Abstract
Despite progress in developing defined conditions for human embryonic stem cell (hESC) cultures, little is known about the cell-surface receptors that are activated under conditions supportive of hESC self-renewal. A simultaneous interrogation of 42 receptor tyrosine kinases (RTKs) in hESCs following stimulation with mouse embryonic fibroblast (MEF) conditioned medium (CM) revealed rapid and prominent tyrosine phosphorylation of insulin receptor (IR) and insulin-like growth factor-1 receptor (IGF1R); less prominent tyrosine phosphorylation of epidermal growth factor receptor (EGFR) family members, including ERBB2 and ERBB3; and trace phosphorylation of fibroblast growth factor receptors. Intense IGF1R and IR phosphorylation occurred in the absence of MEF conditioning (NCM) and was attributable to high concentrations of insulin in the proprietary KnockOut Serum Replacer (KSR). Inhibition of IGF1R using a blocking antibody or lentivirus-delivered shRNA reduced hESC self-renewal and promoted differentiation, while disruption of ERBB2 signaling with the selective inhibitor AG825 severely inhibited hESC proliferation and promoted apoptosis. A simple defined medium containing an IGF1 analog, heregulin-1β (a ligand for ERBB2/ERBB3), fibroblast growth factor-2 (FGF2), and activin A supported long-term growth of multiple hESC lines. These studies identify previously unappreciated RTKs that support hESC proliferation and self-renewal, and provide a rationally designed medium for the growth and maintenance of pluripotent hESCs.
Introduction
In guiding human embryonic stem cell (hESC) technology toward the clinic, 1 key issue to be addressed is a lack of standardization in the culture and maintenance of hESCs. In the absence of mouse embryonic fibroblast (MEF) feeder layers, many researchers rely on “conditioning” in which medium is first exposed to MEFs to acquire soluble factors that support the propagation of undifferentiated hESCs in culture. It has been difficult to discern how MEF conditioning enables hESCs to maintain an undifferentiated state. Other common features of more recently developed hESC culture conditions include the presence of fibroblast growth factor-2 (FGF2), the absence of serum, and the presence of a serum substitute such as KnockOut Serum Replacer (KSR, proprietary formulation; Invitrogen, Carlsbad, CA).1-3 Other factors suggested to play a role in supporting the maintenance of hESCs include transforming growth factor β1 (TGFβ1),4 activin A (ActA),5,6 platelet-derived growth factor (PDGF) and sphingosine-1-phosphate,7 BIO, a small-molecule inhibitor of GSK3β,8 and neurotrophins.9 Several defined medium systems have been described for hESCs and are based upon FGF2 in combination with nodal,10 TGFβ1, GABA, and pipecolic acid, plus lithium chloride,11 Wnt3a plus April/BAFF,12 or the N2/B27 supplements.13
Although these studies have focused on identifying growth factors and conditions that support the proliferation of undifferentiated hESCs, little is known about the cell-surface receptors that are activated when hESCs are exposed to conditions favorable for self-renewal. A number of receptor tyrosine kinases (RTKs) are expressed at high levels on hESCs,14 including insulin-like growth factor-1 receptor (IGF1R), fibroblast growth factor receptor (FGFR1), and EPHA1, as well as ERBB2 and ERBB3 (which are members of the epidermal growth factor receptor [EGFR] family), while expression of FGFR2 (EGFR) FGFR4, vascular endothelial growth factor receptor-2 (VEGFR2), IGFR2, KIT, and RET has also been detected.15,16 RTKs are likely to be central signaling effectors17 that influence survival, apoptosis, proliferation, or differentiation decisions in pluripotent cells. To determine if any of these RTKs are involved in self-renewal, we simultaneously interrogated the tyrosine phosphorylation status of 42 RTKs in hESCs grown in MEF–conditioned medium (CM) and developed a defined medium for hESC culture.
Materials and methods
Cell culture
The National Institutes of Health (NIH)–registered H1, BG01, BG02, and BG03 hESC lines, as well as CyT49, an hESC line isolated using human feeder cells under good manufacturing process (GMP) conditions (Novocell, San Diego, CA), were used in these experiments. Nonconditioned medium (NCM) and CM were prepared as described previously,2 and hESCs were maintained in CM on plates coated with Matrigel (BD Biosciences, San Jose, CA) diluted 1:30, or in NCM on MEF feeder layers where indicated. Cultures were routinely passaged with collagenase IV2 or dispase.3 Karyotype analyses of hESCs grown in CM or defined conditions (DC) HAIF (Heregulin-1B, Activin A, Insulin like growth factor-1, FGF2) were performed using standard G-banding techniques.
Culture of hESCs in defined conditions
DC-HAIF consisted of DMEM/F12 (Invitrogen), 2% fatty acid-free Cohn fraction V bovine serum albumin (BSA; Serologicals, Norcross, GA), 1 × nonessential amino acids, 50 U/mL penicillin, 50 μg/mL streptomycin, 50 μg/mL ascorbic acid, 10 μg/mL bovine or human transferrin, 0.1 mM β-mercaptoethanol (all from Invitrogen), 1 times trace elements A, B, and C (Mediatech, Manassas, VA), 10 ng/mL HRG1β (Peprotech, Rocky Hill, NJ), 10 ng/mL ActA (R&D Systems, Minneapolis, MN), 200 ng/mL LR3-IGF1 (JRH Biosciences, Lenexa, KS), and 8 ng/mL FGF2 (Sigma, St Louis, MO) or R&D Systems). HESCs were cultured in DC-HAIF on growth factor–depleted Matrigel (BD Biosciences) diluted 1:200. Near-confluent hESC cultures were passaged by treating with 10 mg/mL (approximately 2000 U/mL) collagenase IV for 3 minutes, washing with 0.2% BSA in DMEM/F12, and gently scraping to lift and break up the colonies. Colony pieces were centrifuged at 200g, aspirated, gently resuspended in DC-HAIF, and typically split 1:3 to new plates. The formulation of DC-HAIF is shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article); a detailed protocol for the maintenance of hESCs in DC-HAIF is available in Document S1. Cell- and colony-counting experiments, the generation and analysis of teratomas, embryoid bodies, and defined differentiation to endodermal lineages are detailed in Document S1.
Human RTK arrays
Proteome Profiler human phospho-RTK antibody arrays (R&D Systems) were used according to the manufacturer's instructions (Figure S1; Document S1). A total of 500 μg fresh protein lysates were incubated overnight with nitrocellulose membranes dotted with duplicate spots for 42 anti-RTK and control antibodies. Bound phospho-RTKs were detected with a pan antiphosphotyrosine antibody conjugated to horseradish peroxidase using chemiluminescence.
Receptor inhibition studies
For experiments using the IGF1R blocking antibody, H1 hESCs were incubated with 10 μg/mL A12 (ImClone, New York, NY) or control human IgG (Sigma) in CM. IGF1R expression was evaluated by flow cytometry after 4 hours of incubation with A12 or control antibody. Colonies were counted and graded morphologically on the indicated days (Document S1; Figure S2). Both differentiation and apoptosis were evaluated 3 days after the addition of A12. Morphologic assessment of differentiation was confirmed by SSEA3 staining using flow cytometry, and apoptosis was assessed by TUNEL staining. Inhibition of ERBB2 with AG825 (Calbiochem, San Diego, CA) used hESCs growing in CM on Matrigel diluted 1:30 (Document S1). Cell-cycle and apoptosis assays were performed after 4 days of culture in CM plus either AG825 or DMSO (control).
Flow cytometry, RT-PCR analysis, and immunofluorescence
Flow cytometry analyses or sorting were performed using a FACScan, FACSCanto, or FACS Aria cell sorter (BD Biosciences). The FlowTACS kit (R&D Systems) was used to detect apoptotic cells, according to the manufacturer's instructions. Antibodies used to detect cell-surface antigens for flow and other immunofluorescence analyses are described in Document S1. Cell-cycle and DNA content analysis was performed using propidium iodide staining and analysis with the Modfit 3.0 software (Verity House Software, Topsham, ME). Immunostaining of cell cultures used secondary antibodies conjugated with Alexa-488 (green) or -594 (red) (Invitrogen). Nuclei were counterstained with DAPI (Sigma). Stained slides were mounted with Aqua Poly/Mount (Polysciences, Warrington, PA) and images were captured using Nikon TE2000-S and -E inverted microscopes (Nikon, Tokyo, Japan), Q-Imaging Retiga cameras (Qimaging, Surrey, BC), and the Q-Capture imaging system. Histological sections were stained with hematoxylin and eosin (Sigma), or with primary antibody followed by detection with alkaline phosphatase tagged secondary and counterstaining with hematoxylin. Slides were mounted with Permount (Biomedia, Foster City, CA), and images were captured with an Olympus BX41 light microscope (Olympus, Center Valley, PA), an Olympus DP70 digital camera, and Image Pro Plus 6.0 software (MediaCybernetics, Bethesda, MD). Images were processed with Adobe Photoshop 6.0 (Adobe, San Jose, CA). Methods for RNA isolation, reverse transcription–polymerase chain reaction (RT-PCR) analyses, and primer sets are described in Document S1.
ShRNA inhibition of IGF1R
Design and construction of the IGF1R-targeting shRNA lentivirus vector, plasmid transfection, generation of viral supernatants, evaluation of titer, and transduction of H1 hESCs are all described in Document S1. Transduced cells were cultured for 22 days and evaluated periodically for green fluorescent protein (GFP) and IGF1R expression by flow cytometry.
Illumina bead array and transcriptional analysis
Methods for the preparation of samples, labeling, hybridization, washing, detection, and analysis have been described previously,18 and were carried out according to the manufacturer's instructions (Document S1). Analysis was performed using parallel approaches described for other hESC samples18 with Illumina Sentrix Human-6 Expression Beadchips containing 47 296 transcript probes (Illumina, San Diego, CA). NIH gene expression omnibus (GEO) accession number-GSE6645.
Results
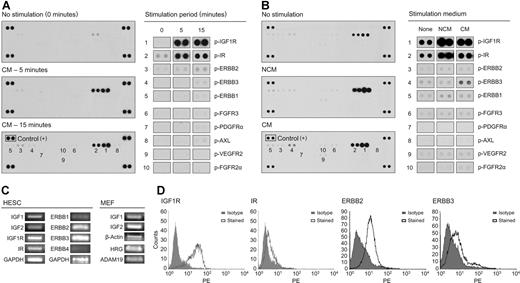
The phosphorylation status of 42 RTKs was simultaneously interrogated in hESCs using a membrane array of primary antibodies and detection with a pan-antiphosphotyrosine antibody (Figure S1). H1 cells cultured on Matrigel were starved overnight with DMEM/F12 and 0.5% BSA and then stimulated with CM for 5 or 15 minutes. Following overnight starvation, variable faint or moderate phosphorylation of several receptors, including IR, IGF1R, ERBB1 (EGFR), ERBB2, ERBB3, VEGFR2, and FGFR3, was detected (Figure 1A,B). Striking induction of IGF1R and IR phosphorylation occurred within 5 minutes of stimulation with CM, and persisted at 15 minutes (Figure 1A). Similarly, phosphorylation of ERBB2 increased at 5 minutes, and rose further at 15 minutes. ERBB1 and ERBB3 were also phosphorylated, but to a lesser extent, at 15 minutes. CM stimulation also elicited faint but consistent phosphorylation of FGFR3. A handful of receptors, including Axl, VEGFR2, PDGFRα, EPHA2, FGFR2α, MER, and Tie2 were phosphorylated faintly and inconsistently in response to CM. Of note, phosphorylated derivatives of FGFR1, and the neurotrophin receptors TRKB and TRKC, were not detected following stimulation with CM.
CM triggers IGF1R/IR and ERBB-family tyrosine phosphorylation in hESCs. (A) RTK array analysis of H1 cells stimulated by CM for 5 or 15 minutes. Tyrosine phosphorylation of IGF1R/IR and lower-intensity phosphorylation of ERBB (EGFR) family members was observed consistently. (B) RTK analysis of H1 cells stimulated by NCM and CM for 15 minutes. Increased ERBB3 phosphorylation was observed with CM stimulation. (C) RT-PCR analysis of IGF1, IGF2, IGF1R, and IR expression in H1 cells, and IGF1 and IGF2 expression in MEFs; ERBB1-4 expression in BG03 cells (and BG01 and BG02 cells; Figure S4); and HRG and ADAM19 expression in MEFs. Full-length gels are presented in Figure S4 online. (D) Flow cytometry of IGF1R, IR, ERBB2, and ERBB3 expression in H1 cells.
CM triggers IGF1R/IR and ERBB-family tyrosine phosphorylation in hESCs. (A) RTK array analysis of H1 cells stimulated by CM for 5 or 15 minutes. Tyrosine phosphorylation of IGF1R/IR and lower-intensity phosphorylation of ERBB (EGFR) family members was observed consistently. (B) RTK analysis of H1 cells stimulated by NCM and CM for 15 minutes. Increased ERBB3 phosphorylation was observed with CM stimulation. (C) RT-PCR analysis of IGF1, IGF2, IGF1R, and IR expression in H1 cells, and IGF1 and IGF2 expression in MEFs; ERBB1-4 expression in BG03 cells (and BG01 and BG02 cells; Figure S4); and HRG and ADAM19 expression in MEFs. Full-length gels are presented in Figure S4 online. (D) Flow cytometry of IGF1R, IR, ERBB2, and ERBB3 expression in H1 cells.
To discriminate between effects mediated by components of NCM and effects resulting from conditioning, receptor phosphorylation was compared after stimulating overnight-starved H1 cells with either NCM or CM. Both media triggered prominent phosphorylation of IR and IGF1R, which implied that a component of the base medium was responsible. This was attributable to insulin, which exceeds 10 μg/mL in 20% KSR (International Patent Publication WO 98/30679; Invitrogen), and at 10 μg/mL can activate IGF1R (Figure S3).19,20 While low-level phosphorylation of ERBB2, FGFR3, and VEGFR2 was also observed, the most apparent differences in the phospho-RTK profiles of hESCs stimulated with CM versus NCM was an increase in ERBB3 phosphorylation in response to CM in 1 experiment (Figure 1B), and elevated ERBB1 phosphorylation in another (data not shown). These results suggest that the elaboration of an EGFR family ligand(s) may be a functionally important consequence of MEF conditioning. Different EGF family members preferentially bind to and activate specific homo- or heterodimers of ERBB1 to ERBB4.21 While EGF and TGFα signal via homodimers of ERBB1, or heterodimers of ERBB1/2, activation of ERBB2/3 suggests the presence of neuregulins,22 of which heregulin-1β (HRG; also called neuregulin 1) is the best-characterized example.
The expression of IGF1R, IR, and ERBB receptors on hESCs and expression of their ligands in MEFs was examined. RT-PCR confirmed expression of transcripts for IR, IGF1R, ERBB2, and ERBB3 in hESCs, whereas ERBB1 appeared to be expressed at a lower level, and ERBB4 was undetectable (Figure 1C; Figure S4). Flow cytometry confirmed homogeneous cell-surface expression of IGF1R, IR, ERBB2, and ERBB3 on hESCs (Figure 1D). Coexpression of OCT4, SSEA4, or SSEA3 with either IGF1R or ERBB2 was demonstrated (Figure S5A,B). In a small percentage of spontaneously differentiated cells (< 2% SSEA4−, or OCT4− cells), the intensity of IGF1R or ERBB2 expression remained constant (Figure S5A,B). MEFs expressed transcripts for IGF1, IGF2, and HRG, as well as ADAM19 (Figure 1C; Figure S4), which is capable of proteolytic shedding of bioactive ERBB2/3 ligands.23 IGF1 and IGF2 transcripts were also detected in hESCs by RT-PCR (Figure 1C; Figure S4). Expression of components of the IGF1R/IR- and ERBB-signaling pathways, including receptors, certain ligands, and downstream processing and signaling molecules of the PI3 and MAP kinase pathways, was a common feature of 7 hESC lines (Table S2). The PI3 and MAP kinase pathways can also be activated by FGF receptor signaling.24 In addition, consistent expression of SMAD1 to SMAD7 suggests a role for ActA/TGFβ/BMP signaling in regulating hESCs4,6,10 (Table S2).
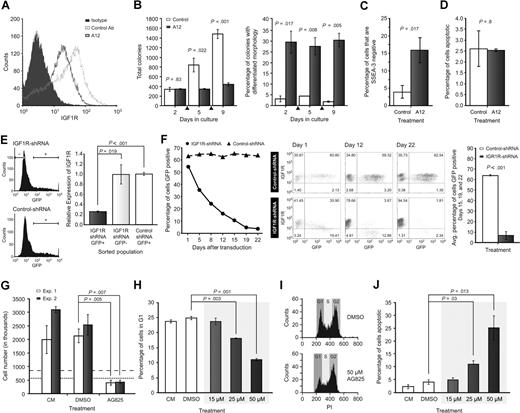
To investigate IGF1R signaling in hESC self-renewal, the effects of the IGF1R-blocking monoclonal antibody A1225 were tested. Treatment with A12 reduced IGF1R expression on the cell surface as described25 (Figure 2A). A12 blocked the expansion of hESC cultures and induced a significant rise in differentiation (Figure 2B,C; Figure S2). While treatment with A12 markedly inhibited the ability of hESCs to expand in culture, it did not induce apoptosis as determined by TUNEL assay (Figure 2D). The role of IGF1R was also examined in hESCs using lentiviral vectors expressing shRNAs (Document S1). A total of 2 lentiviral vectors containing a GFP reporter linked to either an IGF1R-targeted shRNA or a noninhibitory control shRNA were used. Transfection of the IGF1R-targeted vector caused a significant reduction in IGF1R mRNA in GFP+ H1 cells 48 hours after transfection compared with either the GFP− population, or GFP+ cells transfected with the control vector (Figure 2E). To examine the effects of IGF1R inhibition on long-term culture, H1 cells were transduced with either the IGF1R-targeted or control vector lentivirus, and the expression of both GFP and IGF1R was examined in transduced cultures over a 3-week period (Figure 2F). At 24 hours after transduction, both cultures contained a similar proportion of GFP+ cells. The activity of the IGF1R knockdown vector was confirmed by an inverse correlation between GFP intensity and IGF1R levels on the cell surface. In comparison, IGF1R expression remained constant over a range of GFP intensities in hESCs transduced with the control vector. The proportion of GFP+, GFP+/IGF1R+, GFP−/IGF1R+, and GFP−/IGF1R− cells in the control culture remained constant over the course of the experiment. Conversely, in cultures containing the IGF1R-targeted shRNA, the proportion of GFP+ cells fell progressively and significantly. These results indicate that inhibition of IGF1R with a shRNA led to a competitive disadvantage in transduced cells, confirming a requirement for IGF1R signaling in hESC survival and/or self-renewal.
Disruption of IGF1R and ERBB2 signaling inhibits hESC self-renewal. (A) Treatment of H1 cells with the A12 anti-IGF1R blocking antibody led to decreased IGF1R expression on the cell surface as measured by flow cytometry. (B) Colony counting showed that the A12 antibody inhibited H1 cell proliferation (left). Arrows indicate that cultures were passaged on day 3 and 6. Cumulative colony counts increased in the presence of control antibody but not in the presence of A12. A12 also induced hESC differentiation (right) as measured by colony morphology (also see Figure S2). Error bars here and in panels C-H,J are standard deviation (SD). Flow cytometry for (C) SSEA-3 expression confirmed A12 treatment caused increased differentiation, but (D) not an increase in apoptosis. (E) Significant reduction of IGF1R mRNA in the GFP+ population of cultures transfected with an IGF1R-targeted shRNA, compared with either the GFP- population, or GFP+ cells transfected with a control shRNA. (F) Decline in the percentage of GFP+ cells in H1 cultures (left) transduced with a lentiviral vector containing a shRNA targeting IGF1R, rather than a control shRNA. Middle panel shows flow cytometry profiles that indicate stable expression of IGF1R and percentages of cells that are GFP+ in control shRNA–transduced cultures on days 1, 12, and 22, while cultures transduced with the IGF1R-targeted shRNA vector exhibited a decline in IGF1R expression and a reduction in GFP+ and GFP+/IGF1R+ cells. Right panel shows the average percentage of GFP+ cells at days 15, 19, and 22 was significantly lower in the IGF1R-targeted shRNA transduced culture. (G) A total of 50 μM AG825 inhibited proliferation of BG02 hESCs growing in CM. Triplicate cell counts from 2 independent experiments are shown. … and - - - indicate pretreatment cell counts from experiments 1 and 2, respectively. (H,I) Increasing concentrations of AG825 caused a dose-dependent reduction in the proportion of H1 hESCs in the G1 phase of the cell cycle, and (J) a moderate dose-dependent rise in apoptosis.
Disruption of IGF1R and ERBB2 signaling inhibits hESC self-renewal. (A) Treatment of H1 cells with the A12 anti-IGF1R blocking antibody led to decreased IGF1R expression on the cell surface as measured by flow cytometry. (B) Colony counting showed that the A12 antibody inhibited H1 cell proliferation (left). Arrows indicate that cultures were passaged on day 3 and 6. Cumulative colony counts increased in the presence of control antibody but not in the presence of A12. A12 also induced hESC differentiation (right) as measured by colony morphology (also see Figure S2). Error bars here and in panels C-H,J are standard deviation (SD). Flow cytometry for (C) SSEA-3 expression confirmed A12 treatment caused increased differentiation, but (D) not an increase in apoptosis. (E) Significant reduction of IGF1R mRNA in the GFP+ population of cultures transfected with an IGF1R-targeted shRNA, compared with either the GFP- population, or GFP+ cells transfected with a control shRNA. (F) Decline in the percentage of GFP+ cells in H1 cultures (left) transduced with a lentiviral vector containing a shRNA targeting IGF1R, rather than a control shRNA. Middle panel shows flow cytometry profiles that indicate stable expression of IGF1R and percentages of cells that are GFP+ in control shRNA–transduced cultures on days 1, 12, and 22, while cultures transduced with the IGF1R-targeted shRNA vector exhibited a decline in IGF1R expression and a reduction in GFP+ and GFP+/IGF1R+ cells. Right panel shows the average percentage of GFP+ cells at days 15, 19, and 22 was significantly lower in the IGF1R-targeted shRNA transduced culture. (G) A total of 50 μM AG825 inhibited proliferation of BG02 hESCs growing in CM. Triplicate cell counts from 2 independent experiments are shown. … and - - - indicate pretreatment cell counts from experiments 1 and 2, respectively. (H,I) Increasing concentrations of AG825 caused a dose-dependent reduction in the proportion of H1 hESCs in the G1 phase of the cell cycle, and (J) a moderate dose-dependent rise in apoptosis.
A highly selective inhibitor of the ERBB2 tyrosine kinase, tyrphostin AG825,26 was used to investigate the role of ERBB2 in hESCs. AG825 significantly inhibited proliferation of hESCs growing in CM (Figure 2G). Cell-cycle analysis showed a decline in cells in G1 phase, suggesting a delay in the G2-M transition (Figure 2H,I), as well as a dose-dependent rise in apoptosis (Figure 2J). AG825 had no effect on SSEA3 expression when assayed by cytometry, and viable hESCs could be maintained for more than 5 days (not shown). Western blotting demonstrated that AG825 inhibited autophosphorylation of ERBB2 at tyrosine 1248 in starved/HRG-pulsed hESCs (Figure S6), consistent with the literature.26 These findings demonstrated that disruption of ERBB2 signaling severely inhibited hESC proliferation and induced apoptosis.
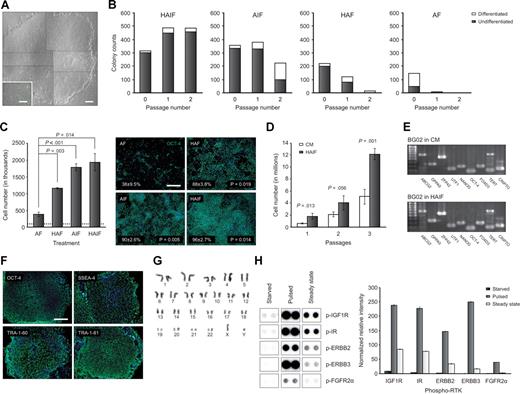
In consideration of the roles of IGF1R and ERBB2 in self-renewal, a defined medium was assembled containing a combination of IGF1R and ERBB2 ligands. Since IGF1 and IGF2 elicit extremely similar profiles of RTK phosphorylation (Figure S7), LONG R3 IGF1 (LR3-IGF1), a GMP-grade recombinant human IGF1 that does not bind insulin-like growth factor–binding proteins (IGFBPs), was added to the medium. The ERBB2 ligand used in the defined medium was the EGF domain of HRG, an EGF family member which induces activation of the ERBB2/3 heterodimer.22 FGF2 and ActA, which have previously been implicated in the maintenance of pluripotent cells,1,3,6 were also included. A simple feeder-, serum-, and KSR-free defined medium providing basal components (Table S1) supplemented with 10 ng/mL HRG, 10 ng/mL ActA, 200 ng/mL LR3-IGF1, and 8 ng/mL FGF2 (DC-HAIF medium) supported the long-term expansion of undifferentiated hESCs. Cultures were initially expanded on Matrigel diluted 1:30, but could be maintained successfully long-term on this substrate diluted 1:200 or 1:1000, or human extracellular matrix (ECM) such as serum or fibronectin (not shown). Undifferentiated BG01 and BG02 hESCs were maintained in DC-HAIF for greater than 7 and 9 months, respectively. CyT49, a hESC line isolated on human feeders under GMP conditions, was also maintained using DC-HAIF for greater than 3 months (not shown). EGF family ligands that activate ERBB1/1 or ERBB1/2 but not ERBB2/3 signaling, namely EGF, heparin-binding EGF, and TGFα, were unable to substitute for HRG in supporting long-term hESC expansion (not shown). In addition, neurotrophin-39 could not substitute for HRG to support hESCs in these defined conditions (not shown). HESC cultures could be passaged directly from feeder layers or CM conditions into DC-HAIF, and exhibited minimal spontaneous differentiation (Figure 3A). Colony and cell-counting assays confirmed that LR3-IGF1 and HRG played the major roles in self-renewal and proliferation in the context of this defined medium (Figure 3B,C), and cultures could not be maintained without HRG beyond approximately 3 weeks (not shown). A direct comparison of serial passaging in CM and DC-HAIF demonstrated that along with providing a substantially simplified approach, expansion of hESC cultures was markedly enhanced in DC-HAIF conditions compared with traditional methods (Figure 3D).
Culture of hESCs in a defined medium designed to stimulate IGF1R/IR and ERBB2/3 signaling. (A) Low (4× objective) and high (40× objective) magnification phase contrast images of morphologically undifferentiated BG02 hESCs growing in DC-HAIF. Overlapping fields (dashed black borders) were aligned to image a representative large colony. Scale bar equals 100 and 25 μm, respectively. (B) Colony counting for serial passaging of CyT49 hESCs in different combinations of growth factors. Starter cultures were growing on MEFs in NCM and the proportion of undifferentiated (■) and differentiated (□) colonies at each stage are indicated. A combination of all 4 factors was necessary to enable long-term maintenance of hESCs. Similar results were obtained with repeated experiments and other hESC lines. Panel B and C abbreviations: H indicates 10 ng/mL HRG1β; A, 10 ng/mL ActA; I, 200 ng/mL LR3-IGF1; and F, 8 ng/mL FGF2. (C) Cell-counting analysis of the role of IGF1 and HRG in hESC proliferation using BG02 cells (left panel). The mean cell number/well before the different growth factor combinations were applied on day 1 is indicated (…). Cultures were disaggregated and counted on day 7, and the mean and standard deviation were plotted. Right panel shows OCT4/DAPI immunostaining of a duplicate repeated experiment (4× objective), which demonstrated that IGF1 and HRG significantly increased the proportion of OCT4+ cells compared with ActA/FGF2 conditions. Scale bar equals 50 μm. (D) Direct comparison of CM and DC-HAIF growth conditions with serial passaging. BG03 cells growing on MEFs were passaged to CM and DC-HAIF conditions in parallel (p0 plates). Triplicate cell counts were performed at p1 to p3 and the split ratio–corrected total cell number was plotted (mean ± SD). Split ratios were (CM, DC-HAIF): p0 (1:3, 1:3); p1 (1:2, 1:3); and p2 (1:2, 1:2). Similar results were obtained with repeat experiments. (E) Maintenance of markers of undifferentiated cells in BG02 DC-HAIF p5 cells compared with BG02 in CM by RT-PCR. (F) Positive immunofluorescence of hESC markers in BG02 DC-HAIF p5 cells (10× objective). Scale bar equals 50 μm. (G) Representative G-banding of BG02 DC-HAIF p26 cells. (H) RTK blotting analysis of BG01 DC-HAIF hESCs starved of growth factors overnight; starved, then pulsed with DC-HAIF for 15 minutes; or steady-state cultures are shown (left panel). The mean and range of normalized relative intensity is plotted (right panel).
Culture of hESCs in a defined medium designed to stimulate IGF1R/IR and ERBB2/3 signaling. (A) Low (4× objective) and high (40× objective) magnification phase contrast images of morphologically undifferentiated BG02 hESCs growing in DC-HAIF. Overlapping fields (dashed black borders) were aligned to image a representative large colony. Scale bar equals 100 and 25 μm, respectively. (B) Colony counting for serial passaging of CyT49 hESCs in different combinations of growth factors. Starter cultures were growing on MEFs in NCM and the proportion of undifferentiated (■) and differentiated (□) colonies at each stage are indicated. A combination of all 4 factors was necessary to enable long-term maintenance of hESCs. Similar results were obtained with repeated experiments and other hESC lines. Panel B and C abbreviations: H indicates 10 ng/mL HRG1β; A, 10 ng/mL ActA; I, 200 ng/mL LR3-IGF1; and F, 8 ng/mL FGF2. (C) Cell-counting analysis of the role of IGF1 and HRG in hESC proliferation using BG02 cells (left panel). The mean cell number/well before the different growth factor combinations were applied on day 1 is indicated (…). Cultures were disaggregated and counted on day 7, and the mean and standard deviation were plotted. Right panel shows OCT4/DAPI immunostaining of a duplicate repeated experiment (4× objective), which demonstrated that IGF1 and HRG significantly increased the proportion of OCT4+ cells compared with ActA/FGF2 conditions. Scale bar equals 50 μm. (D) Direct comparison of CM and DC-HAIF growth conditions with serial passaging. BG03 cells growing on MEFs were passaged to CM and DC-HAIF conditions in parallel (p0 plates). Triplicate cell counts were performed at p1 to p3 and the split ratio–corrected total cell number was plotted (mean ± SD). Split ratios were (CM, DC-HAIF): p0 (1:3, 1:3); p1 (1:2, 1:3); and p2 (1:2, 1:2). Similar results were obtained with repeat experiments. (E) Maintenance of markers of undifferentiated cells in BG02 DC-HAIF p5 cells compared with BG02 in CM by RT-PCR. (F) Positive immunofluorescence of hESC markers in BG02 DC-HAIF p5 cells (10× objective). Scale bar equals 50 μm. (G) Representative G-banding of BG02 DC-HAIF p26 cells. (H) RTK blotting analysis of BG01 DC-HAIF hESCs starved of growth factors overnight; starved, then pulsed with DC-HAIF for 15 minutes; or steady-state cultures are shown (left panel). The mean and range of normalized relative intensity is plotted (right panel).
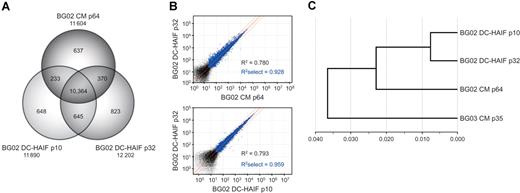
Multiple approaches were used to confirm the maintenance of hESC characteristics in DC-HAIF conditions. Cultures retained expression of transcriptional and cell-surface markers of pluripotency (Figure 3E,F; Figure S8), and G-banding analyses demonstrated the maintenance of euploidy (Figure 3G; Table 1). RTK blotting revealed robust phosphorylation of IGF1R, IR, ERBB2, and ERBB3 in steady-state cultures and a strong response in phosphorylation of these receptors when starved cultures were pulsed with DC-HAIF. FGFR2α was also phosphorylated, but to a lesser extent, in these experiments (Figure 3H). Transcriptional analyses were used to compare global expression in hESCs cells18 maintained in CM and DC-HAIF (Table S3; full dataset). More than 11 600 transcripts were detected in BG02 cells grown in DC-HAIF for 10 and 32 passages, or in CM for 64 passages, with 10 364 transcripts common to all populations (Figure 4A), including known hESC markers such as CD9, DNMT3, NANOG, OCT4, TERT, and UTF1 (not shown). High-correlation coefficients were observed in comparisons of CM and DC-HAIF cultures (R2 select [>0.99 confidence level] = 0.928), as well as in early- and late-passage DC-HAIF cells (R2 select = 0.959) (Figure 4B). Hierarchic clustering analysis demonstrated that BG02 cells maintained in DC-HAIF grouped tightly and retained a close similarity to BG02 and BG03 cells maintained in CM (Figure 4C). These data are consistent with previous analyses showing that undifferentiated hESCs clustered tightly compared with embryoid bodies or fibroblasts.18 Analysis of micro-RNAs (miRNAs) demonstrated expression of a core set of markers that form a characteristic signature in undifferentiated hESCs (J.C. and M.R., submitted manuscript) at the same or higher levels in cells grown in DC-HAIF compared with CM (not shown). Finally, methylation-specific PCR of a subset of CpG sites that distinguish hESCs from other cell types27 indicated that BG02 cells maintained in CM or DC-HAIF exhibited a similar methylation profile (not shown), which was clearly different from cancer cells and differentiated cell types.27 To assess the developmental potential of hESCs maintained in DC-HAIF, cultures were differentiated to representatives of the 3 germ layers both in vivo and in vitro. BG02 cells cultured in DC-HAIF for 6 months (25 passages) maintained the potential to form complex teratomas containing ectoderm, mesoderm, and endoderm (Figure 5A). Differentiation to cells expressing βIII tubulin, smooth-muscle actin, or α-fetoprotein was demonstrated in embryoid bodies in vitro (Figure 5B). Finally, modification of a directed differentiation approach28 enabled the generation of definitive endoderm and foregut endoderm from hESCs using defined medium conditions (Figure 5C,D). These analyses demonstrated the stable maintenance of key characteristics of pluirpotency and validated DC-HAIF as a simple medium for supporting self-renewal.
Summary of cytogenetic analyses of BG01 and BG02 DC-HAIF cells
| Line . | Passage no. . | Karyotype (normal/total) . | |
|---|---|---|---|
| Total . | HAIF . | ||
| BG01 | 32 | 6 | 46,XY (20/20) |
| 41 | 18 | 46,XY (19/20)* | |
| BG02 | 47 | 5 | 46,XY (20/20) |
| 61 | 19 | 46,XY (20/20) | |
| 71 | 26 | 46,XY (20/20) | |
| Line . | Passage no. . | Karyotype (normal/total) . | |
|---|---|---|---|
| Total . | HAIF . | ||
| BG01 | 32 | 6 | 46,XY (20/20) |
| 41 | 18 | 46,XY (19/20)* | |
| BG02 | 47 | 5 | 46,XY (20/20) |
| 61 | 19 | 46,XY (20/20) | |
| 71 | 26 | 46,XY (20/20) | |
One metaphase exhibited an additional human chromosome 16.
Comparison of the transcriptome of hESCs maintained in DC-HAIF or CM. (A) Venn diagram of the distribution of transcripts detected using high-density Illumina Sentrix Human-6 Expression Beadchips containing 47 296 transcript probes in BG02 cells maintained in CM (64 passages) or DC-HAIF (10 or 32 passages in defined medium). A large proportion of the expressed transcripts were detected in all samples. (B) Scatterplot analysis demonstrated that the transcriptional profile of BG02 DC-HAIF p32 cells was highly similar to that of BG02 cells maintained in CM (top panel), and was not substantially altered in early- and late-passage cultures in DC-HAIF (bottom panel). Correlation coefficients (R2) were generated using all detected transcripts with an expression level of more than 0 (black and blue dots), or with transcripts exhibiting a detection confidence level of more than 0.99 (R2 select, blue dots). Red lines delineate the mean and limits of a 2-fold difference. (C) Hierarchic clustering dendrogram of relative gene expression in different populations generated using the Beadstudio software (Illumina, San Diego, CA). Early- and late-passage BG02 cells maintained in DC-HAIF clustered tightly (approximately 0.0075) and retained a close similarity to BG02 and BG03 cells maintained in CM (approximately 0.037). Previous analyses showed that different undifferentiated hESC lines clustered tightly (approximately 0.032) compared with differentiated embryoid body populations (approximately 0.0875) or fibroblasts (approximately 0.160).18
Comparison of the transcriptome of hESCs maintained in DC-HAIF or CM. (A) Venn diagram of the distribution of transcripts detected using high-density Illumina Sentrix Human-6 Expression Beadchips containing 47 296 transcript probes in BG02 cells maintained in CM (64 passages) or DC-HAIF (10 or 32 passages in defined medium). A large proportion of the expressed transcripts were detected in all samples. (B) Scatterplot analysis demonstrated that the transcriptional profile of BG02 DC-HAIF p32 cells was highly similar to that of BG02 cells maintained in CM (top panel), and was not substantially altered in early- and late-passage cultures in DC-HAIF (bottom panel). Correlation coefficients (R2) were generated using all detected transcripts with an expression level of more than 0 (black and blue dots), or with transcripts exhibiting a detection confidence level of more than 0.99 (R2 select, blue dots). Red lines delineate the mean and limits of a 2-fold difference. (C) Hierarchic clustering dendrogram of relative gene expression in different populations generated using the Beadstudio software (Illumina, San Diego, CA). Early- and late-passage BG02 cells maintained in DC-HAIF clustered tightly (approximately 0.0075) and retained a close similarity to BG02 and BG03 cells maintained in CM (approximately 0.037). Previous analyses showed that different undifferentiated hESC lines clustered tightly (approximately 0.032) compared with differentiated embryoid body populations (approximately 0.0875) or fibroblasts (approximately 0.160).18
In vivo and in vitro differentiation of hESCs maintained in DC-HAIF. (A) Analysis of teratomas from BG02 DC-HAIF p25 cells demonstrated pluripotent differentiation potential to ectoderm, mesoderm, and endoderm. NSE indicates neuron-specific enolase; SMA, smooth-muscle actin; and pCK, pan-cytokeratin. Scale bars equal 20 μm (NSE, p63, 100× oil objective), 50 μm (SMA, Ducts, pCK, 20× objective), and 200 μm (Bone, 10× objective). (B) Differentiation of BG02 DC-HAIF p5 cells to ectoderm (βIII tubulin+), mesoderm (SMA+), and endoderm (alphafetoprotein+ [αFP]) lineages in embryoid bodies (20× objective, same scale as panel D). (C,D) Directed differentiation of BG02 DC-HAIF p48 cells (HAIF) to definitive endoderm (d3) and foregut endoderm (d6) using defined medium differentiation conditions (Document S1). (C) qPCR analyses showed down-regulation of OCT4 expression and minimal PAX6 or CDX2 expression in differentiated samples, suggesting differentiation of pluripotent cells and lack of substantial neuroepithelial or trophectodermal differentiation. The generation of definitive endoderm at day 3 was confirmed by up-regulation of SOX17, CXCR4, and CER expression, and foregut endoderm at day 6 by up-regulation of FOXA2, HNF1β, and HNF4α. Similar results were observed with CyT49 cells. Error bars are plus or minus SD. (D) Immunostaining analyses confirmed homogenous expression of OCT4 and lack of SOX17 expression in undifferentiated cultures (HAIF). After 3 days, most cells express SOX17 with only pockets of OCT4+ cells remaining (d3). After 6 days, the differentiation to foregut endoderm was confirmed by expression of HNF1β and HNF4α (d6). Only rare OCT4+ cells were present at d6 (not shown). Scale bar equals 50 μm (20× objective).
In vivo and in vitro differentiation of hESCs maintained in DC-HAIF. (A) Analysis of teratomas from BG02 DC-HAIF p25 cells demonstrated pluripotent differentiation potential to ectoderm, mesoderm, and endoderm. NSE indicates neuron-specific enolase; SMA, smooth-muscle actin; and pCK, pan-cytokeratin. Scale bars equal 20 μm (NSE, p63, 100× oil objective), 50 μm (SMA, Ducts, pCK, 20× objective), and 200 μm (Bone, 10× objective). (B) Differentiation of BG02 DC-HAIF p5 cells to ectoderm (βIII tubulin+), mesoderm (SMA+), and endoderm (alphafetoprotein+ [αFP]) lineages in embryoid bodies (20× objective, same scale as panel D). (C,D) Directed differentiation of BG02 DC-HAIF p48 cells (HAIF) to definitive endoderm (d3) and foregut endoderm (d6) using defined medium differentiation conditions (Document S1). (C) qPCR analyses showed down-regulation of OCT4 expression and minimal PAX6 or CDX2 expression in differentiated samples, suggesting differentiation of pluripotent cells and lack of substantial neuroepithelial or trophectodermal differentiation. The generation of definitive endoderm at day 3 was confirmed by up-regulation of SOX17, CXCR4, and CER expression, and foregut endoderm at day 6 by up-regulation of FOXA2, HNF1β, and HNF4α. Similar results were observed with CyT49 cells. Error bars are plus or minus SD. (D) Immunostaining analyses confirmed homogenous expression of OCT4 and lack of SOX17 expression in undifferentiated cultures (HAIF). After 3 days, most cells express SOX17 with only pockets of OCT4+ cells remaining (d3). After 6 days, the differentiation to foregut endoderm was confirmed by expression of HNF1β and HNF4α (d6). Only rare OCT4+ cells were present at d6 (not shown). Scale bar equals 50 μm (20× objective).
Discussion
This study highlights IGF1R and ERBB2 as 2 previously unappreciated RTKs that are essential for hESC self-renewal, enabling the development of a defined medium for hESC culture. While previous reports have suggested several factors that may influence self-renewal in the background of KSR-containing medium, relatively few studies have demonstrated the successful expansion of undifferentiated hESCs under defined conditions.10,11,13,29 These defined approaches also contained high concentrations of insulin, present in the N2 or B27 medium supplements, or added as a generic “survival factor.” Insulin is a potent growth factor that can bind either IR or IGF1R and signal through the PI3 kinase/AKT pathway, and the presence of insulin in the vast majority of hESC growth conditions highlights the importance of IGF1R/IR signaling for the maintenance of hESCs. While the roles of IR and IGF1R signaling were not distinguished in this study, our findings suggest that IGF1R plays an important role because (1) the specific IGF1R blocking antibody A12, which does not inhibit IR,25 reduced hESC proliferation and promoted differentiation; and (2) hESCs transduced with a shRNA specific for IGF1R could not sustain self-renewal.
While both NCM and CM contain high concentrations of insulin, only CM supports the long-term culture of hESCs,2 suggesting that insulin-initiated IR and IGF1R signaling and low levels of FGF2 are not sufficient to maintain self-renewal. CM elicited a profile of RTK phosphorylation in hESCs that suggested MEFs produce EGF family ligands. This is consistent with the capacity of HRG to support pluripotency in defined conditions. Like other EGF family members, HRG is first expressed as a transmembrane protein, and is cleaved to various bioactive soluble ectodomains by ADAM proteases.23 HRG binds to ERBB3 via its EGF domain, and ERBB3 heterodimerizes with ERBB2. ERBB3 does not have a functional tyrosine kinase domain, and is phosphorylated in trans by ERBB2.22 The role of HRG-ERBB2/3 signaling in self-renewal points to other potential features of pluripotent cells. Because hESCs grow as polarized epithelia30 and are regulated by EGF family pathways, other mechanisms in the regulation of epithelia may also be relevant. Interestingly, inappropriate activation of ERBB2 is associated with transformation of multiple types of epithelial tissues.22 Duplication of ERBB2 at 17q21.1, trisomies of chromosome 17q, or overexpression of ERBB2 or HRG are transforming events in breast cancer and other tumors. Selective advantage and overgrowth of hESCs bearing trisomies of chromosomes 17 (ERBB2) and 12 (ERBB3 at 12p13) have been reported by several laboratories,31 raising the possibility that similar mechanisms of transformation occur in hESCs and epithelial tissues. ERBB2 is also part of a cohort of genes that display a diagnostic methylation profile that distinguishes pluripotent cells from other cell types,27 implying tight epigenetic regulation of this gene in the undifferentiated state. Finally, HRG has been shown to support the expansion of mouse primordial germ cells, a pluripotent cell type highly related to ESCs.32 These data are all suggestive of a wider functional role of HRG and ERBB2/3 signaling in the self-renewal of pluripotent cells.
Signaling through the PI3 kinase/AKT pathway has recently been implicated in the self-renewal of mouse, primate,33 and human ESCs.34,35 Critically, inhibition of signaling through this pathway using the small-molecule antagonists LY294002 (PI3 kinase inhibitor) and AKT1-II (AKT1 inhibitor) induced differentiation of hESCs.34 Treatment of hESCs with LY294002 was associated with a rapid collapse in phosphorylation of AKT1, pS6, pS6K/p70, and GSK3β.35 We have confirmed the importance role of this pathway by demonstrating that 1 to 20 nM rapamycin, an inhibitor of FRAP1 (mTOR), severely restricted the proliferation of hESCs in DC-HAIF and induced differentiation in short term culture (not shown). Because the PI3 kinase pathway delivers both strong proliferative and antiapoptotic signals, IR/IGF1R signaling should not be overlooked when interpreting the effects of factors that feed into this common pathway, including FGF2, PDGF, and neurotrophins.3,7,9,11 PI3 kinase signaling leads to the activation of AKT and AKT-mediated inhibition of GSK3β, and therefore the presence of insulin also complicates the interpretation that small-molecule inhibitors of GSK3β are mimicking the effects of WNT signaling.8 It is possible that multiple inputs are required for appropriate signaling and regulation of self-renewal through PI3 kinase in hESCs, which in this study was driven by activation of IR, IGF1R, and ERBB2/3.
This study identified IGF1R and ERBB2 as RTKs involved in self-renewal of hESCs and enabled the development of a defined medium based around activation of these receptors. We have used this strategy to demonstrate the capacity for large-scale expansion of undifferentiated hESCs. A starting confluent culture of BG02 cells in one 60-mm plate was expanded in DC-HAIF through 4 passages over 20 days to generate more than 1010 cells (T.S., A.R., unpublished data, December 2006). The ability to massively expand undifferentiated hESC cultures, coupled with effective differentiation approaches that also use defined conditions to generate clinically relevant populations,28 lays important groundwork for eventual clinical applications.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Note added in proof:
A requirement for IGF1R signaling in hESC self-renewal was also reported by another group36 while this manuscript was under review.
Acknowledgments
We would like to thank Dr Steve Plymate for the A12 antibody; members of the Blau Lab for helpful discussions; Paul Fields for expert assistance; and Dr Clifton Baile, Diane Hartzel, and the Animal Facility, Animal and Dairy Science Department, University of Georgia, for assisting with generating teratomas.
This work was supported by grants from the National Institute of General Medical Sciences at the NIH (1P01GM081619-01; C.A.B. and C.W.) and the National Institute of Research Resources (9R24RR021313–04 to T.S.). Federal funding was not used for non–NIH-registered hESC lines.
National Institutes of Health
Authorship
Contribution: L.W. designed, performed, and supervised experiments, interpreted findings, and assisted in writing the manuscript. T.C.S. designed, performed, and supervised experiments, interpreted findings, and wrote and assembled the manuscript. E.S.S., S.N.B., A.M.L., and M.J.G. performed defined medium studies and other experiments. D.S.D. assisted with experiments and constructed figures. S.S. performed and interpreted microarray and other molecular analyses. A.M.N. and C.B.W. assisted with the generation of hESC cultures. M.Z. and C.-Z.S. designed and constructed lentiviral shRNA vectors. X.C. performed qPCR analysis of shRNA experiments. E.W.U. performed histologic analyses of teratomas. K.A.D. performed qPCR analysis of endoderm differentiations. J.D.C. supervised and interpreted microarray and other molecular analyses. M.S.R. conceived, directed, and interpreted microarray and molecular analyses. C.A.B. conceived, designed, and supervised investigation of MEF-CM, interpreted findings, and assisted in writing the manuscript. A.J.R. conceived the defined medium study, assisted design and supervision of experiments, interpreted findings, and assisted in writing manuscript. L.W. and T.C.S. contributed equally to this manuscript.
Conflict-of-interest disclosure: T.C.S., E.S.S., S.N.B., M.J.G., K.A.D., and A.J.R. are employees and have stock options in a company that is not publicly traded (Novocell). S.S., J.D.C., and M.S.R. are employees and have ownership interest in a publicly traded company (Invitrogen).
Correspondence: Allan J. Robins, Novocell Inc, 111 Riverbend Rd, Athens, GA 30602; e-mail: arobins@novocell.com; or C. Anthony Blau, Mailstop 357710, K260 Health Sciences Building, University of Washington, Seattle, WA 98195; e-mail: tblau@u.washington.edu.

![Figure 5. In vivo and in vitro differentiation of hESCs maintained in DC-HAIF. (A) Analysis of teratomas from BG02 DC-HAIF p25 cells demonstrated pluripotent differentiation potential to ectoderm, mesoderm, and endoderm. NSE indicates neuron-specific enolase; SMA, smooth-muscle actin; and pCK, pan-cytokeratin. Scale bars equal 20 μm (NSE, p63, 100× oil objective), 50 μm (SMA, Ducts, pCK, 20× objective), and 200 μm (Bone, 10× objective). (B) Differentiation of BG02 DC-HAIF p5 cells to ectoderm (βIII tubulin+), mesoderm (SMA+), and endoderm (alphafetoprotein+ [αFP]) lineages in embryoid bodies (20× objective, same scale as panel D). (C,D) Directed differentiation of BG02 DC-HAIF p48 cells (HAIF) to definitive endoderm (d3) and foregut endoderm (d6) using defined medium differentiation conditions (Document S1). (C) qPCR analyses showed down-regulation of OCT4 expression and minimal PAX6 or CDX2 expression in differentiated samples, suggesting differentiation of pluripotent cells and lack of substantial neuroepithelial or trophectodermal differentiation. The generation of definitive endoderm at day 3 was confirmed by up-regulation of SOX17, CXCR4, and CER expression, and foregut endoderm at day 6 by up-regulation of FOXA2, HNF1β, and HNF4α. Similar results were observed with CyT49 cells. Error bars are plus or minus SD. (D) Immunostaining analyses confirmed homogenous expression of OCT4 and lack of SOX17 expression in undifferentiated cultures (HAIF). After 3 days, most cells express SOX17 with only pockets of OCT4+ cells remaining (d3). After 6 days, the differentiation to foregut endoderm was confirmed by expression of HNF1β and HNF4α (d6). Only rare OCT4+ cells were present at d6 (not shown). Scale bar equals 50 μm (20× objective).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/12/10.1182_blood-2007-03-082586/4/m_zh80240710510005.jpeg?Expires=1769305849&Signature=VVzob9zHSQH~wIkbHggly1w4IWJ5bEeV1Wruo0WMCjIjAq1wZf87f7v4l1HeX~XI14ocOEleh~obAfdQZ8g54H134~C0jZoy8EhxHGdLP3csGaaIa88z2oBvjwE5Z6x5mKV1Zvqa6BifFeEt59qNu3tuihWvrEThThTVcS72NgcmyPsHKNNpPoA1ogqrg5o3z9VNYyS8lNvNrHoJmF88BoULEnCZaziCK2IyRjnYRW3zccRCZPtylnE4qgKjca4b7BJVLkxiT~NmUpz4DLpHCwyVV6MRyXwDFfZBwnO1KFhziRhiqSAVT06Xo8iR8SjMx6w3HlMR816tPZVtojKzxQ__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal