To the editor:
The CXC chemokine receptor 3 (CXCR3) regulates migration and function of T cells during inflammation. Resting T cells express low levels of CXCR3 but they up-regulate CXCR3 upon activation. Several studies suggest that T helper 1 (Th1)/Th2-associated cytokines regulate levels of CXCR3 on T cells.1–3 Interferon-γ (IFN-γ), which signals via STAT1, enhances CXCR3 expression on T cells,2 but its role in regulating CXCR3 on CD4+ versus CD8+ T cells is not clear. We therefore examined the role of IFN-γ and STAT1 in regulating CXCR3 levels on CD4+ and CD8+ T cells.
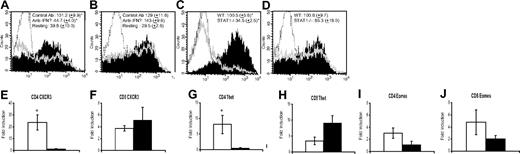
Naive CD4+ and CD8+ T cells from C57BL/6 mice were stimulated as described previously2 with anti-CD3e (3 μg/mL; Biolegend, San Diego, CA) and anti-CD28 (4 μg/mL; Biolegend) in the presence of anti–IFN-γ neutralizing or control antibody, and CXCR3 expression was analyzed by flow cytometry. CD4+ T cells activated in the presence of anti–IFN-γ Ab (clone no. XMG1.2; Pharmingen, San Diego, CA) expressed less CXCR3 than controls (Figure 1A). However, IFN-γ blockade had no effect on CXCR3 levels in CD8+ T cells (Figure 1B). Control CD4+ and CD8+ T cells up-regulated CXCR3 (Figure 1A,B) and produced comparable IFN-γ (data not shown).
IFN-γ and STAT1 are required for efficient induction of CXCR3 on CD4+ but not CD8+ T cells and T-bet, but not Eomes, controls CXCR3 induction. After stimulation with anti-CD3 and anti-CD28 antibody, cells were rested 24 hours without stimulation. CXCR3 surface protein expression was measured by flow cytometric staining of wild-type CD4 (A) and CD8 (B) cells in the presence of 50 μg/mL IFN-γ–neutralizing antibody or an isotype control. Stained cells treated with anti–IFN-γ are represented by hollow gray peaks, cells stimulated in the presence of control antibody are depicted by solid peaks, and dotted lines represent a PE-labeled isotype control. Surface CXCR3 was also analyzed on STAT1−/− CD4 (C) and CD8 (D) T cells. STAT1+/+ cells are represented by solid peaks, STAT1−/− cells are represented by hollow gray peaks, and isotype control staining is repressed by a dotted line. Levels of CXCR3 (E,F), T-bet (G,H), and Eomes (I,J) mRNA in activated CD4 and CD8 T cells from WT (□) as well as STAT1−/− mice (■) were measured by real-time RT-PCR. Data were normalized to the housekeeping gene GAPDH, and the results are presented as fold-induction of gene expression over nonactivated cells. Histograms in panels A and B are representative of 3 independent experiments, while panels C and D represent 1 of 5 independent experiments. Mean fluorescence intensity (MFI) data represent the mean (± SEM) for all experiments. Resting WT CD4+ and CD8+ T cells in these experiments yielded average MFIs of 39.8 (± 10.0) and 29.5 (± 2.6), respectively, while STAT1−/− CD4+ and CD8+ T cells resulted in 32.6 (± 10.7) and 34.1 (± 11.4), respectively, prior to activation. (E-J) Averaged results (± SEM) of 5 independent experiments. Statistical analysis was performed using the Mann-Whitney rank-sum test (SigmaStat; Systat Software, San Diego, CA). *P < .05 was considered significant.
IFN-γ and STAT1 are required for efficient induction of CXCR3 on CD4+ but not CD8+ T cells and T-bet, but not Eomes, controls CXCR3 induction. After stimulation with anti-CD3 and anti-CD28 antibody, cells were rested 24 hours without stimulation. CXCR3 surface protein expression was measured by flow cytometric staining of wild-type CD4 (A) and CD8 (B) cells in the presence of 50 μg/mL IFN-γ–neutralizing antibody or an isotype control. Stained cells treated with anti–IFN-γ are represented by hollow gray peaks, cells stimulated in the presence of control antibody are depicted by solid peaks, and dotted lines represent a PE-labeled isotype control. Surface CXCR3 was also analyzed on STAT1−/− CD4 (C) and CD8 (D) T cells. STAT1+/+ cells are represented by solid peaks, STAT1−/− cells are represented by hollow gray peaks, and isotype control staining is repressed by a dotted line. Levels of CXCR3 (E,F), T-bet (G,H), and Eomes (I,J) mRNA in activated CD4 and CD8 T cells from WT (□) as well as STAT1−/− mice (■) were measured by real-time RT-PCR. Data were normalized to the housekeeping gene GAPDH, and the results are presented as fold-induction of gene expression over nonactivated cells. Histograms in panels A and B are representative of 3 independent experiments, while panels C and D represent 1 of 5 independent experiments. Mean fluorescence intensity (MFI) data represent the mean (± SEM) for all experiments. Resting WT CD4+ and CD8+ T cells in these experiments yielded average MFIs of 39.8 (± 10.0) and 29.5 (± 2.6), respectively, while STAT1−/− CD4+ and CD8+ T cells resulted in 32.6 (± 10.7) and 34.1 (± 11.4), respectively, prior to activation. (E-J) Averaged results (± SEM) of 5 independent experiments. Statistical analysis was performed using the Mann-Whitney rank-sum test (SigmaStat; Systat Software, San Diego, CA). *P < .05 was considered significant.
Next, we compared CXCR3 surface expression and mRNA levels in stimulated CD4+ and CD8+ T cells from wild-type (WT) and STAT1−/− C57BL/6 mice. Activated STAT1−/− CD4+ T cells failed to up-regulate CXCR3 as efficiently as WT CD4+ T cells (Figure 1C). In contrast, STAT1−/− CD8+ T cells showed a significant induction of CXCR3 similar to WT CD8+ T cells (Figure 1D). Low CXCR3 expression on STAT1−/− CD4+ T cells also correlated with low CXCR3 mRNA levels (Figure 1E,F). Activated WT and STAT1−/− CD4+ as well as CD8+ T cells produced significant IFN-γ, but levels were lower in STAT1−/− T cells (WT CD4, 19908.17 [± 2582.868] pg/mL; WT CD8, 27361.05 [± 3484.116] pg/mL; STAT1−/− CD4, 8012.928 [± 1627.75] pg/mL; STAT1−/− CD8, 17152.14 [± 3680.819] pg/mL). Blockade of IFN-γ did not inhibit CXCR3 expression on STAT1−/− CD8+ T cells, suggesting that IFN-γ was not inducing CXCR3 via a STAT1-independent pathway (data not shown).
We also determined whether the transcription factors T-bet and eomesodermin (Eomes) are involved in CXCR3 induction on STAT1−/− CD8+ T cells by measuring mRNA levels by real-time reverse transcription–polymerase chain reaction (RT-PCR). Both these factors control CD4+ and CD8+ T-cell activity by regulating expression of many genes, including Cxcr3,4 and their expression in T cells can be induced via STAT1-depdendent and -independent mechanims.5,6 STAT1−/− CD4+ T cells showed less induction of T-bet mRNA than WT CD4+ T cells (Figure 1G). In contrast, STAT1−/− CD8+ T cells contained more T-bet mRNA than WT CD8+ T cells, but the difference was not significant (Figure 1H). STAT1−/− CD4+ and CD8+ T cells also contained less Eomes mRNA than WT T cells, but these differences were not significant (Figure 1I,J).
These findings show that IFN-γ and STAT1 are critical for efficient induction of CXCR3 on CD4+, but not CD8+ T cells upon activation. To best of our knowledge, this is the first study to demonstrate differential requirement for IFN-γ and STAT1 in up-regulation of CXCR3 on CD4+ versus CD8+ T cells. These findings are important in designing therapeutic strategies to control CXCR3 expression in T cells for treatment of autoimmune and inflammatory diseases.
Authorship
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr. Abhay R. Satoskar, Department of Microbiology, The Ohio State University, 484 West 12th Ave, Columbus, OH 43221; e-mail: satoskar.2@osu.edu.
This work was supported by the National Institutes of Health (NIH) grant RO1 51328 to A.R.S.
We thank Drs Virginia Sanders, Michael Caliguiri, and Joan Durbin for critical review of the paper and for suggestions.
Contribution: J.B. and S.O. performed experiments and wrote the paper. C.L.D. analyzed data and wrote the paper. A.R.S. designed experiment, analyzed data, and wrote the paper.
References
National Institutes of Health

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal