Salinosporamide A (also called NPI-0052), recently identified from the marine bacterium Salinispora tropica, is a potent inhibitor of 20S proteasome and exhibits therapeutic potential against a wide variety of tumors through a poorly understood mechanism. Here we demonstrate that salinosporamide A potentiated the apoptosis induced by tumor necrosis factor α (TNF), bortezomib, and thalidomide, and this correlated with down-regulation of gene products that mediate cell proliferation (cyclin D1, cyclooxygenase-2 [COX-2], and c-Myc), cell survival (Bcl-2, Bcl-xL, cFLIP, TRAF1, IAP1, IAP2, and survivin), invasion (matrix metallopro-teinase-9 [MMP-9] and ICAM-1), and angiogenesis (vascular endothelial growth factor [VEGF]). Salinosporamide A also suppressed TNF-induced tumor cell invasion and receptor activator of nuclear factor κB ligand (RANKL)-induced osteoclastogenesis. We also found that it suppressed both constitutive and inducible NF-κB activation. Compared with bortezomib, MG-132, N-acetyl-leucyl-leucyl-norleucinal (ALLN), and lactacystin, salinosporamide A was found to be the most potent suppressor of NF-κB activation. Further studies showed that salinosporamide A inhibited TNF-induced inhibitory subunit of NF-κB α (IκBα) degradation, nuclear translocation of p65, and NF-κB-dependent reporter gene expression but had no effect on IκBα kinase activation, IκBα phosphorylation, or IκBα ubiquitination. Thus, overall, our results indicate that salinosporamide A enhances apoptosis, suppresses osteoclastogenesis, and inhibits invasion through suppression of the NF-κB pathway.
Introduction
An extensive experience in drug development indicates that “mother nature” is the richest source of new medicines. As many as 48 anticancer drugs of 65 that were approved by the FDA between 1981 and 2002 were either derived from natural sources or based on natural sources.1 Salinosporamide A (renamed as NPI-0052) is one such compound; it was first discovered during the fermentation of Salinispora tropica, a new seawater-requiring actinomycete found in marine sediments in the Bahamas.2,–4 Salinosporamide A, which has an unusual bicyclic β-lactone γ-lactam structure, was found to be a potent inhibitor of all the 3 proteolytic activities of the mammalian 20S proteasome, having a concentration that inhibits response by 50% (IC50) of 3.5 nM for chymotrypsin-like (CT-L), 28 nM for trypsin-like (T-L), 430 nM for caspase-like (CL) activities.5,6 A limited number of studies have revealed that it can induce apoptosis of multiple myeloma cells,6 colon cancer cells,7 chronic lymphocytic leukemia,8 acute lymphocytic leukemia, and acute myeloid leukemia.9 In animal model systems also, this proteasome inhibitor was found to suppress the growth of multiple myeloma6 and colon cancer.7
To date, the mechanism of action of this molecule has not been established. Because proteasomal activity is critical for the activation of the transcription factor nuclear factor κB (NF-κB), we chose it as a potential target likely to be modulated by this agent. Our hypothesis was that modulation of NF-κB expression by NPI-0052 accounts for much of its activity. An additional consideration is that NF-κB plays an important role in tumorigenesis, apoptosis, inflammation, immunomodulation, angiogenesis, and bone remodeling.10 NF-κB consists of Rel domain-containing proteins present in the cytoplasm of all cells, where they are kept in an inactive state by a family of anchorin domain-containing proteins that includes the inhibitory subunit of NF-κB α (IκBα), IκBβ, IκBγ, IκBϵ, bcl-3, p105, and p100. Under resting conditions, NF-κB consists of a heterotrimer of p50, p65, and IκBα in the cytoplasm11 ; only when activated and translocated to the nucleus is the sequence of events leading to activation initiated. The activation of NF-κB involves the phosphorylation, ubiquitination, and degradation of IκBα and phosphorylation of p65, which in turn lead to the translocation of NF-κB to the nucleus where it binds to specific response elements in the DNA.12 NF-κB has been shown to regulate the expression of a number of genes whose products are involved in tumorigenesis.10 These include antiapoptotic genes (eg, ciap, survivin, traf, cflip, bcl-2, and bcl-xl); cox2; mmp-9; VEGF; genes encoding adhesion molecules, chemokines, and inflammatory cytokines; and cell cycle regulatory genes (eg, cyclin d1 and cmyc). Because of the critical role of NF-κB in cancer, we investigated the effect of salinosporamide A (NPI-0052) on apoptosis induced by tumor necrosis factor α (TNF) and chemotherapeutic agents, on invasion, on osteoclastogenesis, on NF-κB-regulated gene products, and on the NF-κB activation pathway.
Materials and methods
Materials
Salinosporamide A (renamed as NPI-0052) was kindly supplied by Nereus Pharmaceuticals (San Diego, CA). A 1-mM stock solution was dissolved in 100% dimethyl sulfoxide, stored at -20°C, and then diluted as needed in cell culture medium. Bacteria-derived recombinant human TNF, purified to homogeneity with a specific activity of 5 × 107 U/mg, was kindly provided by Genentech (South San Francisco, CA). Penicillin, streptomycin, Iscove Modified Dulbecco Medium (IMDM), Dulbecco modified Eagle medium (DMEM), and RPMI 1640 medium were obtained from Invitrogen (Grand Island, NY). Fetal bovine serum (FBS) was obtained from ATLANTA Biologicals (Lawrenceville, GA). Bortezomib (PS-341) was obtained from Millennium (Cambridge, MA), and thalidomide was obtained from Tocris Cookson (St Louis, MO). Anti-β-actin and monoclonal antiubiquitin antibodies were obtained from Sigma-Aldrich (St Louis, MO), and antibodies against p65, IκBα, cyclin D1, matrix metalloproteinase-9 (MMP-9), poly(adenosine 5′-diphosphate-ribose)polymerase (PARP), IAP1, IAP2, Bcl-2, Bcl-xL, c-Myc, ICAM-1, lactate dehydrogenase (LDH), and Lamin B and the annexin V staining kit were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antivascular endothelial growth factor (VEGF) antibody was purchased from NeoMarkers (Fremont, CA), and anticyclooxygenase-2 (COX-2) antibody was purchased from BD Biosciences (San Diego, CA). Phospho-specific anti-IκBα (serine 32) antibody was purchased from Cell Signaling (Beverly, MA). Anti-IκBα kinase (IKK)-α, anti-IKK-β, and anti-cFLIP antibodies were kindly provided by Imgenex (San Diego, CA). Survivin antibody was purchased from R&D Systems (Minneapolis, MN). Suc-LLVY-AMC peptide substrate was purchased from Calbiochem (San Diego, CA).
Cell lines
KBM-5 (human myeloid leukemia), RAW 264.7 (mouse macrophage), H1299 (human lung adenocarcinoma), DU145 (human prostate cancer), U266 (human multiple myeloma), PC-3 (human prostate adenocarcinoma), RPMI-8226 (human multiple myeloma), and A293 (human embryonic kidney) were obtained from American Type Culture Collection (Manassas, VA). KBM-5 cells were cultured in IMDM medium supplemented with 15% FBS. Raw 264.7 cells and PC-3 cells were cultured in DMEM/F-12 medium; H1299 cells, DU145 cells, RPMI-8226, and U266 were cultured in RPMI 1640 medium; and A293 cells were cultured in DMEM supplemented with 10% FBS. All media were also supplemented with 100 U/mL penicillin and 100 μg/mL streptomycin. The mouse embryonic fibroblast (MEF) derived from p65−/− C57Bl/6J mice and its wild type were kindly provided by Dr. David Baltimore (California Institute of Technology, Pasadena, CA). Cells were cultured in DMEM supplemented with 10% FBS, 100 U/mL penicillin, and 100 μg/mL streptomycin.
Cytotoxicity assay
Cytotoxicity was assayed by the modified tetrazolium salt 3-(4-5-dimethylthiozol-2-yl)2-5-diphenyl-tetrazolium bromide (MTT) assay as described previously.13
Live and dead assay
To measure apoptosis, we used the Live and Dead assay (Molecular Probes, Eugene, OR), which determines intracellular esterase activity and plasma membrane integrity. Briefly, 106 cells were incubated with NPI-0052 for 4 hours and then treated with TNF, thalidomide, and bortezomib for 16 hours at 37°C. Cells were stained with the Live and Dead reagent (5 μM ethidium homodimer, 5 μM calcein-AM) and then incubated at 37°C for 30 minutes. Cells were analyzed under a fluorescence microscope (Labophot-2; Nikon, Tokyo, Japan). Cells plated with Mounting Media (Sigma-Aldrich) were analyzed under a fluorescence microscope (Lapshot-2) equipped with a CFWN 10 /1.5 NA oil-immersion objective lens and a Photometrics Coolsnap CF color camera (Nikon, Lewisville, TX). Images were acquired with MetaMorph 4.6.5 software (Universal Imaging, Downingtown, PA).
Annexin V assay
One of the early indicators of apoptosis is the rapid translocation and accumulation of the membrane phospholipid phosphatidylserine from the cell's cytoplasmic interface to the extracellular surface. This loss of membrane asymmetry can be detected using the binding properties of annexin V. To detect apoptosis, we used annexin V antibody conjugated with the fluorescent dye fluorescein isothiocyanate (FITC). Briefly, 106 cells were pretreated with NPI-0052 for 4 hours, treated with TNF for 18 hours, and then subjected to annexin V and propidium iodide (PI) staining. Cells were washed, stained with FITC-conjugated anti-annexin V antibody, and then analyzed with a flow cytometer (FACSCalibur; BD Biosciences).
Terminal deoxynucleotidyl transferase-mediated deoxyuridine triphosphate nick end-labeling (TUNEL) assay
We assayed cytotoxicity by the TUNEL method, which examines DNA strand breaks during apoptosis, using an in situ cell death detection reagent (Roche Molecular Biochemicals, Mannheim, Germany). Briefly, 106 cells were pretreated with NPI-0052 for 4 hours and treated with TNF for 18 hours at 37°C. Thereafter, cells were incubated with reaction mixture for 60 minutes at 37°C. Stained cells were analyzed by flow cytometer (FACSCalibur, BD Biosciences).
PARP cleavage assay
For detection of cleavage products of PARP, whole-cell extracts were prepared by subjecting NPI-0052-treated cells to lysis in lysis buffer (20 mM Tris, pH 7.4; 250 mM NaCl; 2 mM [ethylenedinitrilo]tetraacetic acid [EDTA], pH 8.0; 0.1% TritonX-100; 0.01 μg/mL aprotinin; 0.005 μg/mL leupeptin; 0.4 mM phenylmethylsulfonyl fluoride [PMSF]; and 4 mM Na3VO4). Lysates were spun at 20 817g for 10 minutes to remove insoluble material, resolved by 10% sodium dodecyl sulfate polyacrylamide gel electrophoresis (SDS-PAGE), and probed with PARP antibodies.
Western blot analysis
To determine the effect of NPI-0052 on TNF-dependent IκBα phosphorylation, IκBα degradation, and p65 translocation, cytoplasmic and nuclear extracts were prepared as previously described13 from KBM-5 cells (1 × 106/mL) that had been pretreated with NPI-0052 for 4 hours and then exposed to TNF for various times. Cytoplasmic and nuclear proteins (30 μg) were resolved on 10% SDS-PAGE gel, transferred to a nitrocellulose membrane, blocked with 5% nonfat milk, and probed with specific antibodies against IκBα, posphorylated IκBα, and p65. To determine the expression of cyclin D1, COX-2, MMP-9, ICAM-1, c-Myc, IAP-1, IAP-2, Bcl-2, Bcl-xL, VEGF, TRAF1, cFLIP, and survivin in whole-cell extracts of treated cells (106 cells in 1 mL of medium), 30 μg of whole-cell lysate was resolved by SDS-PAGE, electrotransferred to nitrocellulose membrane, sliced based on the molecular weight, and then probed with antibodies against various proteins. The blots were washed, exposed to horseradish peroxidase (HRP)–conjugated secondary antibodies for 1 hour, and finally detected by enhanced chemiluminescence (ECL) reagent (Amersham Pharmacia Biotechnology, Piscataway, NJ). The bands were quantified using a personal densitometer scan v1.30 using Imagequant software version 3.3 (Molecular Dynamics, Piscataway, NJ).
Invasion assay
The membrane invasion culture system was used to assess cell invasion because invasion through the extracellular matrix is a crucial step in tumor metastasis. The BD BioCoat tumor invasion system is a chamber that has a light-tight polyethelyene terephthalate membrane with 8-μm-diameter pores and is coated with a reconstituted basement membrane gel (BD Biosciences). A total of 2.5 × 105 H1299 cells were suspended in serum-free medium and seeded into the upper wells. After incubation overnight, cells were treated with NPI-0052 for 4 hours and then stimulated with TNF for a further 24 hours in the presence of 5% FBS and the NPI-0052. The cells that invaded through the Matrigel (ie, those that migrated to the lower chamber during incubation) were stained with 4 μg/mL calcein-AM (Molecular Probes) in phosphate-buffered saline (PBS) for 30 minutes at 37°C and scanned for fluorescence with a Victor 3 multiplate reader (Perkin Elmer Life and Analytical Sciences, Boston, MA); fluorescent cells were counted.
Osteoclast differentiation assay
To determine the effect of NPI-0052 on receptor activator for nuclear factor κB ligand (RANKL)–induced osteoclastogenesis, we cultured RAW 264.7 cells, which can differentiate into osteoclasts by RANKL in vitro.14 RAW 264.7 cells were cultured in 24-well dishes at a density of 104 cells per well and allowed to adhere overnight. The medium was then replaced, and the cells were pretreated with NPI-0052 for 4 hours and then treated with RANKL. After 5 days of incubation, the cells were stained for tartrate-resistant acid phosphatase (TRAP) expression, as previously described,14 using an acid phosphatase kit (Sigma-Aldrich), and the TRAP-positive multinucleated osteoclasts (> 3 nuclei) per well were counted.
Electrophoretic mobility shift assay
To determine NF-κB activation by TNF, we performed electrophoretic mobility shift assay (EMSA) essentially as previously described.15
IKK assay
To determine the effect of NPI-0052 on TNF-induced IKK activation, we analyzed IKK by a method essentially as described previously.13
Immunolocalization of NF-κB p65
The effect of NPI-0052 on the TNF-induced nuclear translocation of p65 was examined by an immunocytochemical method using an epifluorescence microscope (Labophot-2; Nikon, Tokyo, Japan) and a Photometrics Coolsnap CF color camera (Nikon, Lewisville, TX) as described previously.13
NF-κB–dependent reporter gene transcription
The effect of NPI-0052 on TNF-induced NF-κB–dependent reporter gene transcription in A293 cells was measured as previously described.14
In vitro chymotrypsin-like 20S proteasome activity assay
The chymotrypsin-like activity of the 20S proteasome in cells was determined by measurement of fluorescence generated from the cleavage of the fluorogenic substrate Suc-LLVY-AMC. RPMI-8226 and PC-3 cells were treated with various concentrations of NPI-0052 or bortezomib for 1 hour and washed with cold Dulbecco PBS (D-PBS), resuspended in cold lysis buffer (20 mM HEPES [4-(2-hydroxyethyl)-1-piperazineethane sulfonic acid], 0.5 mM EDTA, 0.05% Triton X-100 and pH 7.3, supplemented with a Complete Mini proetease inhibitor cocktail tablet [1 tablet per 10 mL of lysis buffer]) and lysed on ice for 15 minutes. Cell lysates were cleared by centrifugation at 20 817g for 10 minutes at 4°C. Protein content was determined for each sample using the Bradford reagent (Bio-Rad, Hercules, CA). Supernatants (20 μg) were combined with substrate buffer (20 mM HEPES, 0.5 mM EDTA, pH 8.0 with a final SDS concentration of 0.035%), containing 20 μM peptide substrate in 96-well plate. Fluorescence was read on a Fluorokan Ascent flurometer (Thermo Electron, Waltham, MA) using an excitation of 390 nm and an emission of 460 nm.
Results
The goal of this study was to investigate the effect of NPI-0052 on the transcription factor NF-κB signaling pathway, on NF-κB–regulated gene products, and on NF-κB–mediated cellular responses. The concentration of NPI-0052 used to study its effect on NF-κB pathway and the duration of exposure had minimal effect on the viability of cells, as determined by the trypan blue dye exclusion test (data not shown). To examine the effect of NPI-0052 on the NF-κB activation pathway, we used TNF because the pathway activated by this agent is relatively well understood.
NPI-0052 potentiates the apoptotic effects of TNF and chemotherapeutic drugs
Because NF-κB activation has been shown to suppress the apoptosis induced by various drugs,16,17 we determined the potential of NPI-0052 to enhance apoptosis induced by TNF and chemotherapeutic drugs using the Live and Dead, PARP cleavage, annexin V staining, and TUNEL staining methods in KBM5 cells. The effect of NPI-0052 on TNF- and drug-induced apoptosis was examined by the MTT assay. We found that NPI-0052 enhanced the cytotoxic effects of TNF, thalidomide, and bortezomib (Figure 1A). The Live/Dead assay, which measures intracellular esterase activity and plasma membrane integrity, showed that NPI-0052 up-regulated TNF-induced apoptosis from 20% to 68%. NPI-0052 also potentiated the apoptotic effects of thalidomide and bortezomib (Figure 1B). These effects were not limited to human chronic myeloid leukemia cells because NPI-0052 also enhanced the apoptotic effects of thalidomide and bortezomib in human multiple myeloma (U266) and human prostate DU145 cells (Figure 1C). To determine whether this effect is dose-dependent, cells were treated with different concentrations of NPI-0052 in the presence of either TNF, thalidomide, or bortezomib. The apoptosis by all these agents was enhanced by NPI-0052 in a dose-dependent manner (Tables 1 and 2). Confirming these results, annexin V staining showed that NPI-0052 up-regulated TNF-induced early apoptosis from 17% to 65% (Figure 1D). The TUNEL staining method showed that NPI-0052 enhanced apoptosis from 5% to 31% (Figure 1E). Lastly, using the PARP-cleavage assay to detect TNF-induced caspase activation, again we found that NPI-0052 potentiated TNF-induced apoptosis (Figure 1F). All these assays demonstrate that NPI-0052 enhanced the apoptotic effects of TNF and chemotherapeutic drugs.
NPI-0052 enhances apoptosis induced by TNF and chemotherapeutic drugs. (A) KBM-5 cells (10 000 cells/0.1 mL) were incubated at 37°C with 1 nM TNF, 10 μg/mL thalidomide, or 20 nM bortezomib in the presence and absence of 50 nM NPI-0052 as indicated for 18 hour, and the viable cells were assayed using the MTT reagent. The results are expressed as mean cytotoxicity (± standard deviation [SD]) from triplicate cultures. (B) Cells (106/mL) were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF, 10 μg/mL thalidomide, or 20 nM bortezomib for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are representative of 3 independent experiments showing similar results. (C) U266 (106/mL) or DU145 (105/mL) cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 10 μg/mL thalidomide or 20 nM bortezomib for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are representative of 3 independent experiments showing similar results. Images were acquired as described in “Live and dead assay.” (D) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 18 hours. Afterward, they were incubated with anti-annexin V antibody conjugated with FITC plus PI and analyzed with a flow cytometer for apoptotic effects. (E) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 18 hours. Cells were fixed, stained with TUNEL assay reagent, and then analyzed with flow cytometer. (F) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and subjected to Western blot analysis using anti-PARP antibody.
NPI-0052 enhances apoptosis induced by TNF and chemotherapeutic drugs. (A) KBM-5 cells (10 000 cells/0.1 mL) were incubated at 37°C with 1 nM TNF, 10 μg/mL thalidomide, or 20 nM bortezomib in the presence and absence of 50 nM NPI-0052 as indicated for 18 hour, and the viable cells were assayed using the MTT reagent. The results are expressed as mean cytotoxicity (± standard deviation [SD]) from triplicate cultures. (B) Cells (106/mL) were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF, 10 μg/mL thalidomide, or 20 nM bortezomib for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are representative of 3 independent experiments showing similar results. (C) U266 (106/mL) or DU145 (105/mL) cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 10 μg/mL thalidomide or 20 nM bortezomib for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are representative of 3 independent experiments showing similar results. Images were acquired as described in “Live and dead assay.” (D) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 18 hours. Afterward, they were incubated with anti-annexin V antibody conjugated with FITC plus PI and analyzed with a flow cytometer for apoptotic effects. (E) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 18 hours. Cells were fixed, stained with TUNEL assay reagent, and then analyzed with flow cytometer. (F) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and subjected to Western blot analysis using anti-PARP antibody.
KBM-5 cells treated with NPI-0052, TNF, thalidomide, and bortezomib alone or in combination
| NPI-0052, nM . | NPI-0052, % apoptosis . | TNF, % apoptosis . | Thalidomide, % apoptosis . | Bortezomib, % apoptosis . | |||
|---|---|---|---|---|---|---|---|
| 1 nM . | 5 nM . | 5 μg/mL . | 10 μg/mL . | 10 nM . | 20 nM . | ||
| 0 | 1 ± 0.6 | 12.3 ± 2.1 | 23.3 ± 4.2 | 7 ± 2 | 11.3 ± 3.5 | 6.7 ± 2.1 | 10.7 ± 2.1 |
| 25 | 7 ± 2 | 24 ± 5.3 | 36.7 ± 5.5 | 14 ± 4 | 26 ± 5.3 | 12.7 ± 3.1 | 33.7 ± 4 |
| 50 | 20.7 ± 4 | 65.3 ± 6.5 | 86.3 ± 3.5 | 36 ± 3.6 | 54.3 ± 4 | 29.7 ± 3.5 | 55.7 ± 2.1 |
| 100 | 40.7 ± 4 | 93 ± 2.0 | 98 ± 2.6 | 50 ± 3 | 88 ± 5.6 | 55.3 ± 3.5 | 91.3 ± 1.2 |
| NPI-0052, nM . | NPI-0052, % apoptosis . | TNF, % apoptosis . | Thalidomide, % apoptosis . | Bortezomib, % apoptosis . | |||
|---|---|---|---|---|---|---|---|
| 1 nM . | 5 nM . | 5 μg/mL . | 10 μg/mL . | 10 nM . | 20 nM . | ||
| 0 | 1 ± 0.6 | 12.3 ± 2.1 | 23.3 ± 4.2 | 7 ± 2 | 11.3 ± 3.5 | 6.7 ± 2.1 | 10.7 ± 2.1 |
| 25 | 7 ± 2 | 24 ± 5.3 | 36.7 ± 5.5 | 14 ± 4 | 26 ± 5.3 | 12.7 ± 3.1 | 33.7 ± 4 |
| 50 | 20.7 ± 4 | 65.3 ± 6.5 | 86.3 ± 3.5 | 36 ± 3.6 | 54.3 ± 4 | 29.7 ± 3.5 | 55.7 ± 2.1 |
| 100 | 40.7 ± 4 | 93 ± 2.0 | 98 ± 2.6 | 50 ± 3 | 88 ± 5.6 | 55.3 ± 3.5 | 91.3 ± 1.2 |
Data shown are means plus or minus the standard deviation.Cells (106/mL) were treated with indicated concentrations of NPI-0052, TNF, thalidomide, and bortezomib alone or in combination for 16 hours at 37oC. Cells were stained with a live/dead assay reagent for 30 minutes and then analyzed under a fluorescence microscope as described in ″Live and dead assay.″ The results shown are the percentage of apoptosis and are representative of 3 independent experiments.
U266 cells treated with NPI-0052, thalidomide, and bortezomib alone or in combination
| NPI-0052, nM . | NPI-0052, % apoptosis . | Thalidomide, % apoptosis . | Bortezomib, % apoptosis . | ||
|---|---|---|---|---|---|
| 5 μg/mL . | 10 μg/mL . | 10 nM . | 20 nM . | ||
| 0 | 1.3 ± 0.6 | 6 ± 1 | 11.3 ± 1.5 | 9 ± 1 | 18.7 ± 1.5 |
| 50 | 3 ± 1 | 34 ± 3.6 | 82.3 ± 3.1 | 25 ± 2.6 | 56 ± 2.6 |
| 100 | 20.7 ± 4 | 57 ± 3.6 | 99.7 ± 0.6 | 52.7 ± 2.5 | 90.3 ± 1.5 |
| NPI-0052, nM . | NPI-0052, % apoptosis . | Thalidomide, % apoptosis . | Bortezomib, % apoptosis . | ||
|---|---|---|---|---|---|
| 5 μg/mL . | 10 μg/mL . | 10 nM . | 20 nM . | ||
| 0 | 1.3 ± 0.6 | 6 ± 1 | 11.3 ± 1.5 | 9 ± 1 | 18.7 ± 1.5 |
| 50 | 3 ± 1 | 34 ± 3.6 | 82.3 ± 3.1 | 25 ± 2.6 | 56 ± 2.6 |
| 100 | 20.7 ± 4 | 57 ± 3.6 | 99.7 ± 0.6 | 52.7 ± 2.5 | 90.3 ± 1.5 |
Data shown are means plus or minus the standard deviation. Cells (106/mL) were treated as in Table 1.
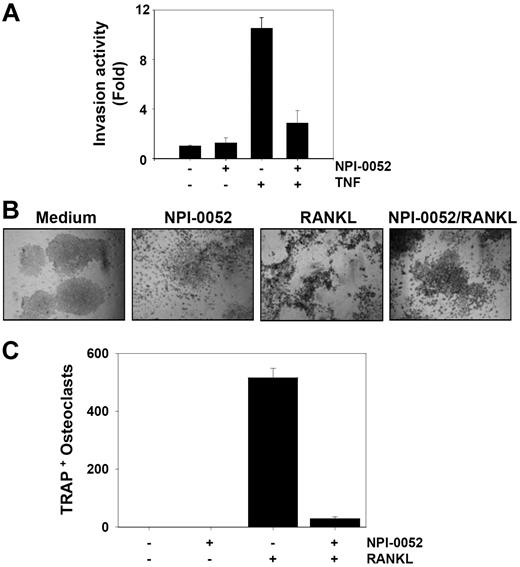
NPI-0052 suppresses TNF-induced tumor cell invasion activity
TNF can induce the expression of genes involved in tumor metastasis.18 MMPs, COXs, and adhesion molecules regulated by NF-κB have been linked with tumor invasion.19 Whether NPI-0052 can modulate TNF-induced tumor cell invasion activity was investigated in vitro. To determine this, tumor cells were seeded to the top chamber of the Matrigel invasion chamber with TNF in the presence or absence of NPI-0052 and then examined for invasion. As shown in Figure 2A, TNF induced tumor cell invasion by almost 10-fold, and NPI-0052 suppressed this activity. NPI-0052 alone had no effect on invasion activity.
NPI-0052 suppresses TNF-induced invasive activity and RANKL-induced osteoclastogenesis. (A) H1299 cells (2.5 × 104) were seeded into the upper wells of a Matrigel invasion chamber overnight in the absence of serum, pretreated with 50 nM NPI-0052 for 4 hours, treated with 1 nM TNF for 24 hours in the presence of 5% serum, and then subjected to invasion assay. The value for no NPI-0052 and no TNF was set to 1.0. (B) RAW 264.7 cells (104) were plated overnight, pretreated with 50 nM NPI-0052 for 4 hours, and then treated with 5 nM RANKL. At 5 days later, cells were stained for TRAP and evaluated for osteoclastogenesis. Cells were analyzed under a phase contrast microscope (Nikon, Tokyo, Japan) and photographs were taken using Photometrics Coolsnap CF color camera (Nikon, Lewisville, TX). (C) Photographs were taken after 5 days of incubation with RANKL. The numbers of TRAP-positive multinucleated osteoclasts (> 3 nuclei) per well were counted. Error bars indicate SD of triplicate value.
NPI-0052 suppresses TNF-induced invasive activity and RANKL-induced osteoclastogenesis. (A) H1299 cells (2.5 × 104) were seeded into the upper wells of a Matrigel invasion chamber overnight in the absence of serum, pretreated with 50 nM NPI-0052 for 4 hours, treated with 1 nM TNF for 24 hours in the presence of 5% serum, and then subjected to invasion assay. The value for no NPI-0052 and no TNF was set to 1.0. (B) RAW 264.7 cells (104) were plated overnight, pretreated with 50 nM NPI-0052 for 4 hours, and then treated with 5 nM RANKL. At 5 days later, cells were stained for TRAP and evaluated for osteoclastogenesis. Cells were analyzed under a phase contrast microscope (Nikon, Tokyo, Japan) and photographs were taken using Photometrics Coolsnap CF color camera (Nikon, Lewisville, TX). (C) Photographs were taken after 5 days of incubation with RANKL. The numbers of TRAP-positive multinucleated osteoclasts (> 3 nuclei) per well were counted. Error bars indicate SD of triplicate value.
NPI-0052 suppresses RANKL-induced osteoclastogenesis
One of the key factors mediating osteoclastogenesis is receptor activator of nuclear factor-κB (NF-κB) ligand (RANKL), a member of the TNF superfamily. The interaction of RANKL with RANK results in a cascade of intracellular events, including the activation of the NF-κB signaling pathway.20,21 We investigated whether NPI-0052 can suppress RANKL-induced osteoclastogenesis. We found that RANKL induced osteoclast differentiation, as indicated by the expression of TRAP, and that NPI-0052 suppressed it (Figure 2B,C). Neither NPI-0052 nor RANKL exhibited significant cytotoxicity in RAW 264.7 cells (data not shown).
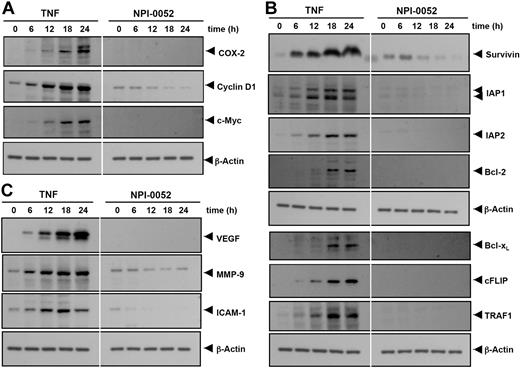
NPI-0052 suppresses TNF-induced NF-κB–dependent cell proliferation gene products
Both cyclin D1 and COX-2 contribute to carcinogenesis by promoting cell proliferation. Cyclin D1 is a protein that controls the transition from G1 to S phase in the cell cycle and is overexpressed in a variety of tumors.22 COX-2 is an enzyme that catalyzes the production of prostaglandin E2 (PGE2) from arachidonic acid, which has been linked to proliferation and metastasis of tumor cells.23 We examined whether NPI-0052 modulates the expression of proliferative gene products induced by TNF. As shown in Figure 3A, TNF induced the expression of COX-2, cyclin D1, and c-myc proteins in a time-dependent manner, and NPI-0052 significantly inhibited their expression.
NPI-0052 represses TNF-induced NF-κB–dependent expression of proliferation-, antiapoptosis-, and metastasis-related gene products. Proliferative (A), antiapoptotic (B), and metastatic (C) gene products are shown. KBM-5 cells were incubated with 50 nM NPI-0052 for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared, and 30 μg of the whole-cell lysate was resolved by SDS-PAGE, electrotransferred to nitrocellulose membrane, sliced from the membrane based on the molecular weight, and then probed with antibodies against survivin, IAP1/2, Bcl-2, Bcl-xL, cFLIP, TRAF1, VEGF, MMP-9, ICAM-1, COX-2, c-Myc, Cyclin D1, or β-actin as described in “Materials and methods.” TNF-treated and TNF plus NPI-0052-treated samples were run on the same gel under identical conditions and probed with the same immunoblotting solutions.
NPI-0052 represses TNF-induced NF-κB–dependent expression of proliferation-, antiapoptosis-, and metastasis-related gene products. Proliferative (A), antiapoptotic (B), and metastatic (C) gene products are shown. KBM-5 cells were incubated with 50 nM NPI-0052 for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared, and 30 μg of the whole-cell lysate was resolved by SDS-PAGE, electrotransferred to nitrocellulose membrane, sliced from the membrane based on the molecular weight, and then probed with antibodies against survivin, IAP1/2, Bcl-2, Bcl-xL, cFLIP, TRAF1, VEGF, MMP-9, ICAM-1, COX-2, c-Myc, Cyclin D1, or β-actin as described in “Materials and methods.” TNF-treated and TNF plus NPI-0052-treated samples were run on the same gel under identical conditions and probed with the same immunoblotting solutions.
NPI-0052 suppresses TNF-induced NF-κB–dependent antiapoptotic gene products
NF-κB regulates the expression of the antiapoptotic proteins such as survivin, IAP1/2, Bcl-2, Bcl-xL, cFLIP, and TRAF1, all of which play a major role in evading apoptosis and prolonging survival of cancer cells.10,24 We examined whether NPI-0052 modulates TNF-induced expression of these genes. As shown in Figure 3B, NPI-0052 significantly inhibited TNF-induced expression of these antiapoptotic proteins in a time-dependent manner.
NPI-0052 suppresses TNF-induced NF-κB–dependent gene products involved in tumor metastasis
VEGF plays a critical role in angiogenesis by promoting the growth of vascular endothelial cells and enhancing vascular permeability,25 whereas MMP-9, by virtue of its property to degrade extracellular matrix, has been implicated in cellular invasion.26 ICAM-1, an adhesion molecule, has also been shown to be required for metastasis of tumors.27 Hence, we determined whether NPI-0052 could modulate TNF-induced expression of VEGF, MMP-9, and ICAM-1 proteins. We found that TNF treatment induced the expression of VEGF, MMP-9, and ICAM-1 gene products in a time-dependent manner and that NPI-0052 inhibited the expression (Figure 3C).
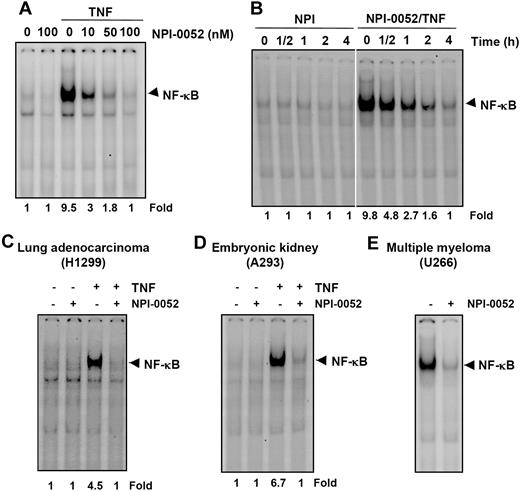
NPI-0052 suppresses NF-κB activation in a dose- and time-dependent manner
We next determined the dose and time of exposure to NPI-0052 required to suppress NF-κB activation. The EMSA results showed that NPI-0052 alone had no effect on NF-κB activation. However, it inhibited TNF-mediated NF-κB activation in a dose-dependent manner (Figure 4A). The suppression of NF-κB activation by NPI-0052 was also found to be time dependent (Figure 4B).
NPI-0052 inhibits both inducible and constitutive NF-κB activation in a dose- and time-dependent manner. (A) KBM-5 cells were incubated with the indicated concentrations of NPI-0052 for 4 hours and then exposed to 0.1 nM TNF for 30 minutes. The nuclear extracts were subjected to EMSA to evaluate NF-κB activation. (B) Cells were preincubated with 50 nM NPI-0052 for the indicated times, treated with 0.1 nM TNF for 30 minutes, and then subjected to EMSA to evaluate NF-κB activation. H1299 (C) or A293 (D) cells were pretreated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for 30 minutes. The nuclear extracts were then prepared and assayed for NF-κB by EMSA as described in “Materials and methods.” (E) U266 cells with constitutive NF-κB activation cells were incubated with (±) 50 nM NPI-0052 for 12 hours. Nuclear extracts were prepared and analyzed for NF-κB activation by EMSA.
NPI-0052 inhibits both inducible and constitutive NF-κB activation in a dose- and time-dependent manner. (A) KBM-5 cells were incubated with the indicated concentrations of NPI-0052 for 4 hours and then exposed to 0.1 nM TNF for 30 minutes. The nuclear extracts were subjected to EMSA to evaluate NF-κB activation. (B) Cells were preincubated with 50 nM NPI-0052 for the indicated times, treated with 0.1 nM TNF for 30 minutes, and then subjected to EMSA to evaluate NF-κB activation. H1299 (C) or A293 (D) cells were pretreated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for 30 minutes. The nuclear extracts were then prepared and assayed for NF-κB by EMSA as described in “Materials and methods.” (E) U266 cells with constitutive NF-κB activation cells were incubated with (±) 50 nM NPI-0052 for 12 hours. Nuclear extracts were prepared and analyzed for NF-κB activation by EMSA.
Inhibition of NF-κB activation by NPI-0052 is not cell type specific
It has been reported that the NF-κB induction pathway in epithelial cells may differ from that in lymphoid cells.28 We therefore investigated whether NPI-0052 inhibited NF-κB activation in different cell types. NPI-0052 completely inhibited TNF-induced NF-κB activation in lung adenocarcinoma (H1299) and embryonic kidney cells (A293) (Figure 4C,D), indicating a lack of cell type specificity.
NPI-0052 inhibits constitutive NF-κB activation
We next tested the effect of NPI-0052 on NF-κB activation in human multiple myeloma (U266), which have been reported to express constitutively active NF-κB.29 U266 cells were treated with NPI-0052 for 12 hours and then analyzed for NF-κB activation. NPI-0052 inhibited constitutively active NF-κB in these cells (Figure 4E).
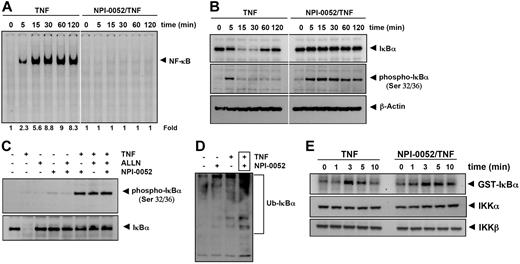
NPI-0052 inhibits TNF-dependent IκBα degradation
To determine whether the NPI-0052 inhibitory activity was due to inhibition of IκBα degradation, we pretreated cells with NPI-0052 for 4 hours, exposed them to TNF for different times, and examined them for NF-κB in the nucleus by EMSA. Increasing activation of NF-κB was detected with increases in incubation times with TNF. The NPI-0052-treated cells showed a complete suppression in activation of NF-κB induced by TNF stimulation (Figure 5A). Because the translocation of NF-κB to the nucleus is preceded by the proteolytic degradation of IκBα,30 we determined whether NF-κB inhibitory activity of NPI-0052 was due to inhibition of IκBα degradation. We pretreated cells with NPI-0052, exposed them to TNF for different times, and examined the IκBα status in the cytoplasm by Western blot analysis. TNF induced IκBα degradation in control cells within 5 minutes and reached maximum at 15 to 30 minutes, but TNF could not induce IκBα degradation in NPI-0052 pretreated cells (Figure 5B upper panel). These results indicate that NPI-0052 inhibits TNF-induced IκBα degradation.
NPI-0052 inhibits TNF-dependent NF-κB activation and IκBα degradation. (A) Cells were preincubated with 50 nM NPI-0052 for 4 hours, treated with 0.1 nM TNF for the indicated times, and then subjected to EMSA and Western blot analysis to evaluate NF-κB activation. (B) Cells were incubated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for the indicated times. Cytoplasmic extracts were prepared, fractionated on SDS-PAGE, and electrotransferred to a nitrocellulose membrane. Western blot analysis was performed with anti-IκBα and anti-phospho-specific anti-IκBα. (C) Cells were preincubated with 50 nM NPI-0052 for 4 hours, incubated with 50 μg/mL N-acetyl-leucyl-leucyl-norleucinal (ALLN) for 30 minutes, and then treated with 0.1 nM TNF for 15 minutes. Cytoplasmic extracts were fractionated and then subjected to Western blot analysis using phospho-specific anti-IκBα antibody. The same membrane was reblotted with anti-IκBα antibody. (D) Cells were preincubated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for 15 minutes. Cytoplasmic extracts were immunoprecipitated with antibody against IκBα then subjected to Western blot analysis using monoclonal anti-ubiquitin antibody. (E) Cells were preincubated with 50 nM NPI-0052 for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts (1 mg/mL) were immunoprecipitated with antibody against IKK-α and analyzed using an immunocomplex kinase assay. Data are for a representative experiment of 3 independent ones showing similar results.
NPI-0052 inhibits TNF-dependent NF-κB activation and IκBα degradation. (A) Cells were preincubated with 50 nM NPI-0052 for 4 hours, treated with 0.1 nM TNF for the indicated times, and then subjected to EMSA and Western blot analysis to evaluate NF-κB activation. (B) Cells were incubated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for the indicated times. Cytoplasmic extracts were prepared, fractionated on SDS-PAGE, and electrotransferred to a nitrocellulose membrane. Western blot analysis was performed with anti-IκBα and anti-phospho-specific anti-IκBα. (C) Cells were preincubated with 50 nM NPI-0052 for 4 hours, incubated with 50 μg/mL N-acetyl-leucyl-leucyl-norleucinal (ALLN) for 30 minutes, and then treated with 0.1 nM TNF for 15 minutes. Cytoplasmic extracts were fractionated and then subjected to Western blot analysis using phospho-specific anti-IκBα antibody. The same membrane was reblotted with anti-IκBα antibody. (D) Cells were preincubated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for 15 minutes. Cytoplasmic extracts were immunoprecipitated with antibody against IκBα then subjected to Western blot analysis using monoclonal anti-ubiquitin antibody. (E) Cells were preincubated with 50 nM NPI-0052 for 4 hours and then treated with 1 nM TNF for the indicated times. Whole-cell extracts (1 mg/mL) were immunoprecipitated with antibody against IKK-α and analyzed using an immunocomplex kinase assay. Data are for a representative experiment of 3 independent ones showing similar results.
NPI-0052 has no effect on TNF-dependent IκBα phosphorylation
We next determined whether NPI-0052 affected the TNF-induced IκBα phosphorylation needed for IκBα degradation. TNF induced IκBα phosphorylation in control cells within 5 minutes, but NPI-0052-treated cells sustained the phosphorylation of IκBα in overall interval times (Figure 5B middle panel). To confirm this result, we used N-acetyl-leucyl-leucyl-norleucinal (ALLN), which suppresses the degradation of IκBα by 26S proteasome. Western blot analysis indicated that both ALLN- and NPI-0052-treated cells detected TNF-induced IκBα phosphorylation, (Figure 5C). Thus NPI-0052 accumulated TNF-induced phosphorylation of IκBα through suppression of proteasome function.
Whether NPI-0052 stabilizes TNF-induced ubiquitinated IκBα was examined. As shown in Figure 5D, high-molecular-mass bands appeared after stimulation with TNF in presence of NPI-0052, indicating stabilization. NPI-0052 treatment alone did not induce any substantial IκBα ubiquitination.
NPI-0052 did not affect TNF-induced IKK activation
We tested the effect of NPI-0052 on TNF-induced IKK activation, which is required for TNF-induced phosphorylation of IκBα. As shown in Figure 5E (upper panel), NPI-0052 did not suppress TNF-induced activation of IKK. TNF or NPI-0052 had no direct effect on the expression of IKK protein (Figure 5E bottom panel).
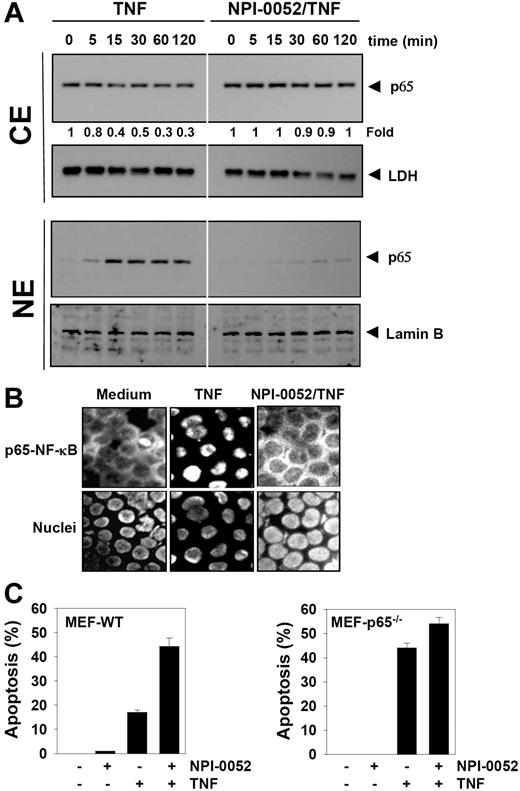
NPI-0052 inhibits TNF-induced nuclear translocation of p65
The degradation of IκBα leads to nuclear translocation of p65. Whether NPI-0052 affects the TNF-induced nuclear translocation of p65 was tested by 2 independent methods. As shown by Western blot analysis in Figure 6A (CE panel), NPI-0052 abolished TNF-induced nuclear translocation of p65 significantly. Likewise, TNF induced nuclear translocation of p65 in a time-dependent manner, and NPI-0052 blocked its translocation (Figure 6A NE panel). Immunocytochemical analysis confirmed that TNF induced nuclear translocation of p65 and NPI-0052 inhibited it in KBM-5 cells (Figure 6B).
NPI-0052 inhibits TNF-induced p65 nuclear translocation. (A) KBM-5 cells were incubated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for the indicated times. Cytoplasmic and nuclear extracts were prepared, fractionated on SDS-PAGE, and electrotransferred to a nitrocellulose membrane. The analysis was performed using p65 antibodies. (B) Immunocytochemical analysis of TNF-induced p65 nuclear translocation. KBM-5 cells were incubated with 50 nM NPI-0052 for 4 hours, treated with 1 nM TNF for 15 minutes, and then subjected to immunocytochemical analysis as described in “Immunolocalization of NF-κB p65.” Cells plated with Mounting Media (Sigma-Aldrich) were analyzed under a fluorescence microscope (Lapshot-2, Nikon) equipped with a CFWN 10 /1.5 NA oil-immersion objective lens and a Photometrics Coolsnap CF color camera (Nikon). Images were acquired with MetaMorph 4.6.5 software (Universal Imaging). (C) The wild-type and p65−/− (105/mL) cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are for a representative experiment of 3 independent ones showing similar results. Error bars represent SD of triplicate values.
NPI-0052 inhibits TNF-induced p65 nuclear translocation. (A) KBM-5 cells were incubated with 50 nM NPI-0052 for 4 hours and then treated with 0.1 nM TNF for the indicated times. Cytoplasmic and nuclear extracts were prepared, fractionated on SDS-PAGE, and electrotransferred to a nitrocellulose membrane. The analysis was performed using p65 antibodies. (B) Immunocytochemical analysis of TNF-induced p65 nuclear translocation. KBM-5 cells were incubated with 50 nM NPI-0052 for 4 hours, treated with 1 nM TNF for 15 minutes, and then subjected to immunocytochemical analysis as described in “Immunolocalization of NF-κB p65.” Cells plated with Mounting Media (Sigma-Aldrich) were analyzed under a fluorescence microscope (Lapshot-2, Nikon) equipped with a CFWN 10 /1.5 NA oil-immersion objective lens and a Photometrics Coolsnap CF color camera (Nikon). Images were acquired with MetaMorph 4.6.5 software (Universal Imaging). (C) The wild-type and p65−/− (105/mL) cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are for a representative experiment of 3 independent ones showing similar results. Error bars represent SD of triplicate values.
NPI-0052 did not potentiate TNF-induced apoptosis in p65−/− cells
To determine whether inhibition of NF-κB is a major mechanism by which NPI-0052 mediates its effects, we used the wild-type and p65−/− cells to compare the apoptosis induced by TNF. Results showed that NPI-0052 potentiated TNF-induced apoptosis from 1% to 44% in wild-type cells. In p65−/− cells that lack functional NF-κB, TNF alone could induce apoptosis. We found that NPI-0052 neither had any effect alone nor could it significantly potentiate the apoptotic effect induced by TNF in p65−/− cells (Figure 6C).
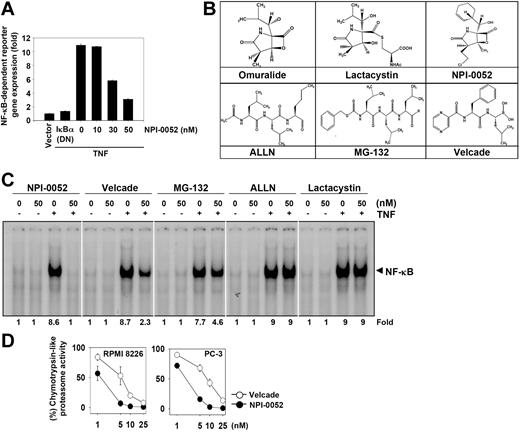
NPI-0052 represses TNF-induced NF-κB–dependent reporter gene expression
Our results up to this point showed that NPI-0052 inhibited the translocation of p65 inside the nucleus. To further show that NPI-0052 inhibited NF-κB–dependent gene transcription, we transiently transfected cells with the NF-κB–regulated secretory alkaline phosphatase (SEAP) reporter construct and then stimulated them with TNF. We found that TNF produced an almost 11-fold increase in SEAP activity over vector control (Figure 7A), which was inhibited by dominant-negative IκBα, indicating specificity. When the cells were pretreated with NPI-0052, TNF-induced NF-κB–dependent SEAP expression was inhibited in a dose-dependent manner. These results demonstrated that NPI-0052 inhibits the NF-κB–dependent reporter gene expression induced by TNF.
NPI-0052 inhibits TNF-induced NF-κB–dependent reporter gene (SEAP) expression and suppresses chymotrypsin-like activity of 20S proteasome. (A) A293 cells were transiently transfected with an NF-κB–containing plasmid linked to the SEAP gene and then pretreated with the indicated concentrations of NPI-0052 for 4 hours. After 24 hours in culture with 1 nM TNF, cell supernatants were collected and assayed for SEAP activity as described in “Materials and methods.” Results are expressed as fold activity over the activity of the vector control. (B) Chemical structures of various proteasome inhibitors. (C) KBM-5 (1 × 106) cells were incubated with 50 nM each of various proteasome inhibitors for 4 hours and then exposed to 0.1 nM TNF for 30 minutes. The nuclear extracts were subjected to EMSA to evaluate NF-κB activation. (D) RPMI 8226 and PC-3 cells were treated with various concentrations of NPI-0052 and bortezomib for 1 hour, prepared the cell lysates, and assayed for the chymotrypsin-like activity of the 20S proteasome as described in “Materials and methods.” Results are presented as percentage inhibition compared with the chymotrypsin-like activity observed in untreated cells. Error bars represent SD of triplicate values.
NPI-0052 inhibits TNF-induced NF-κB–dependent reporter gene (SEAP) expression and suppresses chymotrypsin-like activity of 20S proteasome. (A) A293 cells were transiently transfected with an NF-κB–containing plasmid linked to the SEAP gene and then pretreated with the indicated concentrations of NPI-0052 for 4 hours. After 24 hours in culture with 1 nM TNF, cell supernatants were collected and assayed for SEAP activity as described in “Materials and methods.” Results are expressed as fold activity over the activity of the vector control. (B) Chemical structures of various proteasome inhibitors. (C) KBM-5 (1 × 106) cells were incubated with 50 nM each of various proteasome inhibitors for 4 hours and then exposed to 0.1 nM TNF for 30 minutes. The nuclear extracts were subjected to EMSA to evaluate NF-κB activation. (D) RPMI 8226 and PC-3 cells were treated with various concentrations of NPI-0052 and bortezomib for 1 hour, prepared the cell lysates, and assayed for the chymotrypsin-like activity of the 20S proteasome as described in “Materials and methods.” Results are presented as percentage inhibition compared with the chymotrypsin-like activity observed in untreated cells. Error bars represent SD of triplicate values.
NPI-0052 is the most potent NF-κB blocker among the different proteasome inhibitors
Besides NPI-0052, several other proteasome inhibitors have been described including bortezomib, MG-132, ALLN, and lactacystin. The latter is a precursor of omuralide (Figure 7B). Bortezomib has been approved for the treatment of multiple myeloma. Whether these proteasome inhibitors are as effective as NPI-0052 in suppressing TNF-induced NF-κB activation was investigated. At the same concentration, EMSA results showed that NPI-0052 was maximally effective in inhibiting TNF-mediated NF-κB activation; bortezomib and MG-132 showed a partial inhibition. At the dose tested, ALLN and lactacystin had no effect on TNF-induced NF-κB activation (Figure 7C). Thus NPI-0052 was the most potent inhibitor among the known proteasome inhibitors.
NPI-0052 is more potent than bortezomib in inhibiting 20S proteasome in tumor cells
Whether enhanced NF-κB suppressive activity of NPI-0052 and bortezomib is linked to inhibition of the chymotrypsin-like activity of 20S proteasome in tumor cells was investigated. The results indicated that NPI-0052 was a more potent inhibitor of 20S proteasome activity than bortezomib in 2 different cell types (Figure 7D), indicating that suppression of NF-κB activation by NPI-0052 is associated with proteasome inhibition.
Discussion
The present study was designed to investigate the effects of NPI-0052 on TNF-induced NF-κB signaling pathway. We found that NPI-0052 potentiated the apoptosis induced by TNF and chemotherapeutic drugs, suppressed TNF-induced tumor cell invasion, and inhibited RANKL-induced osteoclastogenesis. This effect of NPI-0052 correlated with down-regulation of various gene products that mediate cell proliferation, cell survival, invasion, and angiogenesis, all known to be associated with NF-κB. NPI-0052 suppressed not only inducible but also constitutive NF-κB activation. We also found that the NPI-0052 blocked TNF-induced IκBα degradation; suppressed p65 translocation to the nucleus; and inhibited NF-κB–dependent reporter gene expression. We showed that suppression of NF-κB activation by NPI-0052 was due to accumulation of phosphorylated IκBα, ubiquitinated IκBα, and inhibition of proteasome activity.
Our studies are the first to demonstrate that NPI-0052 can potentiate the apoptotic effects of TNF against tumor cells. Because NF-κB activation has been linked with suppression of TNF-induced apoptosis,31,32 it is very likely that these effects of NPI-0052 are mediated through down-regulation of NF-κB. Our studies also show that NPI-0052 can enhance the apoptotic effects of thalidomide and bortezomib against human myeloid leukemia and multiple myeloma. Multiple myeloma cells that are resistant to bortezomib have been shown to be sensitive to NPI-0052,6 indicating the 2 inhibitors may have different mechanisms of action. Although NPI-0052, bortezomib, and thalidomide all suppressed NF-κB activation, that alone is not sufficient to account for the potentiation or chemoresistance.
We also found that NPI-0052 can suppress TNF-induced tumor cell invasion. MMP-9 that is regulated by NF-κB, has been linked with invasion.33 Thus, it is very likely that suppression of NF-κB by NPI-0052 leads to suppression of invasion. For the first time, we also show that NPI-0052 can suppress RANKL-induced osteoclastogenesis. RANKL mediates osteoclastogenesis in part through activation of NF-κB.34 Thus, it is very likely that the suppression of NF-κB by NPI-0052 is what leads to suppression of osteoclastogenesis.
NF-κB is known to regulate the expression of survivin, IAP1/2, Bcl-2, Bcl-xL, cFLIP, and TRAF1, and their overexpression in numerous tumors has been linked to tumor survival, chemoresistance, and radioresistance. We showed that NPI-0052 down-regulated most of these gene products, which may account for the potentiation of the cytotoxic effects of TNF, thalidomide, and bortezomib by NPI-0052.
The genes that are involved in the proliferation and metastasis of cancer have been shown to be regulated by NF-κB.10 We also found for the first time that NPI-0052 can suppress TNF-induced NF-κB-regulated gene products. We showed in this report that NPI-0052 inhibits the expression of cyclin D1, COX-2, and c-myc, all regulated by NF-κB. Our results also showed that the expression of MMP-9, ICAM-1, and VEGF, which are also regulated by NF-κB, are down-regulated by NPI-0052. Similarly, our findings that NPI-0052 suppresses NF-κB-regulated VEGF expression are in agreement with a previous report that NPI-0052 suppressed VEGF-triggered multiple myeloma cell migration.6 MMP-9 plays a pivotal role in tumor invasion and angiogenesis by mediating the degradation of the extracellular matrix, and inhibition of MMP-9 activity has been shown to suppress lung metastasis.35 In agreement with these observations, we found that TNF-induced invasion is inhibited by NPI-0052.
We found that NPI-0052 inhibited not only inducible NF-κB activation but also constitutively activated NF-κB in multiple myeloma cells. Constitutively active NF-κB activation has been found to be critical for the survival and proliferation of various tumor cell types10,36 ; however, the mechanism of constitutive NF-κB activation is not fully understood. Some of the potential mechanisms are overexpression of IκBα without inhibition of NF-κB activity, mutations in the iκbα gene, enhanced IκBα degradation, and constitutive expression of TNF and IL-1.10,36
We found that NPI-0052 blocked TNF-induced NF-κB activation without directly interfering with the DNA binding of NF-κB (data not shown) and did not suppress the TNF-induced IKK activation and Akt activation (data not shown). We found that this inhibition was mediated through the accumulation of phospho-IκBα and ubiquitinated-IκBα rather than its degradation by the proteasome. These results agree with those of previous studies that suggest that NPI-0052 increases the phospho-IκBα induced by TNF.7 We also found that NPI-0052 suppressed chymotrypsin-like proteasome activity, thereby indicating that suppression of NF-κB activation is associated with inhibition of proteasome activity.
We also found that NPI-0052 was the most potent NF-κB inhibitor among various proteasome inhibitors. A comparison of NPI-0052 and bortezomib has been reported previously6,7 and like our study it was found that the IC50 value of NPI-0052 for inhibition of NF-κB activity was lower than that of bortezomib. Whether these differences will translate to greater clinical efficacy of NPI-0052 remains to be seen.
Overall, this study showed for the first time that the antiproliferative, proapoptotic, antiosteoclastogenic, and anti-invasive effects of NPI-0052 might be mediated through the suppression of NF-κB and NF-κB–regulated gene products. Together, these findings provide a novel opportunity to exploit NPI-0052 proteasome inhibitor, not only in the treatment of cancer through the modulation of NF-κB activation pathway. Thus, use of NPI-0052 might provide a novel approach to the treatment of myeloid leukemia and potentially other types of cancers resistant to chemotherapy and radiotherapy.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Clayton Foundation for Research (B.B.A.), a PO1 grant (CA91844) from the National Institutes of Health (NIH) on lung chemoprevention (B.B.A.), and NIH core grant 5P30 CA016672-32 for flow cytometric analysis.
We would like to thank Walter Pagel for a careful review of the manuscript. B.B.A. is the Ransom Horne Jr Professor of Cancer Research.
National Institutes of Health
Authorship
Contribution: K.S.A., G.S., T.H.C., and S.T.C.N. conducted all of the experiments; M.M.C., M.A.P., and A.Y. analyzed the data; and B.B.A. supervised and wrote the paper.
Conflict-of-interest disclosure: M.A.P. is employed by Nereus Pharmaceuticals, which produces NPI-0052. All other authors declare no competing financial interests.
Correspondence: Bharat B. Aggarwal, The University of Texas M. D. Anderson Cancer Center, 1515 Holcombe Boulevard, Box 143, Houston, TX 77030; e-mail:aggarwal@mdanderson.org.

![Figure 1. NPI-0052 enhances apoptosis induced by TNF and chemotherapeutic drugs. (A) KBM-5 cells (10 000 cells/0.1 mL) were incubated at 37°C with 1 nM TNF, 10 μg/mL thalidomide, or 20 nM bortezomib in the presence and absence of 50 nM NPI-0052 as indicated for 18 hour, and the viable cells were assayed using the MTT reagent. The results are expressed as mean cytotoxicity (± standard deviation [SD]) from triplicate cultures. (B) Cells (106/mL) were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF, 10 μg/mL thalidomide, or 20 nM bortezomib for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are representative of 3 independent experiments showing similar results. (C) U266 (106/mL) or DU145 (105/mL) cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 10 μg/mL thalidomide or 20 nM bortezomib for 16 hours. Cell death was determined by the calcein-AM based live/dead assay as described in “Live and dead assay.” Data are representative of 3 independent experiments showing similar results. Images were acquired as described in “Live and dead assay.” (D) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 18 hours. Afterward, they were incubated with anti-annexin V antibody conjugated with FITC plus PI and analyzed with a flow cytometer for apoptotic effects. (E) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for 18 hours. Cells were fixed, stained with TUNEL assay reagent, and then analyzed with flow cytometer. (F) Cells were pretreated with 50 nM NPI-0052 for 4 hours and then incubated with 1 nM TNF for the indicated times. Whole-cell extracts were prepared and subjected to Western blot analysis using anti-PARP antibody.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/110/7/10.1182_blood-2007-04-084996/2/m_zh80200708210001.jpeg?Expires=1769160282&Signature=ypfS~RT-eCP2PYJDixMYkzMqYM6QkfqB-B2E4iUvo7LK47E8tJTuy1MGTTvT~6sJe~JF196HDJLWe0tItK9r8h4Gfq6NnLr3w~h5mesXWIgXGPUgedW3BhdYYZJGTYYG~QT0qdEx0BdhGAR~y2IDhd8CST794r7yUUK-6FRJFiU29eTqZMUsL9RUp16xvMZBNMvLmhS0lGJFlIN4tjuN5lGLR9nEpz23dXXO8kwXhUzC9vW5BCC5nx6~uco4w1UWj4kNweqsToTNwnr6xJRmoPySpsKz7d-hMT31hMkRkUePynVc2QhqifdRyYlrLHYoMvpPrs9IQCZ9nS08N0vFAA__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal