Abstract
Recent evidence indicates that natural killer (NK) cells can negatively regulate T-cell responses, but the mechanisms behind this phenomenon as a consequence of NK–T-cell interactions are poorly understood. We studied the interaction between the NKG2D receptor and its ligands (NKG2DLs), and asked whether T cells expressed NKG2DLs in response to superantigen, alloantigen, or a specific antigenic peptide, and if this rendered them susceptible to NK lysis. As evaluated by FACS, the major histocompatibility complex (MHC) class I chain-related protein A (MICA) was the ligand expressed earlier on both CD4+ and CD8+ T cells in 90% of the donors tested, while UL16-binding protein-1 (ULBP)1, ULBP2, and ULBP3 were induced at later times in 55%–75% of the donors. By carboxyfluorescein diacetate succinimidyl ester (CFSE) labeling, we observed that NKG2DLs were expressed mainly on T cells that had gone through at least one division. Real-time reverse-transcription polymerase chain reaction confirmed the expression of all NKG2DLs, except ULBP4. In addition, T-cell activation stimulated phosphorylation of ataxia-telangiectasia mutated (ATM), a kinase required for NKG2DLs expression after DNA damage, and ATM/Rad3-related kinase (ATR) inhibitors blocked MICA induction on T cells with a mechanism involving NF-κB. Finally, we demonstrated that activated T cells became susceptible to autologous NK lysis via NKG2D/NKG2DLs interaction and granule exocytosis, suggesting that NK lysis of T lymphocytes via NKG2D may be an additional mechanism to limit T-cell responses.
Introduction
The negative regulation of adaptive immunity is relevant to maintain lymphocyte homeostasis and to prevent inappropriate T-cell activation, which can ultimately result in autoimmune or lymphoproliferative diseases. Although it is well-documented that natural killer (NK) cells are important effectors of innate immunity, and their role against virally infected and tumor cells has been studied over the years,1,2 much attention has also been focused on their ability to promote adaptive immunity by secretion of immunomodulatory cytokines and chemokines.3-5 More recently, however, a previously unappreciated negative immunoregulatory role has emerged. In fact, NK cells can downregulate T-cell–mediated immune responses by their killing capacity and by secreting inhibitory cytokines such as interleukin (IL)-10 and transforming growth factor beta (TGF)-β.6-8 In vitro experiments have shown that activated human NK cells can kill dendritic cells (DCs),9,10 probably contributing to inhibition of T-cell activation in inflamed tissues. During murine cytomegalovirus (MCMV) infection, the presence of NK cells limits CD4+ and CD8+ IFN-γ production and proliferation.11 In addition, in a major histocompatibility complex (MHC) class I-positive host grafted with MHC class I-negative bone marrow, development of MHC class I-deficient thymocytes is delayed as a result of NK-cell cytotoxicity.12 Furthermore, studies in humans and animal models have demonstrated that NK cells can prevent the initiation and progression of autoimmune diseases:13 lack of NK cells correlates with severe forms of experimental autoimmune encephalomyelitis and CD4+ T-cell-mediated colitis, suggesting that NK cells can actively inhibit proliferation and cytokine production by autoreactive T cells.14,15 In accordance, reduced NK-cell numbers and compromised NK-cell functions are found in patients with multiple sclerosis, systemic lupus erythematosus, and type I diabetes.13
The molecular mechanisms of T-cell suppression by NK cells, however, remain elusive. NK cells may interfere with antigen-presenting cells, preventing their maturation and functions.9 However, NK cells have been shown to induce cell-cycle arrest of activated T cells in a contact-dependent, reversible, antigen-nonspecific manner.16 In addition, IL-2 activated mouse NK cells kill syngeneic T-cell blasts in a perforin-dependent manner and through the NK activating receptor NKG2D.17 In humans, NKG2D is constitutively expressed on all NK cells and CD8+ T cells, and on most γδ T cells.18 It delivers a potent activating signal, resulting in cytotoxicity, release of cytokines and chemokines, and upregulation of activation antigens.19 NKG2D binds to a family of MHC class I-related molecules: MICA, MICB, UL16-binding protein (ULBP)1, ULBP2, ULBP3, ULBP4/RAET1E, and RAET1G.20-23 Expression of NKG2D ligands (NKG2DLs) has been well-documented in response to neoplastic transformation, viral and bacterial infections, and the interaction between NKG2D and its ligands has been implicated in the NK-cell resistance against tumors and infections.1,2 More recently, expression of NKG2DLs has been observed in diseased tissues from patients affected by autoimmune disorders24 and on some normal hematopoietic cells, suggesting a more general NKG2D-dependent immunoregulatory role of NK cells. In the mouse, NKG2DLs expression has been described on activated T cells, activated macrophages, bone marrow cells, and on some thymocyte subsets.17,25-27 In humans, mature DCs express ULBP1 and MICA, and ULBP1–3 were found on B cells, monocytes, and granulocytes, but not on resting T and NK cells.28-30 Intracellular MICA was detected on CD3 or CD28/PMA stimulated T lymphocytes,31 and ULBP1–3 mRNA has been observed in CD8+ activated T cells cultured with IL-7 and IL-15.32
In this study, we investigated the expression and function of NKG2DLs on human CD4+and CD8+ T cells in response to antigen stimulation. We studied MICA, ULBP1, ULBP2, ULBP3, and ULBP4, and the impact of their expression on the lysis of activated T cells by autologous NK cells, as a possible mechanism contributing to negative regulation of T-cell responses. We also investigated some signaling events responsible for the induction of MICA expression by using the inhibitors caffeine and KU-55933, which target ataxia-telangiectasia mutated/Rad3-related (ATM/ATR) kinases, recently shown to be involved in the induction of NKG2DLs following DNA damage.33
Materials and methods
Antibodies and reagents
The following monoclonal antibodies (mAbs) were used: anti-MICA (M673), anti-MICB (M362), anti-ULBP1 (M295), anti-ULBP2 (M311), and anti-ULBP3 (M550), kindly provided by Dr David Cosman (Amgen, Seattle, WA). Anti-MICA (MAB159227), anti-ULBP1 (MAB170818), anti-ULBP2 (MAB165903), anti-ULBP3 (MAB166510), anti-NKG2D (MAB149810), and recombinant NKG2DL proteins (NKG2DL-Fc) were from R&D Systems (Minneapolis, MN). The following mAbs were from Becton Dickinson (San Diego, CA): purified control IgG1, anti-CD3 allophycocyanin (APC: HIT3a), anti-CD4 peridinin chlorophyll protein (PerCP: SK3), anti-CD8 PerCP (SK1), anti-CD25 APC (2A3), anti-CD69 PE (L78), anti-CD56 (C238), anti-CD3 (OKT3). F(ab′)2 fragments of phycoerythrin (PE)-conjugated goat-antimouse (GAM-PE) IgG, and FITC-conjugated goat-antirabbit IgG (GARB-FITC) were from Jackson Immunoresearch Laboratories (Newmarket, UK). Other antibodies were: F(ab′)2 fragments of goat-antimouse IgG (GAM) (Cappel; Aurora, OH); rabbit anti-phosho-ATM (S1981; R&D Systems); rabbit anti-ATM (Ab-3; Calbiochem; San Diego, CA); anti-phospho-SAPK/JNK (Thr183/Tyr185) mAb and rabbit anti-SAPK/JNK (Cell Signaling; Danvers, MA); and antipaxillin mAb (BD Transduction Laboratories; San Jose, CA). Other reagents used were 5,6-carboxyfluorescein diacetate succinimidyl ester (CFSE), enterotoxin B from Staphylococcus aureus (SEB), phorbol-12-myristate-13-acetate (PMA), ionomycin, caffeine, rapamycin, KU-55993, and etoposide (VP16) (all from Sigma-Aldrich); recombinant human IL-2 (Peprotech; Rocky Hill, NJ); and phytohemagglutinin (PHA-L; Biochrom AG; Berlin, Germany).
Peripheral blood mononuclear cells and T lymphocytes: purification and stimulation
Peripheral blood mononuclear cells (PBMCs), obtained by Ficoll separation of peripheral blood samples from healthy donors, were plated at a concentration of 1.5 × 106 cells/mL in the presence of the superantigen SEB (100 ng/mL), or phorbol-12-myristate-13-acetate (50 ng/mL) plus ionomycin (500 ng/mL), or (PHA) (1 μg/mL). In MLR experiments, CD8+and CD4+ T cells (> 95% pure) were isolated by positive selection using magnetic beads (Dynal, Invitrogen, Carlsbad, CA). Responder T cells were CFSE-labeled, and then plated at a concentration of 1.5 × 106 cells/mL in the presence of the same number of irradiated (3000 rad) allogeneic PBMCs, for 5 and 7 days. CFSE-labeling was performed at day 0 on total PBMCs or on purified CD4+or CD8+ T lymphocytes. Cells were washed with phosphate-buffered saline, resuspended at 50 × 106 cells/mL, and labeled for 10 minutes with CFSE (2.5 μM). Sometimes, PBMCs were pre-incubated for 2 hours at 37°C with caffeine, KU-55933, or rapamycin, used at a final concentration of 5 mM, 10 μM, and 1 μM, respectively, if not otherwise specified. Cells were then plated with the indicated stimuli and inhibitors were present throughout the experiments. In some experiments, an HLA-A2–restricted NS31406-1415-specific CD8+ T-cell clone34 (kindly provided by Dr D. Accapezzato and Prof V. Barnaba, University of Rome “La Sapienza,” Italy) was stimulated with an autologous Epstein-Barr virus-immortalized B lymphoblastoid cell line, pulsed with the NS3 peptide.
Immunofluorescence and FACS analysis
For the simultaneous detection of NKG2DLs on proliferating T lymphocytes, a 4-color fluorescence activated cell sorter (FACS) analysis was used. CFSE-labeled PBMCs were stained with saturating concentrations of different anti-NKG2DLs mAbs, washed and incubated with GAM-PE. Cells were then incubated with normal mouse IgG, followed by labeling with APC-conjugated anti-CD3 and PerCP-conjugated anti-CD8 mAbs. Cells were analyzed by flow cytometry on a FACScalibur (Becton Dickinson). The median of fluorescence intensity (MFI) value of the isotype control antibody was subtracted from the MFI relative to each molecule, except for Figures 1 and 3, in which the MFI value of IgG control is shown. For intracellular staining of phosphorylated ATM (pATM), PBMCs were fixed in 1% formaldehyde and then incubated in methanol. After washing, cells were incubated with 0.2μg of anti-ATM-pS1981, washed, and incubated with GARB-FITC.
Cytotoxicity assay
A standard 4-hour chromium-release assay was used as described.35 PMA-ionomycin activated-CD4+ and CD8+ T cells or SEB-activated PBMCs were used as target cells and were labeled (100-200 μCi 51Cr/106 cells; Amersham BioSciences, Piscataway, NJ) for 90 minutes at 37°C, washed, and 5 × 103 cells/well were plated. Autologous polyclonal NK cells were used as effector cells and were generated as previously described.35 After 8-10 days, the percentage of CD3 − /CD56+cells was more than 90%. These cells were stimulated with 200 IU/mL of IL-2 for 18 hours. In some experiments, effector cells were pre-incubated with saturating amounts of anti-CD56 or anti-NKG2D mAb, for 20 minutes at room temperature, washed, and then used in the assay. Sometimes the Ca++ chelator EGTA (4 mM) was added to the wells. The percentage of specific lysis was calculated by counting an aliquot of supernatant and using the formula: 100 × [(sample release − spontaneous release)/total release − spontaneous release)].
SDS-PAGE and Western blot
Freshly isolated PBMCs were starved in RPMI 1640 with 0.1% fetal calf serum overnight at 4°C, and then washed in serum-free medium. Cells were incubated with anti-CD3 mAb (OKT3) (1 μg/106 cells) for 20 minutes at +4°C, washed, resuspended in serum-free medium at the concentration of 107 cells/250 μL, and stimulated for different times with soluble GAM (1 μg/106 cells) at 37°C. Sometimes, cells were resuspended at the same concentration in serum-free medium, and PMA (50 ng/mL) plus ionomycin (500 ng/mL) were added. Stimulation was stopped by adding cold serum-free medium and pelleting the cells. In some experiments, cells were pretreated with VP16 (10 μM) for 2 hours at 37°C and lysed immediately thereafter. Cells were lysed for 20 minutes at 4°C in ice-cold lysis buffer containing 0.2% Triton X-100, 0.3% NP-40, 50 mM Tris-HCl pH 7.6, 1 mM EDTA, 150 mM NaCl, 1 mM PMSF, 2 μg/mL aprotinin, 2 μg/mL leupeptin, 10 mM NaF, and 1 mM Na3VO4. Protein concentration was measured with the Bio-Rad Protein Assay. Lysates (100 μg) were resolved by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to polyvinylidene diflouride membranes (Millipore; Milan, Italy). Blocked polyvinylidene diflouride membranes were probed with specific antibodies. Immunoreactivity was revealed using an enhanced chemiluminescence kit (Amersham).
Polymerase chain reaction
One μg of total RNA, isolated by Trizol (GIBCO, Invitrogen), was used for cDNA first-strand synthesis in a 25-μL reaction volume, and 1 μL of the resulting cDNA was used in a 25 μL PCR reaction in the presence of FastStart Taq DNA polymerase (Roche Diagnostics). Forward and reverse primers for polymerase chain reaction (PCR) amplification were, respectively: 5′-TGCTTCTGGCTGGCATCTTCC-3′ and 5′-TAGTTCCTGCAGGCAGTC-TGC-3′ for MICA, and 5′-ACCACAGTCCATGCCATCAC-3′ and 5′-TCCACCACCCTGTTGCTGTA-3′ for GAPDH. Primers for RAET1E/ULBP4 have been described.22 PCR conditions were as follows: 94°C for 50 seconds, 58°C for 50 seconds, and 72°C for 50 seconds for 28-36 cycles. Real-time PCR was performed using the ABI Prism 7700 Sequence Detection system (Applied Biosystems, Foster City, CA); cDNA was amplified in triplicate with primers for MICA (Hs00792952_m1), MICB (Hs00792952_m1), ULBP1 (Hs00360941_m1), ULBP2 (Hs00607609_mH), and ULBP3 (Hs00225909_m1), all conjugated with fluorochrome FAM, and β-actin (4326315E) conjugated with fluorochrome VIC (Applied Biosystems). The cycling conditions were 50°C for 10 minutes, followed by 40 cycles of 95°C for 30 seconds, and 60°C for 2 minutes. Data were analyzed using the Sequence Detector v1.7 analysis software (Applied Biosystems).
Electrophoretic mobility-shift assay
Nuclear proteins (10 μg), prepared as described,36 were incubated with radiolabeled DNA probes in a 20 μL reaction mixture containing 20 mM Tris pH 7.5, 60 mM KCl, 2 mM EDTA, 0.5 mM dithiothreitol, 4% Ficoll, and 0.15 μg (for MICA NF-κB) or 1.5 μg (for NF-κB Ig) of poly(dI-dC). Nucleoprotein complexes were resolved as described.36 Oligonucleotides were from Invitrogen. Complementary strands were annealed and end-labeled as described.36 Approximately 3 × 104 cpm of labeled DNA was used in electrophoretic mobility-shift assay reactions. The following double-strand oligomers were used as specific labeled probes or cold competitors (sense strand): MICA NF-κB, 5′-GAGTAGGGGCCCTCCTTTCT-3′37 ; NF-κB Ig, 5′-GATCACAAGGGACTTTCCGCT-3′; Octamer-(h-Histone H2b), 5′-AGCTCTTCACCTTATTTGCATAAGCGAT-3′.
Results
NKG2DLs are induced on CD4+ and CD8+ T lymphocytes in response to SEB stimulation
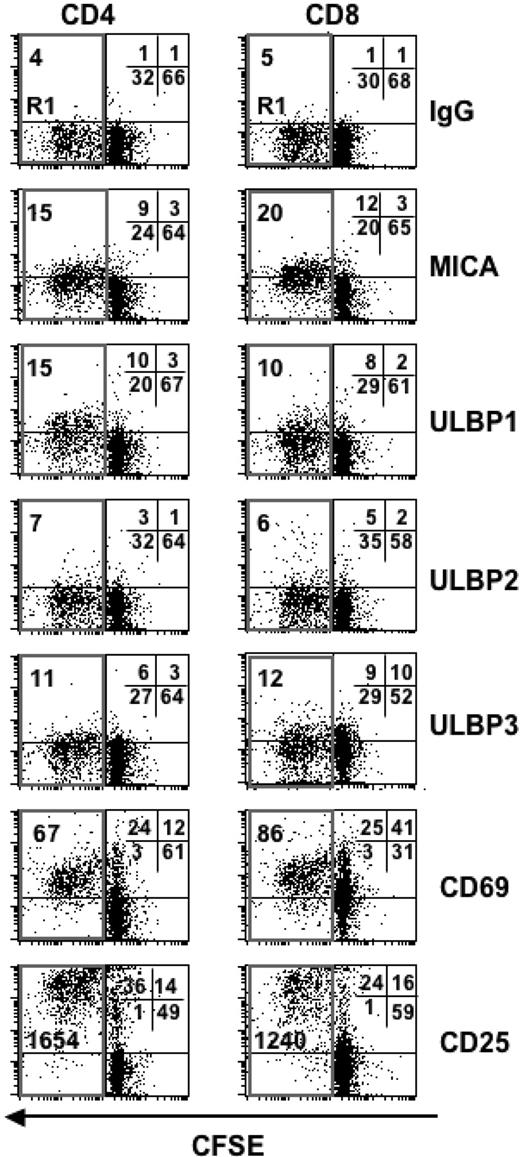
To study if the NKG2D/NKG2DLs interaction could play a role in the negative regulation of T-cell responses mediated by NK cells, we first investigated cell surface expression of NKG2DLs, namely MICA, ULBP1, ULBP2, and ULBP3 on activated T lymphocytes. To obtain the simultaneous activation of CD4+and CD8+ T lymphocytes, PBMCs were stimulated with the superantigen SEB, which binds to some regions of T-cell receptor (TCR) Vβ chains on both CD4+and CD8+ T cells, and to MHC class II molecules on APCs.38 PBMCs were also labeled with CFSE to follow the proliferation of responder T cells. NKG2DLs expression as well as the percentage of dividing cells were monitored on both CD4+and CD8+ T lymphocytes after 3, 5, and 7 days, by immunofluorescence and flow cytometry. As shown in Figure 1, the majority of T cells proliferated in response to SEB, as quantified by the sequential loss of CFSE fluorescence intensity (R1 gate), while in the absence of SEB, the percentage of dividing T cells was very low (< 5%; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Time-course analysis of MICA and ULBP1–3 expression on both CD4+and CD8+ T cells showed that MICA was highly expressed at day 3, persisted at day 5, and then declined at day 7 after stimulation (see also Figure S5). Expression of ULBPs was instead delayed, because ULBP1 and ULBP2 started to be expressed at day 5 and were still persistent at day 7; in this particular donor, only very low levels of ULBP3 were detected (Figures 1, 2). No major differences were observed between CD4+and CD8+ T cells. We detected a specific staining, because the mAb binding to the cells was completely competed by the corresponding recombinant NKG2DL-Fc, but not by a control-Fc (Figure S2). Unstimulated T cells were negative for all these molecules (Figure S1). Interestingly, expression of NKG2DLs was observed on the majority of T cells that had divided (R1 gate, CFSElow; Figure S3), and only on a small fraction of not-divided T lymphocytes (cells outside the R1 gate). Among proliferating cells (R1 gate, CFSElow), up to 50% were NKG2DL+ (Figure S4). No major differences were observed between CD4+and CD8+ T cells. In addition, when we analyzed the data on the basis of CFSE fluorescence intensity by dividing the dot plots in 3 parts (CFSEhigh, CFSEmedium, CFSElow), CFSEhigh cells had the lowest levels of NKG2DL expression and no major differences were observed between CFSElow and CFSEmedium proliferating cells (Tables S1 and S2).
SEB-activated CD4+ and CD8+ T cells express NKG2D ligands on their cell surface. (A) PBMCs were CFSE-labeled and then stimulated with SEB. After 3, 5, and 7 days cells were harvested and stained with anti-CD8, anti-CD3, and antibodies specific for NKG2D ligands (MICA, ULBP1, ULBP2, and ULBP3), in a 4-color FACS analysis. Subpopulations of CD8+ and CD4+ T lymphocytes were identified by gating on CD8+ CD3+ or CD8− CD3+ cells, respectively, and further analyzed for the expression of NKG2D ligands. Progressive loss of CFSE fluorescence intensity in CD4+ or CD8+ T lymphocytes is indicated by the R1 region. The median of MFI relative to each ligand and calculated on cells gated in R1 (which includes both quadrants to the left) is shown. The percentage of cells in all the quadrants is also indicated. X-axis, CFSE fluorescence intensity. Y-axis, NKG2D ligand fluorescence intensity. A representative donor of 13 tested is shown. (B) MICA, MICB, ULBP1, ULBP2, and ULBP3 transcripts in SEB activated CD3+ T cells, quantitated by real-time PCR. PBMCs were stimulated with SEB, cells were harvested at different times, and CD3+ T lymphocytes were further purified by immunomagnetic positive selection and total RNA was isolated. Real-time PCR was performed as described in Materials and methods. Data were normalized by the amount of β-actin mRNA. The range of CT values for each ligand was as follows: 27 to 29 for MICA, 25 to 27 for MICB, 30 to 34 for ULBP1, 30 to 33 for ULBP2, 33 to 35 for ULBP3, and 15 to 18 for β-actin. (C) Reverse-transcriptase PCR analysis of ULBP4 and GAPDH in SEB-activated T cells. The number of PCR cycles was 36 (ULBP4) or 28 (GAPDH).
SEB-activated CD4+ and CD8+ T cells express NKG2D ligands on their cell surface. (A) PBMCs were CFSE-labeled and then stimulated with SEB. After 3, 5, and 7 days cells were harvested and stained with anti-CD8, anti-CD3, and antibodies specific for NKG2D ligands (MICA, ULBP1, ULBP2, and ULBP3), in a 4-color FACS analysis. Subpopulations of CD8+ and CD4+ T lymphocytes were identified by gating on CD8+ CD3+ or CD8− CD3+ cells, respectively, and further analyzed for the expression of NKG2D ligands. Progressive loss of CFSE fluorescence intensity in CD4+ or CD8+ T lymphocytes is indicated by the R1 region. The median of MFI relative to each ligand and calculated on cells gated in R1 (which includes both quadrants to the left) is shown. The percentage of cells in all the quadrants is also indicated. X-axis, CFSE fluorescence intensity. Y-axis, NKG2D ligand fluorescence intensity. A representative donor of 13 tested is shown. (B) MICA, MICB, ULBP1, ULBP2, and ULBP3 transcripts in SEB activated CD3+ T cells, quantitated by real-time PCR. PBMCs were stimulated with SEB, cells were harvested at different times, and CD3+ T lymphocytes were further purified by immunomagnetic positive selection and total RNA was isolated. Real-time PCR was performed as described in Materials and methods. Data were normalized by the amount of β-actin mRNA. The range of CT values for each ligand was as follows: 27 to 29 for MICA, 25 to 27 for MICB, 30 to 34 for ULBP1, 30 to 33 for ULBP2, 33 to 35 for ULBP3, and 15 to 18 for β-actin. (C) Reverse-transcriptase PCR analysis of ULBP4 and GAPDH in SEB-activated T cells. The number of PCR cycles was 36 (ULBP4) or 28 (GAPDH).
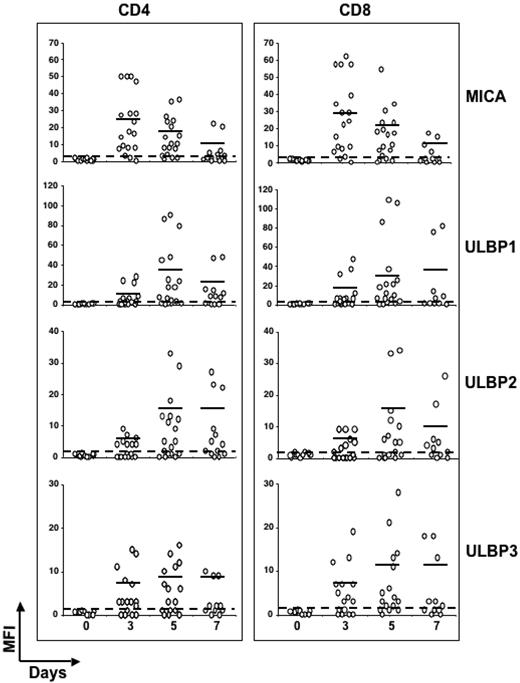
Kinetics of NKG2D ligands expression on SEB-activated CD4 + and CD8 + T cells in several healthy donors. Expression levels of NKG2D ligands were analyzed on SEB-activated CD4+or CD8+ T lymphocytes from different healthy donors, as described in the legend of Figure 1. Each donor is represented by ○. The dashed line delimits negative donors (MFI values < 2). Mean values of MFI are indicated by horizontal bars and were calculated on positive donors (MFI values > 2). Thirteen donors were examined at days 3, 5, and 7, and 6 donors were examined at days 3 and 5.
Kinetics of NKG2D ligands expression on SEB-activated CD4 + and CD8 + T cells in several healthy donors. Expression levels of NKG2D ligands were analyzed on SEB-activated CD4+or CD8+ T lymphocytes from different healthy donors, as described in the legend of Figure 1. Each donor is represented by ○. The dashed line delimits negative donors (MFI values < 2). Mean values of MFI are indicated by horizontal bars and were calculated on positive donors (MFI values > 2). Thirteen donors were examined at days 3, 5, and 7, and 6 donors were examined at days 3 and 5.
Altogether, these results show that NKG2D ligands are mainly expressed on cells undergoing proliferation, independently from the number of cell divisions, but that not all dividing cells are NKG2DL-positive.
Kinetics of NKG2DLs mRNA expression, performed by real-time PCR analysis, confirmed the expression of MICA and ULBP1–3 on SEB-activated T cells (Figure 1B). MICB mRNA was upregulated on activated T cells, but we could not detect its cell surface expression with the available mAbs (data not shown). Quantitative PCR demonstrated that MICA and MICB were the most abundant mRNA while ULBP1, 2, and 3 mRNA were generally present in lower amounts. ULBP4 expression was examined only at the mRNA level, as specific mAbs are not commercially available. SEB-activated T cells did not express ULBP4 mRNA though it was detectable in the Jurkat T-cell leukemia (Figure 1C).
Because gene polymorphism has been described for MIC and ULBP molecules,39-42 we analyzed the expression of NKG2DLs on SEB-activated T lymphocytes in several healthy donors (n = 19). To compare NKG2DLs expression on responding T lymphocytes, a gate including only those cells with reduced CFSE intensity was created, and the MFI value for each ligand was calculated (gate R1, in Figure 1). The kinetics of MICA induction was similar among the different donors, and at day 3 after stimulation it was expressed at the highest levels in the majority of the donors (89%; Figure 2 and Table 1). The greatest percentage of T cells positive for ULBP1 and ULBP2 was instead observed after 5 days, when approximately 75% and 55% of donors expressed ULBP1 and ULBP2, respectively, with similar percentages for both CD4+and CD8+ T cells. Similarly to MICA, MFI values for ULBPs were highly variable, as they comprised from 3 to 109 for ULBP1, and from 3 to 34 for ULBP2 (Table 1). The levels of ULBP3 were quite stable between 3 and 5 days after stimulation, with approximately 63% of donors positive for this ligand and the highest MFI values were reached at day 5. These results indicate that T cells from different donors express different levels of ULBPs and MICA, with minor differences between CD4+and CD8+ T cells. The delayed kinetics in the induction of ULBPs, compared with MICA, suggests that different mechanisms regulate NKG2DL expression on activated T cells.
Kinetics of NKG2D ligands expression on SEB-activated CD4+ and CD8+ T cells
| Cells . | Day . | % CFSE* . | Mean MFI ± SD (range) . | |||
|---|---|---|---|---|---|---|
| MICA . | ULBP1 . | ULBP2 . | ULBP3 . | |||
| CD4 | 3 | 46 ± 16 | 24 ± 16 (3-50) | 11 ± 10 (3-28) | 5 ± 2 (4-9) | 8 ± 5 (3-15) |
| CD4 | 5 | 70 ± 17 | 17 ± 11 (3-36) | 35 ± 33 (3-90) | 14 ± 10 (3-33) | 9 ± 4 (3-16) |
| CD4 | 7 | 73 ± 19 | 10 ± 9 (3-22) | 20 ± 17 (7-48) | 14 ± 10 (4-27) | 9 ± 1 (9-10) |
| CD8 | 3 | 40 ± 16 | 29 ± 20 (3-62) | 18 ± 16 (6-47) | 6 ± 2 (4-9) | 8 ± 5 (3-19) |
| CD8 | 5 | 64 ± 19 | 20 ± 13 (4-54) | 33 ± 33 (3-109) | 14 ± 11 (5-34) | 11 ± 8 (3-28) |
| CD8 | 7 | 72 ± 18 | 12 ± 5 (6-17) | 37 ± 37 (7-82) | 10 ± 9 (3-26) | 11 ± 7 (3-18) |
| Cells . | Day . | % CFSE* . | Mean MFI ± SD (range) . | |||
|---|---|---|---|---|---|---|
| MICA . | ULBP1 . | ULBP2 . | ULBP3 . | |||
| CD4 | 3 | 46 ± 16 | 24 ± 16 (3-50) | 11 ± 10 (3-28) | 5 ± 2 (4-9) | 8 ± 5 (3-15) |
| CD4 | 5 | 70 ± 17 | 17 ± 11 (3-36) | 35 ± 33 (3-90) | 14 ± 10 (3-33) | 9 ± 4 (3-16) |
| CD4 | 7 | 73 ± 19 | 10 ± 9 (3-22) | 20 ± 17 (7-48) | 14 ± 10 (4-27) | 9 ± 1 (9-10) |
| CD8 | 3 | 40 ± 16 | 29 ± 20 (3-62) | 18 ± 16 (6-47) | 6 ± 2 (4-9) | 8 ± 5 (3-19) |
| CD8 | 5 | 64 ± 19 | 20 ± 13 (4-54) | 33 ± 33 (3-109) | 14 ± 11 (5-34) | 11 ± 8 (3-28) |
| CD8 | 7 | 72 ± 18 | 12 ± 5 (6-17) | 37 ± 37 (7-82) | 10 ± 9 (3-26) | 11 ± 7 (3-18) |
Alloantigen stimulation induces NKG2D ligands on CD4+ and CD8+ T lymphocytes
Next, we explored the efficacy of alloantigen stimulation on the induction of NKG2DLs on alloreactive T cells. Purified CD4+or CD8+ T lymphocytes were labeled with CFSE and cocultured with irradiated allogeneic PBMCs for 5 days. Similarly to SEB stimulation, to compare NKG2DLs expression in responding T cells, a gate including only proliferating T cells was made (Figure 3, gate R1), and the MFI values for each ligand were calculated. Alloreactive CD4+ and CD8+ T cells expressed MICA, ULBP1, and ULBP3; very low levels of ULBP2 were detected in this donor. Cells were also positive for the activation antigens CD69 and CD25. Similarly to SEB stimulation, expression of NKG2DLs was mainly observed on those cells with reduced CFSE intensity (Figure 3, gate R1). Induction of NKG2DLs after alloantigen stimulation was examined on 5 donors and we observed that MICA was expressed on all the donors tested, while induction of ULBPs was more variable. ULBP1 and ULBP2 were expressed on approximately 40%, and ULBP3 on approximately 80% of the donors, with no major differences observed between CD4+and CD8+ T cells (data not shown).
Alloantigen stimulation induces NKG2D ligands expression on responder CD4+ and CD8+ T cells. Highly purified CFSE-labeled CD4+or CD8+ T lymphocytes were cultured with irradiated allogeneic PBMCs and harvested after 5 days. Cells were stained with mAbs specific for NKG2D ligands (MICA, ULBP1, 2, and 3) or for the activation markers CD69 and CD25. CD4+or CD8+ T lymphocytes showing a progressive loss of CFSE fluorescence intensity are gated in the R1 region. MFI values relative to each molecule and calculated on cells gated in R1, are reported. The percentage of cells in all the quadrants is also indicated. A representative donor of 5 is shown.
Alloantigen stimulation induces NKG2D ligands expression on responder CD4+ and CD8+ T cells. Highly purified CFSE-labeled CD4+or CD8+ T lymphocytes were cultured with irradiated allogeneic PBMCs and harvested after 5 days. Cells were stained with mAbs specific for NKG2D ligands (MICA, ULBP1, 2, and 3) or for the activation markers CD69 and CD25. CD4+or CD8+ T lymphocytes showing a progressive loss of CFSE fluorescence intensity are gated in the R1 region. MFI values relative to each molecule and calculated on cells gated in R1, are reported. The percentage of cells in all the quadrants is also indicated. A representative donor of 5 is shown.
MICA is induced on secondary antigenic stimulation
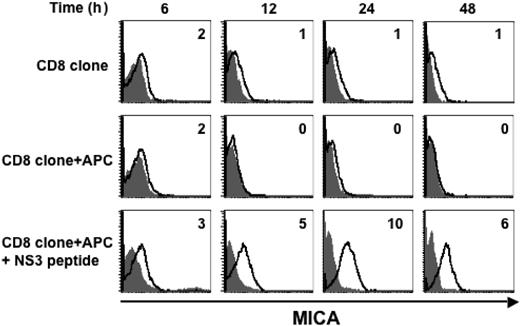
Because MICA was expressed on approximately 90% of donors tested after SEB or alloantigen stimulation, we focused our attention on MICA induction on T cells on a secondary antigenic stimulation. We examined whether MICA expression was induced on a HLA-A2–restricted CD8+ T-cell clone specific for a peptide derived from hepatitis C virus. Antigen stimulation was performed by incubating the CD8+clone for different times with an autologous Epstein-Barr virus-immortalized B lymphoblastoid cell line (APC) pulsed with the hepatitis C virus-derived NS31406-1415 peptide. MICA was induced 12 hours after stimulation, peaked at 24 hours, and declined thereafter (Figure 4). Interestingly, this induction was antigen-specific, because T cells alone, or cocultured with the APC but without peptide, did not express MICA. These results suggest that TCR engagement on Ag-specific T cells induces MICA expression.
MICA is induced on secondary antigenic stimulation. An HLA-A2–restricted CD8+ T-cell clone was cultured alone or with an autologous B lymphoblastoid cell line (APC) pulsed or not with the NS31406-1415 specific peptide derived from hepatitis C virus. Cells were harvested after 6, 12, 24, and 42 hours of culture, and stained with anti-MICA mAb (thick line) or with an isotype control antibody (filled histogram). MFI values relative to MICA, evaluated by gating on CD8+ T cells, are shown. A representative experiment out of 2 is shown.
MICA is induced on secondary antigenic stimulation. An HLA-A2–restricted CD8+ T-cell clone was cultured alone or with an autologous B lymphoblastoid cell line (APC) pulsed or not with the NS31406-1415 specific peptide derived from hepatitis C virus. Cells were harvested after 6, 12, 24, and 42 hours of culture, and stained with anti-MICA mAb (thick line) or with an isotype control antibody (filled histogram). MFI values relative to MICA, evaluated by gating on CD8+ T cells, are shown. A representative experiment out of 2 is shown.
Activated T cells are susceptible to autologous NK- cell–mediated lysis via a mechanism dependent on NKG2D and calcium
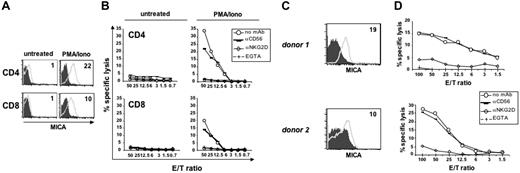
The expression of NKG2DLs on the cell-surface of activated T cells raised the question whether T lymphocytes could be targets of autologous NK-cell–mediated cytotoxicity. In this regard, contrasting results have been reported. In the mouse, a role for NKG2D in NK-cell–mediated lysis of T cells blasts has been demonstrated,17 whereas another study showed that NKG2D was not involved in human T-cell lysis.43 We evaluated the role of NKG2D/NKG2DLs in the NK-cell–mediated lysis of autologous T cells activated by PMA/ionomycin, which mimics TCR signaling (Figure 5A-B) or by SEB (Figure 5C-D). We observed that IL-2–activated NK cells are cytotoxic against activated T cells, and cytotoxicity is blocked by an anti-NKG2D neutralizing antibody (Figure 5B,D), indicating that killing of activated T lymphocytes by autologous NK cells depends on the interaction of NKG2D with its ligands. To investigate if the mechanism responsible of the lysis of activated T cells by NK cells was dependent on the exocytosis of lytic granules, which is a Ca++-dependent event, EGTA was added to the assay. As shown in Figure 5B and 5D, EGTA completely blocked NK-cell–mediated cytotoxicity against PMA/ionomycin-activated CD4+and CD8+ T cells, or against SEB-activated T cells, demonstrating that killing is mediated by the secretory pathway. In conclusion, these results indicate that NK cells can efficiently lyse autologous activated T cells with a mechanism dependent on NKG2D/NKG2DL interaction and release of lytic granules.
NK-cell–mediated cytotoxicity of autologous T-cell blasts is dependent on NKG2D and calcium. (A) Purified CD4+and CD8+ T lymphocytes were cultured with medium alone or stimulated with PMA/ionomycin for 48 hours, and then stained with anti-MICA mAb (thick line) or control Ig (filled histogram). (B) Cytotoxicity assay was performed using untreated or PMA/ionomycin-activated CD4+or CD8+ T lymphocytes as targets, and IL-2–activated autologous NK cells as effectors, at the indicated effector-target ratios. The assay was performed in the presence of an anti-NKG2D neutralizing mAb, of an anti-CD56 mAb used as isotype control, or of the Ca+‖+ chelator EGTA (final concentration of 4 mM). CD4+ T cells from 4 donors and CD8+ T cells from 3 donors were analyzed. Data shown derive from the same donor. (C) PBMCs were stimulated with SEB and after 3 days cells were stained with anti-CD3 and mAbs specific for NKG2D ligands. MFI values relative to MICA (thick line) and evaluated on CD3+ T cells are shown. Control IgG, filled histogram. MFI values of ULBP molecules were as follows: ULBP1 = 0, ULBP2 = 0, ULBP3 = 2 for donor 1; ULBP1 = 1, ULBP2 = 3, ULBP3 = 4 for donor 2 (data not shown). CD3+ T cells in both donors were approximately 80%. (D) The same cells described in panel C were used as targets in a cytotoxicity assay, together with IL-2-activated autologous NK cells as effectors. Data shown derive from 2 of 3 donors.
NK-cell–mediated cytotoxicity of autologous T-cell blasts is dependent on NKG2D and calcium. (A) Purified CD4+and CD8+ T lymphocytes were cultured with medium alone or stimulated with PMA/ionomycin for 48 hours, and then stained with anti-MICA mAb (thick line) or control Ig (filled histogram). (B) Cytotoxicity assay was performed using untreated or PMA/ionomycin-activated CD4+or CD8+ T lymphocytes as targets, and IL-2–activated autologous NK cells as effectors, at the indicated effector-target ratios. The assay was performed in the presence of an anti-NKG2D neutralizing mAb, of an anti-CD56 mAb used as isotype control, or of the Ca+‖+ chelator EGTA (final concentration of 4 mM). CD4+ T cells from 4 donors and CD8+ T cells from 3 donors were analyzed. Data shown derive from the same donor. (C) PBMCs were stimulated with SEB and after 3 days cells were stained with anti-CD3 and mAbs specific for NKG2D ligands. MFI values relative to MICA (thick line) and evaluated on CD3+ T cells are shown. Control IgG, filled histogram. MFI values of ULBP molecules were as follows: ULBP1 = 0, ULBP2 = 0, ULBP3 = 2 for donor 1; ULBP1 = 1, ULBP2 = 3, ULBP3 = 4 for donor 2 (data not shown). CD3+ T cells in both donors were approximately 80%. (D) The same cells described in panel C were used as targets in a cytotoxicity assay, together with IL-2-activated autologous NK cells as effectors. Data shown derive from 2 of 3 donors.
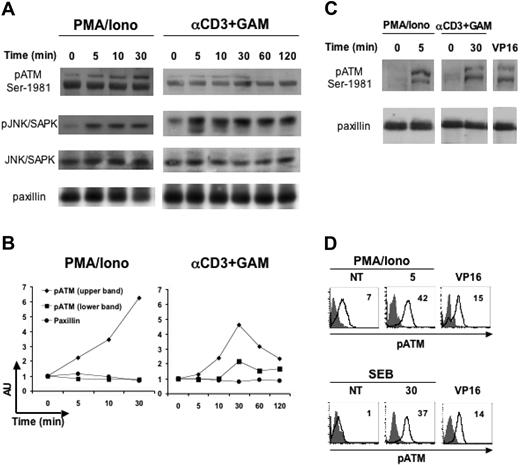
ATM phosphorylation is induced on TCR triggering
Next, we wanted to characterize some of the intracellular pathways leading to the induced expression of MICA on activated T cells. So far, little is known about signaling molecules and transcription factors controlling the expression of this molecule on normal hematopoietic cells.44 Recently, it has been shown that murine and human NKG2DLs are upregulated in nontumor cell lines by genotoxic stress and stalled DNA replication with a mechanism involving ATM or ATR kinases.33 It was previously shown that ATM protein is present in freshly isolated PBMCs and that its expression increases upon PHA stimulation, suggesting a more general role for this kinase than signaling in the response to DNA damage.45 Thus, we asked whether ATM could play a role in the induction of MICA expression on activated T cells. First, we investigated if T-cell activation results in stimulation of ATM phosphorylation by using a phosphospecific Ab directed against serine 1981.46 PBMCs were starved for 18 hours in 0.1% fetal calf serum to decrease the basal level of ATM phosphorylation (data not shown), and then stimulated at different times with PMA/ionomycin or with an anti-CD3 mAb. In response to both stimuli, increased ATM phosphorylation was evident already at 5 minutes after stimulation and remained sustained up to 30 minutes (Figure 6A-B). Cell activation was confirmed by immunoblotting with a mAb specific for the anti-phospho-JNK/SAPK, a kinase activated on TCR engagement,47 while equal protein loading was verified with antitotal JNK or antipaxillin Ab. The double bands we detected probably represent different phosphorylated ATM molecular species exhibiting different electrophoretic mobilities. In fact, it has been reported that ATM can be constitutively phosphorylated48 and undergoes hyperphosphorylation upon DNA damage with at least 3 functionally significant autophosphorylation sites (ie, serine 1981, 1893, and 367).46,49 A similar pattern of phosphorylated bands reacting with anti-pATM Ab was observed in PBMCs treated with VP16 (Figure 6C), a genotoxic agent previously shown to induce DNA damage-dependent ATM-Ser1981 phosphorylation.50 T-cell activation-dependent and VP16-dependent increase of phosphorylated ATM was detected also by intracellular staining (Figure 6D).51,52 Thus, despite a basal level of constitutive ATM phosphorylation that we detected in some but not all donors, both by immunoblotting and FACS analysis, our results indicate that T-cell activation results in stimulation of ATM phosphorylation.
ATM phosphorylation is stimulated on T-cell activation with anti-CD3, PMA/ionomycin or SEB antigen. (A) PBMCs were starved for 18 hours in 0.1% fetal calf serum, and then left untreated or stimulated with PMA (50 ng/mL) plus ionomycin (500 ng/mL), or with anti-CD3 mAb followed by a GAM cross-linking, for the indicated times at 37°C. Cell lysates (100 μg/lane) were immunoblotted with anti-phospho-ATM (specific for the phosphoserine 1981), or with anti-phospho-JNK/SAPK, used as a control of T-cell activation. Protein loading was normalized after stripping and reprobing the latter membrane with antitotal-JNK/SAPK mAb, or with an antipaxillin mAb. Data shown derive from the same experiment and are representative of 1 of 2 (for CD3) or 3 (for PMA/ionomycin) independent experiments. (B) Densitometric analysis of the bands shown in panel A, and relative to pATM (upper and lower bands) and to paxillin (as a control). The relative protein level of stimulated samples with respect to that of unstimulated cells (time 0) is shown. AU, arbitrary units. (C) PBMCs were starved and then stimulated with PMA/ionomycin or anti-CD3+GAM as described in panel A, or incubated with VP16 (10 μM) for 2 hours, and lysed immediately after. Data derive from the same donor analyzed in the same experiment and are representative of 2 donors. (D) PBMCs were starved and stimulated as described, and then stained anti-pATM-Ser1981 (thick line) or cIgG (filled histogram). MFI values relative to pATM are shown. Data are from 2 different donors, one of which (SEB/VP16) is the same as in panel C.
ATM phosphorylation is stimulated on T-cell activation with anti-CD3, PMA/ionomycin or SEB antigen. (A) PBMCs were starved for 18 hours in 0.1% fetal calf serum, and then left untreated or stimulated with PMA (50 ng/mL) plus ionomycin (500 ng/mL), or with anti-CD3 mAb followed by a GAM cross-linking, for the indicated times at 37°C. Cell lysates (100 μg/lane) were immunoblotted with anti-phospho-ATM (specific for the phosphoserine 1981), or with anti-phospho-JNK/SAPK, used as a control of T-cell activation. Protein loading was normalized after stripping and reprobing the latter membrane with antitotal-JNK/SAPK mAb, or with an antipaxillin mAb. Data shown derive from the same experiment and are representative of 1 of 2 (for CD3) or 3 (for PMA/ionomycin) independent experiments. (B) Densitometric analysis of the bands shown in panel A, and relative to pATM (upper and lower bands) and to paxillin (as a control). The relative protein level of stimulated samples with respect to that of unstimulated cells (time 0) is shown. AU, arbitrary units. (C) PBMCs were starved and then stimulated with PMA/ionomycin or anti-CD3+GAM as described in panel A, or incubated with VP16 (10 μM) for 2 hours, and lysed immediately after. Data derive from the same donor analyzed in the same experiment and are representative of 2 donors. (D) PBMCs were starved and stimulated as described, and then stained anti-pATM-Ser1981 (thick line) or cIgG (filled histogram). MFI values relative to pATM are shown. Data are from 2 different donors, one of which (SEB/VP16) is the same as in panel C.
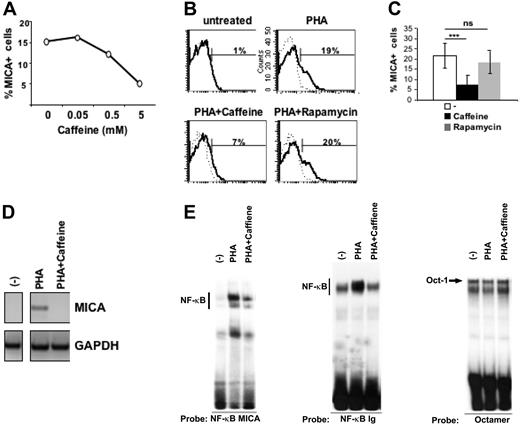
Caffeine treatment inhibits MICA induction on activated T cells in an NF-κB-dependent manner
Caffeine inhibitor is widely used to block ATM and ATR catalytic activity.53 Thus, we asked whether caffeine could interfere with the induction of MICA expression on activated T cells. PBMCs were pretreated with several doses of caffeine and stimulated with PHA for 16 hours. MICA expression was induced on approximately 15% of PHA-stimulated T cells and caffeine inhibited MICA induction in a dose-dependent manner (Figure 7A). Caffeine has also been reported to inhibit mTOR, a protein kinase regulating cell growth and metabolism.53 In an attempt to explore whether caffeine-induced inhibition of MICA expression could be also related to its ability to block mTOR, we analyzed the effect of rapamycin, a specific inhibitor of mTOR but not of ATM/ATR54 on MICA expression. Pretreatment of T lymphocytes with this inhibitor did not significantly affect MICA induction, probably excluding a role of mTOR in the control of TCR-triggered MICA expression (Figure 7B-C). In addition, we found that caffeine-dependent inhibition of MICA was at the mRNA level (Figure 7D).
Caffeine treatment inhibits MICA induction on activated T cells in an NF-κB dependent-manner. (A) PBMCs were pretreated with different doses of caffeine and then stimulated with PHA for 18 hours. Cells were stained with mAbs specific for CD3 and MICA and its expression was evaluated on CD3+ T cells. Data are represented as percentage of MICA-positive cells. A representative experiment of 4 is shown. (B) PBMCs were pretreated with caffeine (5 mM) or rapamycin (1 μM) and then stimulated with PHA for 18 hours. Cells were stained with mAbs specific for CD3, MICA (thick line), or control Ig isotype (dashed line). Expression of MICA was evaluated on CD3+ T cells. (C) PBMCs were prepared as described in panel B. Data are represented as the mean plus or minus one SD of the percentage of CD3+MICA+cells of 8 different healthy donors. Significant differences, as calculated by paired t test, are indicated: ***P < .001; ns, not significant. (D) PBMCs were pretreated with 5 mM caffeine and then stimulated with PHA. After 18 hours, CD3+ T cells were purified by positive immunomagnetic selection. Total RNA was isolated and used for reverse-transcription PCR reactions with primers specific for MICA and GAPDH. A representative experiment out of 5 is shown. (E) Electrophoretic mobility-shift assay was performed using the 32P-labeled NF-κB MICA, and the canonical NF-κB Ig oligonucleotide as a probe in the presence of nuclear extracts (10 μg), from unstimulated (−) or PHA-activated PBMCs (3 hours). Where indicated, PBMCs were pretreated with 5 mM caffeine and then stimulated with PHA for 3 hours. The same nuclear extracts were also used with a Octamer factor(s)-specific probe as a control. A representative experiment out of 3 is shown.
Caffeine treatment inhibits MICA induction on activated T cells in an NF-κB dependent-manner. (A) PBMCs were pretreated with different doses of caffeine and then stimulated with PHA for 18 hours. Cells were stained with mAbs specific for CD3 and MICA and its expression was evaluated on CD3+ T cells. Data are represented as percentage of MICA-positive cells. A representative experiment of 4 is shown. (B) PBMCs were pretreated with caffeine (5 mM) or rapamycin (1 μM) and then stimulated with PHA for 18 hours. Cells were stained with mAbs specific for CD3, MICA (thick line), or control Ig isotype (dashed line). Expression of MICA was evaluated on CD3+ T cells. (C) PBMCs were prepared as described in panel B. Data are represented as the mean plus or minus one SD of the percentage of CD3+MICA+cells of 8 different healthy donors. Significant differences, as calculated by paired t test, are indicated: ***P < .001; ns, not significant. (D) PBMCs were pretreated with 5 mM caffeine and then stimulated with PHA. After 18 hours, CD3+ T cells were purified by positive immunomagnetic selection. Total RNA was isolated and used for reverse-transcription PCR reactions with primers specific for MICA and GAPDH. A representative experiment out of 5 is shown. (E) Electrophoretic mobility-shift assay was performed using the 32P-labeled NF-κB MICA, and the canonical NF-κB Ig oligonucleotide as a probe in the presence of nuclear extracts (10 μg), from unstimulated (−) or PHA-activated PBMCs (3 hours). Where indicated, PBMCs were pretreated with 5 mM caffeine and then stimulated with PHA for 3 hours. The same nuclear extracts were also used with a Octamer factor(s)-specific probe as a control. A representative experiment out of 3 is shown.
Recently, ATM has been shown to activate NF-κB signaling in response to genotoxic stimuli.50 Moreover, an NF-κB binding site in the intron 1 of the MICA gene has been identified, and NF-κB has been proposed to be an important regulator of MICA expression both in activated T lymphocytes and tumor cells.37 Thus, we wanted to investigate if inhibition of ATM/ATR kinases mediated by caffeine could have an effect on NF-κB binding to a canonical binding site and to the intronic NF-κB binding site of the MICA gene. Mobility-shift assays were thus performed on nuclear extracts from PHA-activated T cells, pretreated or not with caffeine. As shown in Figure 7E, PHA stimulation of T cells induced the activation of NF-κB, as measured by its increased binding to the MICA-NF-κB binding site and to a canonical NF-κB site used as a control, and treatment with caffeine significantly inhibited DNA-binding complex formation. As control for equal protein loading, the same amount of nuclear extract was tested in the presence of an Octamer factor(s)-specific probe. The DNA binding of Octamer-1 was unaltered in caffeine-treated cells. Collectively, these data suggest that caffeine, an inhibitor of ATM/ATR kinases, blocks MICA induction on activated T cells through a mechanism involving the inhibition of NF-κB. Moreover, in an effort to elucidate the role of ATM versus ATR in the induction of MICA, we used KU-55933, a recently commercialized specific inhibitor of ATM kinase.55 KU-55933 markedly inhibited MICA expression on activated T cells (Figure S6), clearly demonstrating the involvement of ATM in this event.
Discussion
This study is the first broad description of the expression of human NKG2DLs on antigen-activated CD4+and CD8+ T cells. Our results show that MICA and ULBP1, ULBP2, and ULBP3, but not ULBP4, are expressed on T lymphocytes in response to superantigen, alloantigen, or to a specific antigenic peptide. While we did not observe major differences between CD4+and CD8+ T cells, the levels of NKG2DLs cell surface expression were highly variable among different donors (Figure 2 and Table 1). The variability in NKG2DLs expression—recently reported also for surface expression of ULBPs on B cells, monocytes, and granulocytes30 —can be related to their gene polymorphism. More than 50 alleles of MICA have been reported so far, and amino acid differences in the extracellular domain could affect antibody binding resulting in different levels of expression and, more importantly, in different binding affinities for NKG2D.40,56 Moreover, polymorphism in the promoter sequences of ULBP genes has been recently described, probably determining different affinity or specificity of transcription factor binding or kinetics of transcription initiation, and thus resulting in different ULBPs expression.39 NKG2DLs polymorphism could be one major reason for the discrepancy between our results and those obtained by Molinero et al, who reported that T cells activated by PMA/ionomycin for 72 hours or by PHA plus IL-2 for 96 hours, did not show consistent cell-surface MICA and ULBP1–3.43 Thus, stimulation of T cells at different times and in different experimental settings could be responsible for variable levels of cell-surface NKG2DLs. Another explanation for the discrepancy between the 2 studies may be related to different anti-MICA mAbs used. In fact, when we analyzed the same donors with 2 different antibodies, we observed a different degree of reactivity, with the highest MICA MFI values obtained using the mAb M673 (“Materials and methods”).
Previously, expression of NKG2DLs was thought to be mostly restricted to transformed, infected, and/or stressed cells. Today, however, this view is changing, as more and more experimental evidences report that MIC and ULBPs can be expressed also on normal hematopoietic cells, including bone-marrow cells and mature DCs.28,30 Expression of MICA/B on monocyte-derived DCs is induced by IL-15 and type I IFN.28 Expression of ULBPs increases during hematopoietic differentiation on human bone marrow cells and their expression is associated with the loss of the early hematopoietic marker CD34 and the acquisition of the myeloid markers CD33 and CD14.30 In mouse bone marrow grafts, the NKG2DL Rae-1 was detected on most donor proliferating hematopoietic cells, mainly myeloid lineage progenitors, repopulating an irradiated recipient.27 Similarly, we observed that NKG2DLs were mainly expressed on those T cells that had gone through at least one cell division, as tracked by CFSE labeling. We also observed that NKG2DLs were expressed on a large fraction of proliferating cells, ranging from 10% to 50%. Overall these findings indicate that induction of NKG2DLs on normal cells appears to be related to a particular state of activation, proliferation, and differentiation, but the molecular mechanisms controlling their expression in different normal hematopoietic cells are still elusive.
In an effort to investigate the signaling pathways involved in the regulation of MICA expression on antigen-activated T cells, we focused our attention on ATM and NF-κB. ATM belongs to a family of proteins regulating cell-cycle checkpoints and DNA repair and recombination,57 and mutations in the ATM gene are responsible for ataxia-telangiectasia, a rare autosomal recessive inherited disorder characterized by neurodegeneration, immunodeficiency, and cancer predisposition.58 ATM is involved in NKG2DL upregulation in cells exposed to ionizing radiation or hypotonic conditions,33 and promotes NF-κB activation in response to DNA damaging agents.50,59 NF-κB in turn regulates MICA expression in activated T lymphocytes.37 ATM is a protein kinase of 370 kDa and in normal cells is held inactive as a dimer (or a multimer), with the kinase domain bound to a region surrounding serine 1981. On DNA damage, each molecule of the dimer phosphorylates the other one on serine 1981, as well as on serine 367 and 1893, releasing fully active monomers, capable in turn of phosphorylating several mediators of the cell-cycle checkpoint response.46,49,60 Although ATM has been best characterized in irradiation-induced DNA double-strand breaks and consequent p53 activation, a role as a more versatile protein is emerging. In fact, ATM is activated in response to insulin,61 and “by default” during mitosis in undamaged cells.62 In addition, upregulation of ATM protein levels has been observed in PBMCs in response to mitogenic stimuli.45 In this study, we show for the first time, by Western blot and flow cytometry, that T-cell activation by anti-CD3 antibody, SEB, or PMA/ionomycin treatment results in stimulation of ATM phosphorylation on serine 1981. Our results also demonstrate that caffeine, the inhibitor of ATM/ATR kinases, blocks MICA induction on activated T cells through a mechanism involving the inhibition of NF-κB. Activation of NF-κB by ATM has been recently reported by Wu et al, who demonstrated that ATM selectively associates with NEMO to promote NF-κB activation in human PBL treated with VP16.50 Gasser et al33 have shown that induction of NKG2DLs in response to DNA-damaging agents and DNA synthesis inhibitors, requires ATM or ATR depending on the nature of the stimulus. Although further experiments are needed to understand the specific role played by the kinases in the regulation of NKG2DLs expression on T-cell activation, the inhibition mediated by the specific pharmacologic inhibitor of ATM, KU-55933, strongly supports a role for ATM in the regulation of MICA expression. It will also be important to elucidate if similar signaling pathways upstream of ATM activation lead to NKG2DLs expression in response to antigen recognition or DNA damage.
Expression of MICA on the cell surface of activated T lymphocytes rendered them susceptible to lysis mediated by autologous activated NK cells. The observed killing was dependent on the NKG2D/NKG2DLs interaction and on granule exocytosis, because it was blocked by an anti-NKG2D mAb and by EGTA. Our results are in line with previous evidence showing that murine T-cell blasts are killed by syngeneic NK cells in an NKG2D-dependent manner.17 In another study, Pende et al63 showed that an NK-cell clone displayed an NKG2D-dependent lysis against autologous PHA-blasts, only if the engagement of inhibitory receptors by HLA class I was masked by Ab-mediated blocking. Even though it is possible that other NK activating receptors may participate in the recognition of activated T cells, the discrepancies between the different studies may be related to different levels of NKG2DLs expression and on the different activation status of NK cells.
Induction of NKG2DLs on activated T lymphocytes during the course of an immune response might allow the establishment of a cross-talk with NK cells as well as with other cells expressing the NKG2D receptor, such γδ and CD8+ T cells. This interaction may trigger a granule exocytosis (perforin)-dependent lysis of T cells expressing NKG2DLs and act as a negative regulator of T-cell responses. Interestingly, a role for perforin not only as effector mechanism, but also as immune regulator has been demonstrated. Perforin-deficient patients show lymphoproliferative disorders64 and perforin-mediated killing is involved in downregulating T-cell responses in vivo, because perforin-deficient mice show a huge expansion of activated T cells on chronic lymphocytic choriomeningitis virus (LCMV) infection.65 Moreover, mice deficient in both Fas and perforin have a dramatic acceleration of the spontaneous lymphoproliferative disease seen in Fas-deficient (lpr) mice.65,66 Thus, the receptor/ligand interactions that trigger a perforin-mediated cytotoxicity play a key role in controlling T-cell responses during viral infections, autoimmunity and, most likely, transplantation. During allogeneic hematopoietic cell transplantations, donor T cells in the graft can mediate graft-versus-host disease, which is initiated by host DCs presenting alloantigens to donor T cells;67 graft-versus-host disease has been suggested to be prevented through killing of DCs by alloreactive NK cells.68 We envisage that these NK cells could also kill activated donor T cells (expressing NKG2DLs), thus providing an additional mechanism to protect the host from the detrimental effects of graft-versus-host disease.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Daniele Accapezzato and Vincenzo Barnaba for kindly providing the CD8+ T-cell clone, David Cosman and Amgen for the anti-NKG2D ligands antibodies, Alessandra Soriani for her precious advice on real-time PCR, and Michele Ardolino for help.
This work was supported by grants from the Italian Association for Cancer Research (AIRC), the Italian Ministry of University and Research (MIUR), and the Center of Excellence (BEMM).
Authorship
Contribution: C.C. and A.Z. equally contributed to the work, designed research, performed experiments, and wrote the paper. M.C. performed electrophoretic mobility-shift assay and discussed results. M.P. and L.F. supervised the laboratory activities. A.S. designed research and contributed to paper writing.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alessandra Zingoni, Department of Experimental Medicine, University of Rome “La Sapienza,” Policlinico “Umberto I,” Viale Regina Elena 324, 00161 Rome, Italy; e-mail: alessandra.zingoni@uniroma1.it.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal