Abstract
Clonality often defines the diseased state in hematology. Clonal cells are genetically homogenous and derived from the same precursor; their detection is based on genotype or phenotype. Genotypic clonality relies on somatic mutations to mark the clonal population. Phenotypic clonality identifies the clonal population by the expression pattern of surrogate genes that track the clonal process. The most commonly used phenotypic clonality methods are based on the X-chromosome inactivation principle. Clonality detection based on X-chromosome inactivation patterns (XCIP) requires discrimination of the active from the inactive X chromosome and differentiation of each X chromosome's parental origin. Detection methods are based on detection of X-chromosome sequence polymorphisms identified by protein isoforms, transcribed mRNA, and methylation status. Errors in interpreting clonality tests arise from stochastic, genetic, and cell selection pressures on the mechanism of X inactivation. Progressive X-chromosome skewing has recently been suggested by XCIP clonality studies in aging hematopoietic cells. This has led to new insights into the pathophysiology of X-linked and autoimmune disorders. Other research applications include combining XCIP clonality testing with genetic clonality testing to identify clonal populations with yet-to-be-discovered genetic changes.
Introduction
Clonality studies have played an instrumental role in medicine and hematology. They have been used to establish the single origin of tumors and to differentiate nonmalignant from malignant states.1 Most importantly, they have been used to elucidate the hematopoietic hierarchy.2,3 Much confusion exists, however, regarding the application and interpretation of clonality studies. We discuss clonality testing in general, emphasizing X-chromosome inactivation pattern (XCIP)–based assays and the pitfalls in their interpretation. We also discuss new research applications of clonality testing.
Definition of clonality
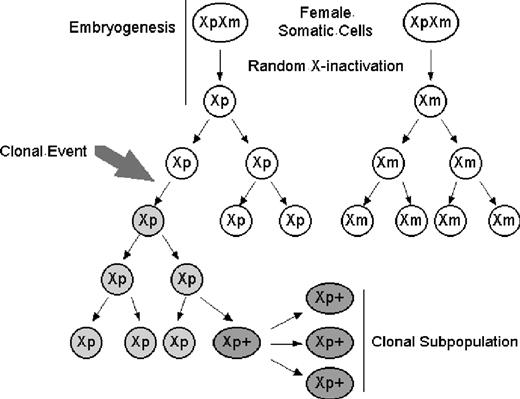
For cells to be clonal means that they are derived from a single precursor that has undergone a somatic mutation. Clonality is a description of lineage as well as homogeneity of the tested tissue. These aspects of clonality can be seen in the acquisition of cytogenetic abnormalities on leukemic transformation in myelofibrosis with myeloid metaplasia.4 Of 28 patients with serial karyotype analysis performed at the time of myelofibrosis with myeloid metaplasia diagnosis and leukemic transformation, 26 demonstrated clonal evolution. Eleven of 26 had evolution of an existing clone with additional cytogenetic abnormalities, whereas 15 of 26 with a normal karyotype acquired one or more cytogenetic abnormalities. Here, a single clonal population could be defined by the first cytogenetic change, whereas a second clonal population could be defined by the additional cytogenetic changes in 11 of 26 patients. These later cells would still have the original clonality marker shared with the original clone, but they would also represent a genetically distinct second clonal population evolving from the original chromosomal aberration (Figure 1).
X-chromosome inactivation and clonal development with further subclone evolution. (A) Clonal populations can acquire multiple genetic abnormalities; each can be used to distinguish subpopulations which remain clonal. Round white Xp, nonclonal cell with active paternal X chromosome; round gray Xp, clonal cell with active paternal X chromosome; oval dark gray Xp+, clonal cell with active paternal X chromosome and new somatic mutation. (B) X-chromosome inactivation. Early in development, both X chromosomes are expressed. Before hematopoietic lineage differentiation, either X chromosome is inactivated. This random choice of inactivated X chromosome is retained through subsequent mitoses. Oval XpXm indicates early embryonic cells expressing both maternal and paternal X chromosomes; round Xp or Xm, nonclonal cells expressing a single X chromosome.
X-chromosome inactivation and clonal development with further subclone evolution. (A) Clonal populations can acquire multiple genetic abnormalities; each can be used to distinguish subpopulations which remain clonal. Round white Xp, nonclonal cell with active paternal X chromosome; round gray Xp, clonal cell with active paternal X chromosome; oval dark gray Xp+, clonal cell with active paternal X chromosome and new somatic mutation. (B) X-chromosome inactivation. Early in development, both X chromosomes are expressed. Before hematopoietic lineage differentiation, either X chromosome is inactivated. This random choice of inactivated X chromosome is retained through subsequent mitoses. Oval XpXm indicates early embryonic cells expressing both maternal and paternal X chromosomes; round Xp or Xm, nonclonal cells expressing a single X chromosome.
Clonality detection
Clonality detection requires a way to mark cells that is retained generation after generation, enabling lineage to be traced by following the marker back to a single progenitor. Cells can be marked by genotype or phenotype (Table 1). In genotypic clonality, an aberrant genetic sequence marks the cell and its progeny. The aberrant sequence is usually related to the disease either as its cause or as a lineage marker of the original precursor that has undergone the disease causing somatic mutation. Examples of genetic clonality markers include the 9q+:22q-translocation/BCR-ABL fusion in chronic myelogenous leukemia and immunoglobulin or T-cell receptor gene rearrangements in lymphoid malignancies.5,6 The aberrant sequences used for marking are somatic mutations, simplifying the methodology for detection and differentiation of the disease clone from normal, nonmutated cells.
Methods of clonality detection
| Marker . | Detection method . | Clinical application . | Research application . |
|---|---|---|---|
| Genotypic | |||
| Disease-associated somatic mutation | PCR; cytogenetics | Differentiation of clonal from reactive polyclonal processes; diagnosis and subtyping of leukemia and lymphoma; MRD monitoring in acute leukemia | Demonstration of clonal processes, lineage, clonal evolution |
| Immunoglobulin rearrangement | PCR | Differentiation of clonal from reactive polyclonal processes; demonstration of clonality, lineage, and differentiation of B-cell lymphoma/leukemia; MRD monitoring in B-cell lymphoma/leukemia | Demonstration of B-cell clonality, lineage, differentiation |
| T-cell–receptor rearrangement | PCR | Demonstration of clonality, lineage, and differentiation of T-cell lymphoma/leukemia; MRD monitoring in T-cell lymphoma/leukemia | Demonstration of T-cell clonality, lineage, differentiation |
| Phenotypic | |||
| Immunophenotype | Flow cytometry | Diagnosis of malignancy; determination of clonality, lineage; differentiation of leukemias/lymphomas; MRD monitoring in acute leukemias | Demonstration of clonality, lineage, clonal evolution |
| X-chromosome inactivation pattern | Protein isoform detection by electrophoretic mobility*; methylation of X-chromosome polymorphic region†; transcription-based assays‡ | Differentiation of clonal from reactive polyclonal processes | Demonstration of clonality, lineage; elucidation of the hematopoietic hierarchy; possible identification of premalignant disease phases; facilitation of demonstration of subclonal development from unrelated somatic processes |
| Marker . | Detection method . | Clinical application . | Research application . |
|---|---|---|---|
| Genotypic | |||
| Disease-associated somatic mutation | PCR; cytogenetics | Differentiation of clonal from reactive polyclonal processes; diagnosis and subtyping of leukemia and lymphoma; MRD monitoring in acute leukemia | Demonstration of clonal processes, lineage, clonal evolution |
| Immunoglobulin rearrangement | PCR | Differentiation of clonal from reactive polyclonal processes; demonstration of clonality, lineage, and differentiation of B-cell lymphoma/leukemia; MRD monitoring in B-cell lymphoma/leukemia | Demonstration of B-cell clonality, lineage, differentiation |
| T-cell–receptor rearrangement | PCR | Demonstration of clonality, lineage, and differentiation of T-cell lymphoma/leukemia; MRD monitoring in T-cell lymphoma/leukemia | Demonstration of T-cell clonality, lineage, differentiation |
| Phenotypic | |||
| Immunophenotype | Flow cytometry | Diagnosis of malignancy; determination of clonality, lineage; differentiation of leukemias/lymphomas; MRD monitoring in acute leukemias | Demonstration of clonality, lineage, clonal evolution |
| X-chromosome inactivation pattern | Protein isoform detection by electrophoretic mobility*; methylation of X-chromosome polymorphic region†; transcription-based assays‡ | Differentiation of clonal from reactive polyclonal processes | Demonstration of clonality, lineage; elucidation of the hematopoietic hierarchy; possible identification of premalignant disease phases; facilitation of demonstration of subclonal development from unrelated somatic processes |
Limited to females of African origin.
Majority of females informative but influenced by environmental factors.
Several polymorphic loci need to be screened, better differentiation of active from inactive X chromosome.
The multitude of lymphoma and leukemia markers using T-cell receptor/immunoglobulin gene rearrangements and chromosomal translocations illustrates the clinical use of genetic clonality markers. However, clonal cell populations without the tested-for genetic markers exist and will not be detected. For detection of clonal populations that do not have known genetic markers, detection of clonality by phenotypic assays is necessary. It is not that genetic changes do not underlie these clonal processes, but rather because the specific mutation is not known, clonality testing will need to depend on those surrogate markers whose expressions are incidentally altered by or track the clonal process.
In contrast to genotypic clonality, which identifies cells using somatic mutations, phenotypic clonality identifies cells using changes in the expression pattern of housekeeping genes. These genes are not mutated from their germline sequences but are differentially expressed. Phenotypic clonality can be assayed by performing flow cytometry for abnormally expressed antigens or by using XCIP-based techniques to detect the loss of heterozygous allele expression. The application of flow cytometry to clonality detection is discussed in the following paragraphs. XCIP-based assays are discussed in “X chromosome inactivation” and the subsections that follow.
In flow cytometry, clonal populations are identified by the constellation of antigens expressed by the cell. These antigens can demonstrate aberrant lineage, asynchronous expression, or overexpression. Aberrant light scattering properties can also be used to identify clonal populations.
Flow cytometry has been applied to the detection of residual clonal populations after treatment (minimal residual disease [MRD]) for acute myelogenous leukemia (AML) and acute lymphoblastic leukemia (ALL). In AML, immunophenotyping can detect clonal populations with a sensitivity as small as one leukemic cell diluted 104 to 105 times (similar to the sensitivity for polymerase chain reaction [PCR]).7 The presence of such populations has been linked to clinically relevant relapses, although the actual cutoffs vary with the clinical study.7,8
Clonality markers, genotypic or phenotypic, are surrogates for clonal processes and must be differentiated from the actual clonal processes. The absence of a clonality marker does not equate to the absence of a clonal population. Clonal evolution frequently occurs in acute lymphoblastic leukemia with concomitant surface antigen shifts.9 In the process, previously detected immunophenotypes may become undetectable, suggesting a loss of clonality when in fact a clonal population still exists, either changed by another clonal somatic event or arising from a previously existent but undetected subclone resulting in a false-negative interpretation. A similar situation arises when PCR-based genetic markers are used.10 False-negative interpretations of MRD can be avoided by following multiple clonality markers so that if one is lost, another can still track the clonal population.
The context of minimal residual clonal population detection affects the interpretation. Whether these small populations result in clinically relevant outcomes depends on the disease. In promyelocytic leukemia, detection of a small residual population by reverse transcription PCR for the normalized quotient of promyelocytic leukemia–retinoic acid receptor (PML-RAR) mRNA/glyceraldehyde-3-phosphate dehydrogenase (GAPDH) mRNA predicts a 7.6-fold increase in relative risk for disease recurrence if greater than 10−6 and a 17.5-fold increase in relative risk if greater than 10−5. At less than 10−6, relative risk for relapse was 0.18.11 In contrast, small residual populations in t(8;21) AML may exist for as long as 8 years without clinical relapse.12 Similar observations have been reported for acute lymphoblastic leukemia in which molecularly detected residual leukemic clones in bone marrow did not result in clinical relapse for as long as 9 years.13 These instances of clinically insignificant MRD may be attributable to eradication of more aggressive subclones with persistence of “premalignant” clones, differences in disease biology, or to control of the clones by immunologic or other homeostatic mechanisms.
The timing of MRD also affects interpretation. In chronic myelogenous leukemia, MRD detected by qualitative reverse transcriptase-PCR of BCR-ABL less than 6 months after transplantation is not predictive for relapse. If detected 6 to 12 months after transplantation, PCR-positive MRD is associated with a 26-fold increase in relative risk for relapse.14 Progressive immune reconstitution after transplantation may make early positive results less significant, and the use of qualitative 2-step nested PCR to amplify BCR-ABL transcripts may introduce variance in the level detected. Indeed, if a quantitative reverse transcriptase-PCR detection method is used, then an early positive result within 3 to 5 months of transplantation coupled with increasing transcript numbers is predictive of relapse.15,16 In summary, interpretation of MRD must consider the disease, timing, and detection method.
The remainder of this article deals with clonality assays based on X-chromosome inactivation patterns. These assays can be classified as phenotypic assays because the change in expression of an X-linked gene is used as the marker of cell origin rather than disease-causing or disease-related genetic changes. We also discuss the methodologies and pitfalls in XCIP interpretation. The relationship between genotypic detection of clonality, phenotypic detection of clonality, and the discovery of disease-causing genes is discussed in “Integrating genotypic and phenotypic clonality detection.”
X-chromosome inactivation
During early female embryonic development, one of the X chromosomes (or more accurately, most of the genes on that X chromosome) is inactivated in each cell. The subsequent progeny of each cell maintain the same inactivated X chromosome, resulting in an organism that is a mosaic of cells expressing genes from one X chromosome or the other (Figure 1).
Historical background
Modern clonality testing originated in the X-chromosome inactivation (XCI) hypothesis of Ernest Beutler.17 This hypothesis postulated that females inactivate one X chromosome to maintain dosage parity with the single X chromosome in males (dosage compensation).18 At approximately the same time, Mary Lyon separately proposed XCI as a mechanism for X-linked coat color variation in mice.19 Seminal observations providing background for the XCI hypothesis came from Ohno, Hauschka, and Makino who demonstrated, first in mice and then in humans, that Barr bodies did not consist of portions of 2 X chromosomes in opposition to each other. Rather, each Barr body was a single X chromosome.20,21 This, with the additional insight that differential chromosome use could result in differential genetic function, led Beutler and Lyon to independently define the XCI phenomenon.
Mary Lyon proposed that the single Barr body X chromosome came randomly from either parent rather than always the father and that it was genetically inactivated. She based her hypothesis on observations of X-linked coat color mutations in heterozygous female mice. In these mice, the phenotype was always a mosaic consisting of patches of normal or mutant color rather than a homogenous blending, suggesting that early in development, the pigmented cells inactivated one or the other X chromosome. Thus, if the X chromosome carrying the mutant allele was inactivated, the patch was of normal color, whereas if the X chromosome carrying the normal allele was inactivated, the patch was of mutant color.19
Beutler and colleagues formulated the XCI hypothesis using studies of the human X chromosome gene glucose 6-phosphate dehydrogenase (G6PD).18 They found that, in females, G6PD activity was not twice that of males and postulated a dosage compensation mechanism. In females heterozygous for G6PD deficiency, dosage compensation results in G6PD expression at half the rate of normal hemizygous males. This could be attributable to either half-level activity in all cells or normal expression in some cells and low expression in other cells, resulting in overall half-level expression. Using a mixture of male cells with deficient G6PD activity and normal G6PD activity, Beutler and colleagues measured G6PD activity (by glutathione stability) and compared it with the response of female erythrocytes. They found that the response curves of the 2 samples were similar in shape and concluded that intermediate activity in females was probably attributable to the same mechanism as in the mixture of male normal and G6PD activity-deficient erythrocytes. Synthesizing these data on X-chromosome dosage compensation with Ohno's cytogenetic data led to the concept of random XCI. Beutler and colleagues later directly demonstrated that cells in an obligate G6PD-deficient heterozygous female were a mixture of erythrocytes with either normal or deficient G6PD function.
Mechanism of X-chromosome inactivation
The exact molecular mechanisms behind XCI are still not fully clarified but involve several steps, including the determination of the number of X chromosomes per cell, selection of either the paternal or maternal X chromosome for subsequent inactivation, and initiation of the actual inactivating process. It has been demonstrated in mice that there are 3 noncoding loci located near the X chromosome X center of inactivation (XCI) that play a pivotal role in the mechanism of X-chromosome inactivation. These loci are X (inactive)-specific transcript (Xist), its antisense partner Tsix, and the intergenic locus Xite.22-25 Xist is necessary for cis inactivation of the X chromosome.26,27 In cell culture systems, autosomally translocated Xist silences the surrounding chromatin, albeit incompletely.28 Tsix and Xite work in parallel to Xist by maintaining X-chromosome transcriptional competence.23,24,29,30
Although the functions of these 3 loci have been deduced using complementary cell lines, the actual physical interactions of these components are less well known. Xist is proposed to achieve cis inactivation of the X chromosome through close interactions between its RNA transcript and the segment of X chromosome to be inactivated.31 The putative trans interactions based on the need to determine one X to be exclusively activated and the other X to be exclusively inactivated remained problematic until recent experiments demonstrated that the 2 X chromosomes undergo interchromosomal pairing.32,33 This is remarkable in that such interchromosomal pairings typically occur in germ cells undergoing meiosis rather than somatic cells undergoing mitosis.
X-chromosome inactivation timing is crucial to the interpretation of XCIP-based clonality assays. It has been assumed that preblastocyst embryos express both X chromosomes and that inactivation did not occur until after implantation and the embryonic stem cells began to differentiate into separate cell lineages.34 Recent experiments, however, demonstrate that XCI occurs as early as the 4-cell stage of the embryo but is variable and leaky and does not become fixed until after implantation but before differentiation of embryonic stem cells into the various cell lineages.35,36 XCI before cell lineage differentiation is crucial for the interpretation of XCIP clonality studies. Hematopoietic cell lines derive not from a single embryonic stem cell but from several progenitors, allowing for the mosaic expression of genes from both X chromosomes.37,38
X-chromosome inactivation–based clonality studies
In normal human females, the hematopoietic organ is a mosaic of 2 populations. Each population expresses alleles from one of the active X chromosomes and the choice of which X chromosome to use is retained mitosis after mitosis. Cell lineage can be traced and subpopulation clonality can be determined by following the activated X chromosome. X chromosome protein products, transcribed mRNA, and DNA methylation status are used to determine which X chromosome is active. Protein isoforms and DNA sequence polymorphisms are used to identify the X chromosomes. These markers must be heterozygous to differentiate between the maternal or paternal origin of the X chromosome. Thus, XCIP clonality assays are limited to females with informative markers.
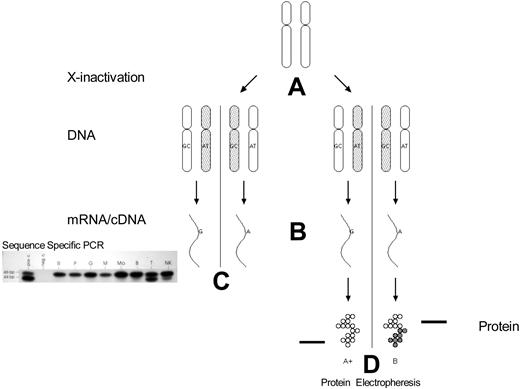
The first clonality studies using X-chromosome inactivation were based on G6PD isoenzymes (Figure 2).17 In the African population, there are 3 main G6PD isoenzymes: G6PD A+ and A− polymorphic variants with faster electrophoretic mobility than wild-type G6PD B. Approximately 35% of black women are heterozygous for the electrophoretically distinguishable isoenzymes. The use of G6PD as a marker of X-chromosome inactivation is limited to females who are heterozygous for these G6PD alleles. Informative females have the A− or A+ allele on one chromosome and the wild B allele on the other.
Clonality detection by mRNA (exonic polymorphisms) and protein polymorphisms. (A) X-chromosome inactivation occurs early in embryonic development. (B) mRNA transcription occurs only from the active X chromosome (with some exceptions). (C) Analysis of peripheral blood mRNA of a female with polycythemia vera. The amplified products can be differentiated by sequence-specific polymerase chain reaction of cDNA followed by ligase reaction detection. bp indicates base pair standards; pos c., positive control; neg c., negative control (no mRNA added to reaction mixture); R, reticulocytes; P, platelets; M, monocytes; Mo, cultured macrophages; B, B lymphocytes; T, T lymphocytes; NK, natural killer cells. (D) G6PD A and B isoforms can be differentiated by electrophoretic mobility.
Clonality detection by mRNA (exonic polymorphisms) and protein polymorphisms. (A) X-chromosome inactivation occurs early in embryonic development. (B) mRNA transcription occurs only from the active X chromosome (with some exceptions). (C) Analysis of peripheral blood mRNA of a female with polycythemia vera. The amplified products can be differentiated by sequence-specific polymerase chain reaction of cDNA followed by ligase reaction detection. bp indicates base pair standards; pos c., positive control; neg c., negative control (no mRNA added to reaction mixture); R, reticulocytes; P, platelets; M, monocytes; Mo, cultured macrophages; B, B lymphocytes; T, T lymphocytes; NK, natural killer cells. (D) G6PD A and B isoforms can be differentiated by electrophoretic mobility.
A natural progression from using protein isoforms to identify the active X chromosome and its parental origin is to directly assay the transcribed mRNA products of the active X chromosome (Figure 2). The parental origin of the activated X chromosome is identified by DNA sequence polymorphisms. This method was first described by Ernest Beutler's laboratory and independently with a modified methodology in our laboratory.39,40 The exonic polymorphisms used to identify the X chromosome are typically nonsynonymous mutations. A variety of genes are used to determine clonal status, including G6PD40 (using a polymorphism different from that used in protein-based clonality assays), iduronate-2-sulfatase,41,42 MPP1 (also known as p55),43 Bruton tyrosine kinase, and FHL-1 (4.5 LIM domain 1).44 In aggregate, approximately 95% of females will be informative for transcriptionally based X-chromosome inactivation clonality assays using these genes.44
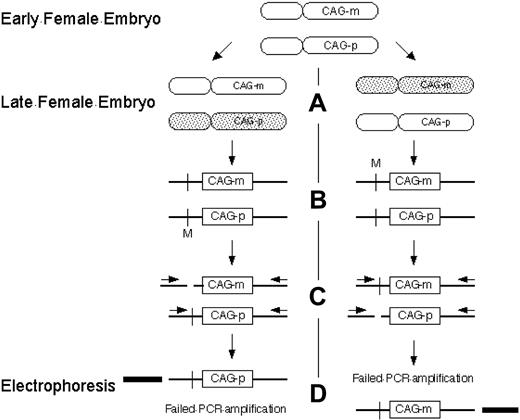
A widely used method for determining clonality using the X- chromosome inactivation principle uses a different approach based on DNA methylation and CAG tandem repeats at the human androgen receptor assay (HUMARA) locus (Figure 3). The number of tandem CAG repeats differentiates the maternal and paternal X chromosomes, and methylation status distinguishes the inactive and active X chromosome. At the HUMARA locus, the inactive X chromosome is methylated, whereas the active X chromosome is unmethylated.45 The HUMARA method is more widely applicable than protein isoform and transcription-based methods because the variable number of CAG nucleotide repeats allows most patients to be informative for the assay.46 Using a variant of the HUMARA method, the clonal origins of AML, leiomyoma, and Wilm tumor have been demonstrated.47
Clonality detection by the HUMARA assay. (A) Methylation of the inactive X chromosome at the HUMARA locus occurs early in embryonic development. Shaded, inactivated X chromosome. (B) Schematic of HUMARA locus. Crossbar indicates methylation sensitive endonuclease site; M, methylated methylation sensitive endonuclease site; CAG-p, paternal CAG tandem repeat sequence; CAG-m, maternal CAG tandem repeat sequence. (C) After digestion by methylation sensitive endonucleases, only inactive (methylated) DNA remains intact. (D) Only intact DNA can be amplified by polymerase chain reaction with flanking primers. Products can be differentiated from each other by the number of tandem CAG repeats in CAG-p and CAG-m and their resultant different electrophoretic mobility.
Clonality detection by the HUMARA assay. (A) Methylation of the inactive X chromosome at the HUMARA locus occurs early in embryonic development. Shaded, inactivated X chromosome. (B) Schematic of HUMARA locus. Crossbar indicates methylation sensitive endonuclease site; M, methylated methylation sensitive endonuclease site; CAG-p, paternal CAG tandem repeat sequence; CAG-m, maternal CAG tandem repeat sequence. (C) After digestion by methylation sensitive endonucleases, only inactive (methylated) DNA remains intact. (D) Only intact DNA can be amplified by polymerase chain reaction with flanking primers. Products can be differentiated from each other by the number of tandem CAG repeats in CAG-p and CAG-m and their resultant different electrophoretic mobility.
Pitfalls in the interpretation of clonality assays based on X-chromosome inactivation
Interpretation of clonality studies can be confounded by technical and methodologic errors as well as errors intrinsic to the mechanism of X-chromosome inactivation.
Technical and methodologic errors
X-chromosome inactivation clonality assays are often performed as in-house research tools. Technical errors can occur during the assay such as incomplete digestion by methylation sensitive enzymes or nonquantitative PCR when allelic fragments of different sizes amplify with variable efficacy.
The methylation status of the HUMARA locus is well characterized and has been reliably used to determine clonality. Ready extension of this method to other X-chromosome loci is confounded by the effect of mitosis, drugs, nutrition, and the in vivo manipulation of cells during diagnostic testing, which can alter methylation patterns. Furthermore, the methylation of inactive genes is not uniform throughout the inactive X-chromosome regions because although many inactivated genes are methylated, some are hypomethylated, and some show no difference in methylation as seen in a region of conserved G6PD polymorphism (G to T at nucleotide 1311) used in XCIP-based assays.40,48
One theoretical concern in X-chromosome inactivation clonality testing is incomplete inactivation of X-chromosome genes. Carrel and Willard have recently shown that 15% of X chromosome genes escape inactivation and 10% of X-chromosome genes are incompletely inactivated and differentially expressed from each chromosome.49 These genes could not be used to determine clonality because they would not accurately reflect the true proportions of clonal versus nonclonal cells. All new X-chromosome polymorphic loci proposed as candidates for determining clonality must be shown to be subject to complete X-chromosome inactivation before they can be used as a valid marker for assessing clonality.43,50
Misinterpretation errors intrinsic to X-chromosome inactivation
These errors do not occur because the assay was incorrectly performed or unable to accurately measure clonality. Rather, the accurate but extreme results are misinterpreted. These errors are false-negatives and false-positives and can occur at 3 steps in the X-chromosome inactivation process: at the time of XCI as a result of (1) genetic and (2) stochastic skewing of the choice of X chromosome, and (3) after XCI when cellular selection pressures for or against one of the X-chromosome allelic gene products alters the apparent ratio of other X-chromosome gene products used as clonality markers.
Stochastic skewing
XCI is a stochastic process and whether a germ cell and its progeny ultimately express one or the other X allele is dependent on chance. Whereas a 50/50 ratio of one allele to the other is ideal and probable in large populations, in the small population of primordial germ cells that undergo X-chromosome inactivation, it is possible to achieve greater representation of one allele than the other (Figure 4). This frequently leads to a normal skewing of the X-chromosome allelic use ratio in terminally differentiated cell populations that are subject to analysis. Using G6PD synonymous alleles that differ from each other by a single nucleotide change of C to T, it has been demonstrated that normal females have variable G6PD allele use ratios as great as 1:7.38 Thus, large proportions of apparently normal women have constitutional skewing of X inactivation. Based on the degree of skewing, it has been calculated that approximately 8 cells contribute to the embryonic pool of cells that go on to form the definitive hematopoietic system.37,38
Stochastic/genetic skewing of X chromosome inactivation. (A) Both X chromosomes are active in early embryonic stem cells. Crosshatching represents the active X chromosomes of maternal (black) and paternal (white) origin. (B) X chromosome inactivation choice is stochastic resulting in (C) variance of the maternal/paternal inactivated X chromosome ratio from the theoretical 1:1. (D) Genetic factors can also affect the choice of X chromosome inactivated, resulting in skewed maternal/paternal inactivated X chromosome ratios. These skewed populations can result in a false determination of clonality.
Stochastic/genetic skewing of X chromosome inactivation. (A) Both X chromosomes are active in early embryonic stem cells. Crosshatching represents the active X chromosomes of maternal (black) and paternal (white) origin. (B) X chromosome inactivation choice is stochastic resulting in (C) variance of the maternal/paternal inactivated X chromosome ratio from the theoretical 1:1. (D) Genetic factors can also affect the choice of X chromosome inactivated, resulting in skewed maternal/paternal inactivated X chromosome ratios. These skewed populations can result in a false determination of clonality.
Genetic skewing defects in the X-chromosome inactivation process
Baseline skewing of X-chromosome inactivation can also be attributable to genetic defects in the X-inactivation process (Figure 4). Among the candidates for genetic mutations is the Xist gene.46 One family has been identified with a mutation in the Xist promoter region and associated nonrandom X-chromosome inactivation.51
Selection in females heterozygous for certain X-linked genetic diseases
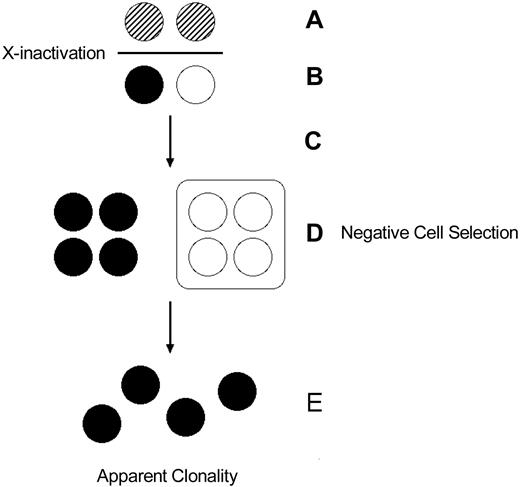
Baseline skewing can also be attributable to selection for or against a particular X-chromosome germ line inherited mutation (Figure 5). This has practical application in the interpretation of clonality studies in females heterozygous for certain X-linked genetic diseases. Wiskott-Aldrich syndrome is characterized by eczema, thrombocytopenia with small platelets, and immune deficits apparent since infancy. Because it is X-linked, males are often afflicted but females generally are not. In females, the cells with the X chromosome bearing the mutant gene are selected against, whereas the cells with the X chromosome bearing the normal gene preferentially survive.52 The majority of heterozygous female carriers of Wiskott-Aldrich syndrome have normal platelet number and size and select against those hematopoietic progenitors that express the Wiskott-Aldrich allele on their active X chromosome.53 However, heterozygous females with Wiskott-Aldrich phenotype, who presumably inactivate the normal allele in the early embryogenesis, exist.54 Pseudoclonality attributable to cell selection has also been reported in females heterozygous for other X-linked disorders such as Lowe Oculo-cerebro-renal syndrome,55 X-linked agammaglobulinemia,56 and G6PD deficiency variants.57,58
Somatic selection after X-chromosome inactivation. (A) Early embryonic cells express both X chromosomes and undergo X-chromosome inactivation. (B) After inactivation, embryonic cells express one or the other X chromosome. Thus, in a disease like Wiskott-Aldrich syndrome, a heterozygous female can express both the normal and mutant alleles, albeit in different cells. (C) The embryonic cells undergo proliferation. (D) Cellular selection against the cells bearing the active X chromosome with the mutant allele results in elimination of that population of cells, resulting in (E) an apparently clonal (“pseudoclonal”) population.
Somatic selection after X-chromosome inactivation. (A) Early embryonic cells express both X chromosomes and undergo X-chromosome inactivation. (B) After inactivation, embryonic cells express one or the other X chromosome. Thus, in a disease like Wiskott-Aldrich syndrome, a heterozygous female can express both the normal and mutant alleles, albeit in different cells. (C) The embryonic cells undergo proliferation. (D) Cellular selection against the cells bearing the active X chromosome with the mutant allele results in elimination of that population of cells, resulting in (E) an apparently clonal (“pseudoclonal”) population.
Aging and X-chromosome inactivation
Skewed lyonization may occur as part of the aging process, although the mechanism is less clear. In a study of peripheral granulocytes using the methylation-based HUMARA assay, skewing was reported to be progressive with age: 1.9% in neonates, 4.5% in women 28 to 32 years old, and 22.7% in women older than 60. Essentially only neutrophils showed skewing with age and T lymphocytes showed less evidence of unbalanced X inactivation. The hematopoietic cells (including the erythroid, granulocytic, monocytic, and megakaryocytic lineages) of 30% to 40% of elderly women were reported to have more than 90% expression of one parental X chromosome.59 Although suggestive of an effect of age on X-chromosome inactivation, this study was limited because it compared the average skew of an older versus younger population instead of the skew in a person after aging. Another study addressed this concern by examining X-chromosome skewing using HUMARA methylation in paired samples taken an average of 16.5 years apart in 178 patients ranging in age from 0 to 74. These investigators found more progression of X-inactivation skewing in women older than 60 when they entered the study. In women younger than 60, no consistent trend toward increased skewing was found. Based on their findings, these investigators proposed that age-related X-chromosome skewing may occur in a discontinuous/catastrophic manner rather than one of gradual accretion.60 Increased X-chromosome inactivation pattern skewing after age 50 has also been found by other investigators.61
The apparent development of clonality related to age has been challenged by investigators using methylation-based clonality assays as well as X-chromosome transcription–based clonality assays.62 When age-related clonality is reported, it is generally correlated with a skewed allelic ratio rather than the exclusive expression of a single X-chromosome allele. Defining clonality more stringently as expression of only a single X-chromosome transcript in the studied tissue, our laboratory has not observed any clonal XCIP in studies of 200 samples of apparently nonclonal tissue taken from heterozygous females, suggesting the rarity of this phenomenon in the general population.38 However, we have not yet systematically studied large numbers of females older than 70 years.
Experimental evidence exists supporting clonal expansion and XCIP skewing with age in cats.63 Abkowitz and colleagues performed serial determinations of XCIP skewing using G6PD isoforms in a feline hematopoiesis model. The cats were derived from 2 different feline strains with divergent X-chromosome polymorphisms. They found that over a period of several years, progenitor cell populations shifted toward the same allelic phenotype secondary to hemizygous selection. Thus, age-related skewing of XCIP occurs in felines and may be related to genetic factors. A similar phenomenon may exist in humans.64 Recent studies comparing the magnitude and direction of presumed acquired skewing in granulocytes compared with T cells in monozygotic versus dizygotic twins suggest a genetic contribution to the observed variance in skewing. The calculated estimate of heritability was 68%, although the confidence interval ranged from 0% to 100%.65
Age-related X-chromosome–inactivation skewing can manifest clinically as late-onset X-linked disorders. An 81-year-old woman who was heterozygous for the erythroid-specific 5-aminolevulnate synthase gene and unaffected in her youth later developed microcytic sideroblastic anemia. She was initially thought to have myelodysplastic syndrome but on true diagnosis was treated to resolution with pyridoxine.66 Whether this was related to acquired clonality associated with myelodysplastic syndrome is not clear.
Recently, age-related X-chromosome–inactivation skewing has been implicated in the pathogenesis of scleroderma, autoimmune thyroid disease (AITD, including Graves disease and Hashimoto thyroiditis), and primary biliary cirrhosis.67-69 All 3 of these diseases are more common in females later rather than earlier in life, providing an epidemiologic basis for further investigation. The increased autoimmune susceptibility in 45 XO Turner syndrome also suggests a role for loss of X-chromosome material through skewing or monosomy in the pathogenesis of autoimmune disorders.70 Using the HUMARA assay to determine XCIP, it was found that 64% of patients with scleroderma versus 8% of control subjects showed X-chromosome skewing less than 80% (P < .001). When XCIP were studied with the HUMARA in AITD, 34% of concordant twins compared with 11% of normal control subjects had X-chromosome skewing less than 80% (P = .003). A similar ratio was found when discordant twin pairs were studied.68 Fluorescent in situ hybridization analysis was used to study the prevalence of monosomy X in primary biliary cirrhosis versus hepatitis C and normal controls. The investigators found that in peripheral blood cells, the age-adjusted frequency of monosomy X was 0.05 in primary biliary cirrhosis, 0.032 in hepatitis C, and 0.028 in normal controls (P < .001).67 These investigators applied the same fluorescent in situ hybridization analysis to determine monosomy X rates in systemic sclerosis and autoimmune thyroid disease. They found a frequency of 6.2% in systemic sclerosis, 4.3% in AITD, and 2.9% in normal female controls (P < .001).71
Several mechanisms have been proposed linking X-chromosome skewing and monosomy with autoimmune disease, including the loss of X-chromosome genes crucial to maintaining sex hormone levels as well as those mediating immune tolerance.72 Linkage studies of autoimmune disorders have identified several non-MHC loci on the X chromosome that may be involved in immune regulation.73,74 Other proposed mechanisms include loss of antigens expressed from the inactivated X chromosome followed by loss of T-cell tolerance and autoimmunity. This mechanism seems less likely given the disparateness of affected organ and tissue types. In contrast to a deficiency of X-chromosome function, a competing theory of pathogenesis is residual microchimerism from previous pregnancies. Although not eliminating microchimerism as a cause of autoimmune disease, experimental evidence obtained by fluorescent in situ hybridization analysis showing the absence of Y-chromosome material in scleroderma and AITD peripheral blood cells that are monosomal for the X chromosome suggests this is less likely.71 Although no studies have formally linked age-related XCIP skewing with the development of autoimmune disease, the available evidence suggests a connection may exist and deserves further research.
Integrating genotypic and phenotypic clonality detection
Genotypic and XCIP clonality represent different ways of looking at the same process. Use of either method to detect a dominant clonal population has been discussed. Use of genotypic clonality to detect a small clonal population in the context of a larger nonclonal population was discussed as minimal residual disease detection. Use of XCIP analysis to detect a small clonal population in the context of a larger nonclonal population is more difficult because the presence of a small clonal population would be indicated by an increase in XCIP skewing above a theoretical baseline. The theoretical baseline is often assumed to be 50%, but recent studies have demonstrated unequal expression of certain X-chromosome genes.49 Both the theoretical baseline and degree of XCIP skew would be subject to statistical variation, which could overshadow the true difference. XCIP clonal population size estimates in essential thrombocytosis have been shown to have wide 95% confidence intervals, more so in older patients, with systemic biases toward overestimating the true population size.75 The large variability in XCIP clonality testing limits its use to dominant clonal populations rather than small clonal populations.
XCIP analysis and concurrent JAK2 617V>F testing of large clonal populations has resulted in new insights into the pathophysiology of myeloproliferative disorders. Not all cases of suspected myeloproliferative disease proven to be clonal by XCIP clonality testing have JAK2 617V>F mutations.76-79 The XCIP markers iduronate-2-sulfatase and MPP1 and the genetic marker JAK2 617V>F were used to characterize the granulocytes of patients with MPD. All the granulocytes were clonal by XCIP, but not all the granulocytes in the same patient had the JAK2 617V>F mutation, suggesting the presence of a JAK2 617V>F-negative population.80 In addition to the concerns about XCIP analysis variability raised by Campbell et al,75 age-related and constitutive XCIP skewing can confound the finding of discordant clonal population sizes. These concerns were refuted by demonstrating the presence of del20q, an independent clonality marker, in all cells and by demonstrating polyclonal T cells, which would not be present if constitutive skewing were present.80 Our laboratory has also reported similar discrepancies in clonal population size. XCIP analysis demonstrated granulocyte clonality in 10 female patients with polycythemia vera (PV), but the JAK2 617V>F mutation frequency was less than 50% (27.5% ± 11%) in 3 patients and more than 50% (75% ± 10.5%) in 7 patients. Because of potential confounding from constitutive or age-related skewing, we confirmed clonality analysis with erythropoietin growth-independent colonies from the same patients. Individual colonies represent clonal processes because they are derived from single progenitors. Of 69 individual colonies, JAK2 was wild type in 10, mutated heterozygous in 6, and mutated homozygous in 23, indicating the presence of a clonal JAK2 617V>F-negative population.81 Other investigators have also reported discordant clonal populations in the essential thrombocythemia and myelofibrosis with myeloid metaplasia when analyzed with the HUMARA assay or XCIP in combination with JAK2 gene analysis.82-84 In aggregate, these studies document the existence of clonal JAK2 617V>F-negative populations in the development of myeloproliferative diseases. Because clonal processes are thought to be initiated by underlying somatic mutations, the clonal JAK2 617V>F-negative populations may harbor as-yet-undiscovered mutations contributing to the pathogenesis of myeloproliferative disease. The majority of JAK2 617V>F-positive patients with polycythemia vera who transform to acute leukemia have leukemic blasts that are JAK2 617V>F-negative. However, it remains to be determined if they originate from the pre–JAK2 617V>F clone.85
Summary
Clonality testing has played an important role in medicine and hematology. Detection of clonality can be based on genotype and phenotype. The most frequently used phenotypic clonality technique is based on the X-chromosome inactivation principle. Proper interpretation of these results can be confounded by technical errors and methodological errors. Methodological errors can occur as a result of stochastic skewing, genetically determined skewing, environmentally induced methylation changes, and somatic selection after X-chromosome inactivation. The primary limitation of XCI clonality studies is the need for informative female patients.
XCIP clonality testing has been applied to demonstrate progressive X-chromosome skewing in aging hematopoietic cells. This has led to new insights into the pathophysiology of X-linked and autoimmune disorders. Other research applications include combining XCIP clonality testing with genotypic clonality testing to identify clonal populations with unknown underlying genetic mutations, allowing the identification of novel disease-associated mutations.
Acknowledgments
This work was supported by 1P01CA108671-O1A2, 1R01HL50077–12, and a VA Merit Review Award (J.T.P., primary investigator).
Authorship
Contribution: G.L.C. wrote and extensively revised the first draft; J.T.P. contributed key portions to the first draft and critically reviewed and revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Josef Prchal, Division of Hematology, Department of Medicine, University of Utah School of Medicine Room 5C402 SOM, 30 North 1900 East, Salt Lake City, UT 84132; e-mail: josef.prchal@hsc.utah.edu.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal