Abstract
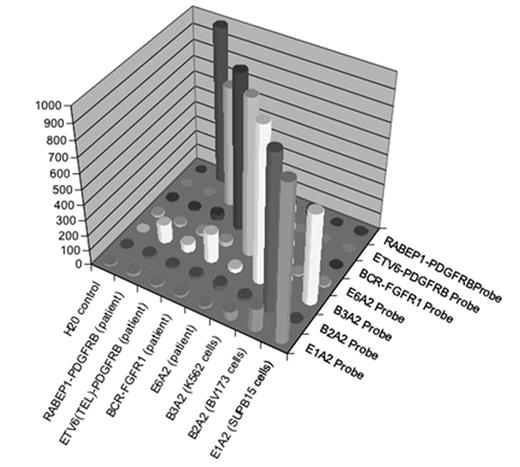
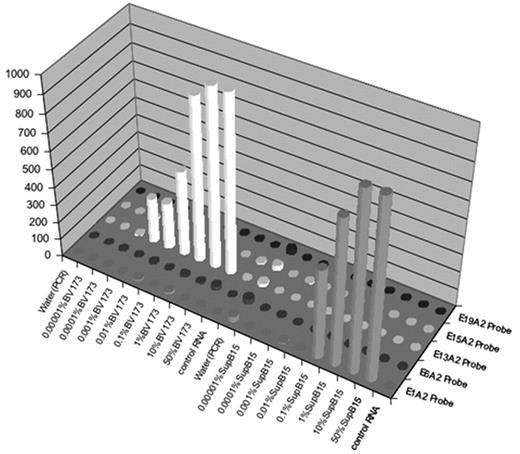
Chronic Myeloproliferative disorders (CMPDs) are malignancies characterized by excessive proliferation of one or more of the myeloid lineages driven by constitutively activated tyrosine kinases (CATKs). CATKs are increasingly serving as targets for cancer-specific therapy and newer and more potent TK inhibitors are becoming available for clinical use. CATKs in CMPDs may result from karyotypically evident or cryptic chromosomal translocations leading to TK gene fusions or from karyotypically silent gain-of-function point mutations. More than ten CATK genes have been shown to result in CMPDs by imparting a demonstrable positive growth drive. CATK inhibitors important in the treatment of CMPDs have different inhibitory profiles, making it critical to identify the specific mutation involved in patients receiving targeted therapy. Of these TK genes, ABL, PDGFRB, PDGFRA, FGFR1, ETV6, JAK2 and MPL are the most frequently involved. Since most of these TK genes can participate in chromosome translocations with a large number of partner genes, the resulting number of possible CATKs is rather large, making detection and identification a challenging task. To date, two classic (B2A2 and B3A2) and seven less common fusions of the ABL and BCR genes have been described. In addition, over twenty non-BCR-ABL mutations have been shown to have pathogenetic relevance in CMPDs. We have developed a simple, high-throughput assay with a fast turnaround time that solves the complex problem of detecting and identifying multiple translocations and point mutations. Two steps are involved in this expandable assay. The first is a multiplex RT-PCR reaction that amplifies fusion transcripts and point mutations described in CMPDs from peripheral blood or bone marrow. This PCR reaction employs internal specific primers targeted to each of eight BCR-ABL mutations (E1A2, E1A3, B2A2, B3A2, E6A2, E8A2, E15A2, E19A2) eight non-BCR-ABL mutations (FIP1L1-PDGFRA, BCR-PDGFRA, BCR-FGFR1, MYO18A-FGFR1, CDK5RAP2-PDGFRA, ETV6-PDGFRB, RABAPTIN-5-PDGFRB, H4-PDGFRB) and two gain of function mutations (JAK2V617F and MPLW515L) and a biotinylated common primer set to increase amplification efficiency. The second is a liquid bead microarray (Luminex) which uses optically encoded beads to detect probes specific for multiple known translocations in a single tube. To date, we have selectively detected four BCR-ABL and three non-BCR-ABL mutations from cloned patient samples and the cell lines SupB15 (E1A2 p190 BCR-ABL fusion), K562 (B3A2 p210 BCR-ABL fusion) and BV173 (B2A2 p210 BCR-ABL fusion). Dilution studies using these cell lines demonstrate detection down to between 1/1000 and 1/100000 cell equivalents without significant cross-talk.
Author notes
Disclosure: No relevant conflicts of interest to declare.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal