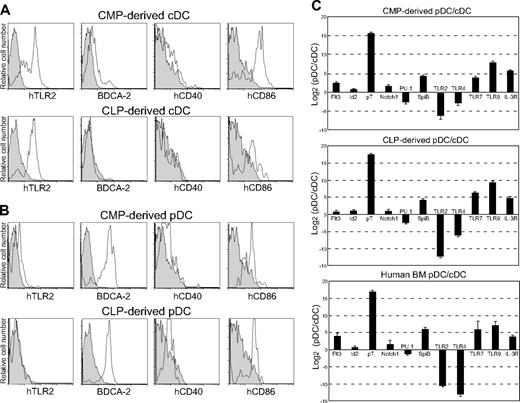
We then evaluated the expression patterns of DC-related antigens in these DC subsets generated from either the myeloid or the lymphoid lineage in vivo. DCs express various costimulatory molecules such as hCD40, hCD80, hCD83, and hCD86 together with MHC antigens, all of which are crucial for efficient activation of T cells.1 As shown in Figure 4A-B, both cDCs and pDCs expressed hCD86 but not hCD40, hCD80 (data not shown), or hCD83 (data not shown), suggesting an immature phenotype of these DCs. BDCA-2, a marker specific for human pDCs,35 was expressed only in pDCs, whereas TLR2 was expressed in cDCs but not in pDCs.10 The expression patterns of these molecules were well preserved in both DC subsets regardless of their lineage origin.
We further tested the level of DC-related transcripts in cDCs and pDCs by real-time PCR assays. Figure 4C shows the relative mRNA levels of representative genes between cDCs and pDCs purified from bone marrow of healthy human volunteers and of mice reconstituted with human CMPs or CLPs. Genes tested included TLRs (TLR2, TLR4, TLR7, TLR9), transcription factors (SPIB, SPII, ID2, and NOTCH1) and PTCRA.
Characterization of human CMP- and CLP-derived DC subsets. Flow-cytometric analysis of surface DC–related antigens in human CMP-derived and human CLP-derived cDCs (A) and pDCs (B). The gray area represents each isotype control. Note that the expression patterns of each DC-related antigen are almost identical in cDCs and pDCs, irrespective of their CMP or CLP origin. (C) The relative expression levels of RNAs isolated from cDCs and pDCs in the bone marrow of mice that received CMPs (top) and CLPs (middle) and in normal human bone marrow (bottom). Representative data from 3 independent experiments using real-time PCR analyses are shown. Error bars indicate the SEM of triplicate cultures.
Characterization of human CMP- and CLP-derived DC subsets. Flow-cytometric analysis of surface DC–related antigens in human CMP-derived and human CLP-derived cDCs (A) and pDCs (B). The gray area represents each isotype control. Note that the expression patterns of each DC-related antigen are almost identical in cDCs and pDCs, irrespective of their CMP or CLP origin. (C) The relative expression levels of RNAs isolated from cDCs and pDCs in the bone marrow of mice that received CMPs (top) and CLPs (middle) and in normal human bone marrow (bottom). Representative data from 3 independent experiments using real-time PCR analyses are shown. Error bars indicate the SEM of triplicate cultures.
Consistent with the FACS data (Figure 4A-B), TLR2 mRNA was expressed at a higher level in cDCs. TLR4 was also expressed strongly in cDCs. pDCs are known to selectively express TLR7 and TLR9, which are normally expressed in the cellular endosomes.10,36 SpiB, an Ets family member, is a key regulator of pDC development because SpiB knockdown in human CD34+ progenitor cells compromises their development into pDCs.37 PU.1, another Ets family transcription factor, is required for development of cDC and CD14+ myeloid cells.38 In mice reconstituted with either CMPs or CLPs, SpiB and PU.1 are expressed at a higher level in pDCs and cDCs, respectively (Figure 4C). Lymphoid-related Notch1 and pTα were expressed in pDCs irrespective of their origin. It should be noted that the expression patterns of these genes in DC subsets generated in the NOD-scid/IL2rγnull xenotransplantation system were consistently observed in DC subsets purified directly from human bone marrow (Figure 4C).
Taken together, these findings suggest that DC subsets exhibit similar phenotypic and transcriptional characteristics regardless of their CMP or CLP origin. In addition, the similar gene-expression pattern in human DC subsets developing in the NOD-scid/IL2rγnull and the human bone marrow suggests that NOD-scid/IL2rγnull bone marrow provides the proper microenvironment that allows human progenitors to recapitulate normal differentiation and maturation into the DC lineage.
Each DC subset possesses an identical transcriptional profile irrespective of its lineage origin, even at the genome-wide level
The fact that each DC subset possessed similar expression patterns of representative genes related to DC functions urged us to overview their mRNA expression at the genome-wide level by using a cDNA microarray. Cells (104) of each DC subset purified from mice reconstituted with human CMPs or CLPs were subjected to this analysis.
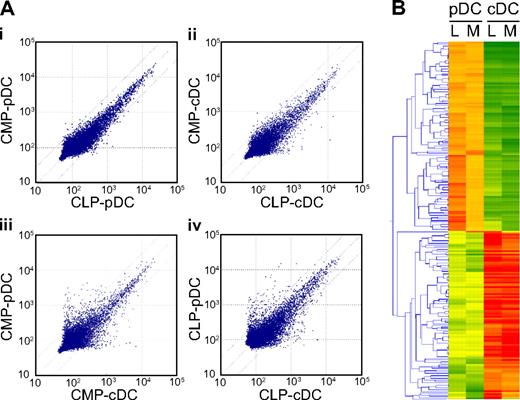
Figure 5A shows scatter plots of the signal intensity of 48 000 transcripts. Signal intensity for each gene was scanned, normalized, and plotted to evaluate the correlation between the 2 groups. Strikingly, the difference in signal intensity of each gene was marginal between CMP- and CLP-derived pDCs (Figure 5Ai) and between CMP- and CLP-derived cDCs (Figure 5Aii) (the correlation coefficient = 0.98 and 0.95, respectively). In contrast, the same analysis between CMP-derived cDCs and pDCs (Figure 5Aiii) or between CLP-derived cDCs and pDCs (iv) showed that a substantial number of genes exhibited dissociated signal intensities (r = 0.917 and 0.925, respectively). These data again suggest that the lineage origin does not affect the transcriptional profile of each DC subset.
A genome-wide representation of DC subsets of myeloid or lymphoid origin by microarray analysis. (A) Scatter plots of signal intensity of 48 000 different transcripts measured on an Illumina beads chip microarray. Data are presented as CMP- versus CLP-derived pDCs (i), CMP- versus CLP-derived cDCs (ii), CMP-derived cDCs versus pDCs (iii), and CLP-derived cDCs versus pDCs (iv). Representative data of 3 independent experiments are shown. (B) The heat map representation of 218 genes that displayed more than 3-fold differences between cDCs and pDCs of each lineage origin. The expression pattern of these genes is well preserved in each DC subset irrespective of its CMP or CLP origin. Representative data of 3 independent experiments are shown. L and M denote lymphoid and myeloid origin, respectively.
A genome-wide representation of DC subsets of myeloid or lymphoid origin by microarray analysis. (A) Scatter plots of signal intensity of 48 000 different transcripts measured on an Illumina beads chip microarray. Data are presented as CMP- versus CLP-derived pDCs (i), CMP- versus CLP-derived cDCs (ii), CMP-derived cDCs versus pDCs (iii), and CLP-derived cDCs versus pDCs (iv). Representative data of 3 independent experiments are shown. (B) The heat map representation of 218 genes that displayed more than 3-fold differences between cDCs and pDCs of each lineage origin. The expression pattern of these genes is well preserved in each DC subset irrespective of its CMP or CLP origin. Representative data of 3 independent experiments are shown. L and M denote lymphoid and myeloid origin, respectively.
We then focused the analysis on a subset of 218 genes that displayed more than 3-fold differences between cDCs and pDCs of each lineage origin. Figure 5B shows the heat map representation of these genes lined up by the K-means clustering method. The differential expression patterns of such genes in each DC subset were generally preserved regardless of CMP or CLP origin.
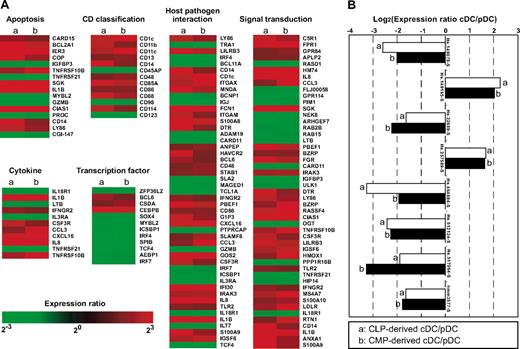
The list of the 218 genes included a number of functional ones relevant to apoptosis, CD classification, transcription factors, cytokines, host-pathogen interaction, and signal transduction (Figure 6A). All of these genes displayed almost identical expression patterns irrespective of their lineage origin. Of note, this list includes CD11c, CD123 (IL-3Rα), CD86, and TLR2 genes. Both CD11c and CD123 proteins are expressed at a high level in human cDCs and pDCs, respectively (Figures 1-3). Pronounced expression of CD86 and TLR2 mRNA in cDCs is consistent with the pattern of the protein expression of these molecules (Figure 4). These data verify that the protein levels of at least these molecules correlate with their mRNA level measured by microarray analysis.
Genes of different expression patterns between each DC subset of myeloid or lymphoid origin. (A) The representative list of 218 genes that displayed significantly different (> 3-fold) expression levels between cDC and pDC subsets. Genes are categorized into groups of genes relevant to apoptosis, CD classification, transcriptional regulation, cytokines, host-pathogen interaction, and signal transduction. All of these functional genes displayed consistent expression patterns irrespective of their lineage origin. (B) List of unknown genes included in the 218 gene subset. Their expression patterns are also preserved in CMP- or CLP-derived DC subsets. Representative data of 3 independent experiments are shown.
Genes of different expression patterns between each DC subset of myeloid or lymphoid origin. (A) The representative list of 218 genes that displayed significantly different (> 3-fold) expression levels between cDC and pDC subsets. Genes are categorized into groups of genes relevant to apoptosis, CD classification, transcriptional regulation, cytokines, host-pathogen interaction, and signal transduction. All of these functional genes displayed consistent expression patterns irrespective of their lineage origin. (B) List of unknown genes included in the 218 gene subset. Their expression patterns are also preserved in CMP- or CLP-derived DC subsets. Representative data of 3 independent experiments are shown.
In addition, the list included several unknown genes whose expression patterns were consistent between the myeloid and lymphoid DC subset (Figure 6B). These data suggest that the lineage origin does not affect the transcriptional signatures of cDCs and pDCs.
Discussion
Hematopoietic cells are commonly classified into 2 major lineages, myeloid and lymphoid, and each lineage contributes mainly to the innate and acquired immune systems, respectively. DCs are unique cell populations that play a central role in connecting each immune system to form a functional immune network.1 For example, DCs express specific types of TLRs to primarily respond to pathogens but also present foreign and self-antigens on their surface for lymphocytes to induce an immune reaction or self-tolerance. pDCs possess ectopic “lymphoid” markers, including pTα and RAG, and the rearrangement of the IgH gene, whereas both cDC and pDC subsets can be produced from lymphoid as well as myeloid progenitors, compromising their lineage definition.17 Thus, previous studies subclassified each DC subset into myeloid and lymphoid and have tried to find subclass-specific phenotypes and functions.17,20-23 Such analysis has been difficult, however, particularly in human hematopoiesis at least, due to lack of a reliable in vivo assay system.
In this paper, by using an efficient xenogeneic transplantation assay, we provide formal proof that both myeloid and lymphoid pathways can contribute to the formation of the DC pool in human hematopoiesis. In the NOD-scid/IL2rγnull system,26 human CMPs and CLPs transiently reconstitute myeloid and lymphoid cells, respectively, and displayed significant cell expansion sufficient to detect their DC progeny. We have shown that murine CMPs contribute toward generation of cDCs and pDCs in the bone marrow approximately 2-fold more than murine CLPs do, per-cell basis, after transplantation.17 We, however, did not see a statistically significant difference of DC reconstitution potential between human CMPs and CLPs in our experiments using 4 different human cord blood samples. Since the expansion of human progenitor populations is limited in our experiments, xenogeneic transplantation using a higher number of human CMPs and CLPs should be performed in a future study to precisely evaluate relative contribution of myeloid and lymphoid pathways toward DC pool formation.
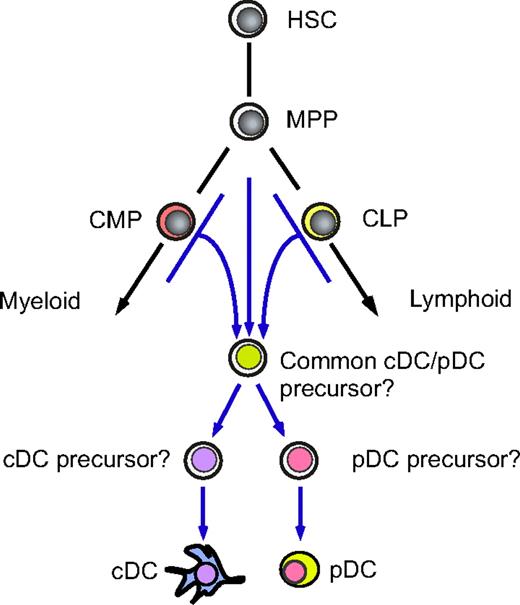
To our surprise, however, an extensive analysis of the phenotype and gene expression in pDCs and cDCs of either CMP or CLP origin revealed that the transcriptional profiles are well preserved regardless of their origin, suggesting that a common DC developmental program is operative even after initiation of myeloid or lymphoid differentiation programs. Importantly, the pattern of representative gene expression in human DC subsets obtained in this xenogeneic system was quite similar to that of primary DC subsets purified from normal, steady-state human bone marrow (Figure 4C). To test the functional maturation of human cDCs generated in our system, we purified CMP- or CLP-derived cDCs and cultured with allogeneic T cells from the blood in adults in vitro for 7 days. In either case, allogeneic T cells displayed a significant proliferation estimated by 3H-thymidine incorporation39 only in the presence of donor-derived cDCs (data not shown), suggesting that both myeloid and lymphoid cDCs developed in the xenogeneic system are capable of presenting alloantigens to T cells. Together, these data suggest that the NOD-scid/IL2rγnull bone marrow provides the proper microenvironment for human progenitors to recapitulate human DC development machinery in the bone marrow. Collectively, human myeloid and lymphoid pathways might naturally give rise to equivalent DC subsets in vivo. The proposed developmental path of DCs is schematized in Figure 7.
A proposed developmental scheme of the DC lineage cells. Human myeloid and lymphoid pathways generate indistinguishable cDC and pDC populations. These DC subsets may use common developmental programs that are independent of the conventional myeloid or lymphoid pathways. It is still unclear whether human cDCs and pDCs develop via the putative common cDC/pDC precursor stage.
A proposed developmental scheme of the DC lineage cells. Human myeloid and lymphoid pathways generate indistinguishable cDC and pDC populations. These DC subsets may use common developmental programs that are independent of the conventional myeloid or lymphoid pathways. It is still unclear whether human cDCs and pDCs develop via the putative common cDC/pDC precursor stage.
Previous papers have shown that the CMP, as a population, has minor B-cell potential.32,40 In fact, in our experiments, injection of 104 CMPs resulted in generation of a few percent of CD19+ B cells in the spleen (data not shown). Such B-cell potential is likely to be derived from rare pro-B cells sharing the phenotype with the CMP population but not from CLP contaminants because T- or NK-cell differentiation from CMPs has not been observed in mouse40 or human hematopoiesis.32 Furthermore, B-cell–committed precursors do not give rise to dendritic cells in mice or humans (unpublished data, F.I. and K.A., December 2006). DC development from the CMPs might occur in association with myelo-monocytic differentiation but not with minor B-cell development. Therefore at least the vast majority of human CMP-derived DCs in our experiments should be categorized into the myeloid lineage.
Although DCs were originally defined as myeloid cells, the alternative lymphoid origin for DCs was demonstrated in murine hematopoiesis by the fact that early thymocyte progenitors are capable of generating CD8α+ cDCs in vivo.41,42 This important finding was further strengthened by the fact that mice deficient for the myeloid transcription factors RelB43 or PU.144 lack CD8α− cDCs, whereas those deficient for “lymphoid” transcription factors Ikaros,45 Id2,46 or IRF-847 lack only CD8α+ cDC subsets. On the other hand, T-cell–deficient c-Kit−/−γc−/−48 or Notch1−/−49 mice possess CD8α+ cDCs, and both CD8α+ and CD8α− cDCs were produced from murine CLPs and CMPs with similar efficiency on a per-cell basis.33,50,51 Thus, results on murine DC ontogeny by using CD8α expression as a lymphoid marker are apparently controversial. Because CD8α+ and CD8α− cDCs could have different functions,4 CD8α expression may reflect a different maturation or activation stage of cDCs.52 The developmental relationship between cDCs and pDCs is also unclear. Circulating common CD11c+MHCII− cDC/pDC progenitors were previously identified in mice, though this population is not completely homogenous since circulating cells of this phenotype were found to include NK cells.53 Either murine CMP or CLP populations can generate the CD11c+MHCII− cDC/pDC progenitor population.17 cDC monopotent progenitors exist in the murine spleen,54 which may be downstream of the common cDC/pDC progenitor (Figure 7). We have reported that murine CMP- or CLP-derived DC subsets possessed similar expression patterns of proteins and genes related to DC development and function.17,50 These mouse data collectively suggest that myeloid and lymphoid DC pathways may merge into the common DC progenitor stage, forming single cDC and pDC pools irrespective of their lineage origin.
In human hematopoiesis, DC subclass markers such as CD8α or the common DC progenitor stage have not been identified, although at least a fraction of human CMPs and CLPs should contain progenitor cells that can clonally give rise to cDCs and pDCs.25 Our data demonstrate that human myeloid and lymphoid pathways can generate indistinguishable cDC and pDC populations, at least based on a wide array of transcriptional profiles, suggesting that these DC subsets use common developmental programs that are independent of the conventional myeloid or lymphoid pathways. The question is why such flexibility of developmental potential is maintained, especially in the DC lineage. An interesting hypothesis is that either myeloid or lymphoid progenitors contribute to efficient and immediate distribution of DCs in specified myeloid or lymphoid organs, respectively. Myeloid progenitors may preferentially contribute to DC development in the bone marrow, whereas lymphoid progenitors may do so in lymphoid organs such as the lymph node and the thymus. Thus, the flexible DC developmental potential may have an advantage to efficiently replenish DC subsets or their precursors into both myeloid and lymphoid organs throughout body. In fact, by congenic mouse transplantation assays, contribution of lymphoid progenitors to formation of the peripheral DC pool was approximately 50% in the thymus, whereas only approximately 10% occurred in the bone marrow and the spleen.17,50 Unfortunately, the xenotransplantation system is not suitable for such analysis for human DCs, since the homing machinery of human progenitors may not properly operate in xenogeneic hosts. For example, after transplantation of bone marrow cells, murine multipotent progenitors immediately immigrate into the spleen to form spleen colony-forming units (CFU-Ss), whereas human bone marrow cells do not form CFU-Ss after transplantation (unpublished data, F.I. and K.A., October 2006). This is probably because the spleen is the major hematopoietic organ in mice but not in humans.
In summary, the NOD-scid/IL2rγnull system provides the proper microenvironment for human DC subsets as well as other blood cells to develop from early hematopoietic stem and progenitor cells in vivo. Human myeloid and lymphoid progenitors can give rise to DC subsets with identical transcriptional profiles in this system, suggesting that both “myeloid” and “lymphoid” DCs use a common developmental machinery, which is normally triggered even after activation of myeloid or lymphoid differentiation programs. Thus, DCs might constitute an independent cell lineage that cannot be appropriately categorized into the conventional lymphoid or myeloid lineages. More detailed studies on human DC development and function, including the identification of a putative human cDC/pDC progenitor stage, should be performed to understand the unique developmental program and pathway in the human DC lineage.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work is supported by grants from the Ministry of Education, Culture, Sports, Science, and Technology of Japan (F.I., H.N., K.A.); the National Institutes of Health (L.D.S., K.A.); and the Juvenile Diabetes Research Foundation (L.D.S.).
The authors are grateful to Dr Dennise-Dalma Weiszhausz for critical reading of this manuscript.
National Institutes of Health
Authorship
Contribution: F.I. and H.N. designed and performed experiments and wrote the manuscript. T.I., S.Y., N.S., S.O., T.M., H.M., and S.F. performed experiments. L.D.S. and M.H. contributed to discussion. K.A. designed experiments and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Koichi Akashi, Center for Cellular and Molecular Medicine, Kyushu University Hospital, 3–1-1 Maidashi, Higashi-ku, Fukuoka 812-8582, Japan; e-mail:akashi@cancer.med.kyushu-u.ac.jp.