E-proteins are widely expressed basic helix-loop-helix (HLH) transcription factors that regulate differentiation in many cell lineages, including lymphoid, muscle, and neuronal cells. E-protein function is controlled by HLH inhibitors such as Id and SCL/TAL1 proteins, which recently have been suggested to play a role in hematopoietic stem cell (HSC) differentiation. However, the precise stages when these proteins are expressed and their specific functions are not entirely clear. Using a knock-in mouse model where the sequence for the enhanced green fluorescent protein (GFP) was inserted downstream of the Id1 promoter, we were able to track Id1 expression on an individual cell basis and detected Id1 expression in long-term repopulating HSCs (LT-HSCs). Functional assays showed that the Id1/GFP+Lin−Sca1+c-kitHi population was highly enriched for LT-HSCs. Consistent with this expression pattern, Id1 deficiency led to a 2-fold reduction in the number of LT-HSCs defined as Lin−Sca1+c-kitHiCD48−CD150+. Primary bone marrow transplantation studies revealed that Id1 is dispensable for short-term engraftment. In contrast, both Id1−/− whole bone marrow and Lin−Sca1+c-kitHiThy1.1Lo-enriched HSCs, but not Id3−/− marrow, displayed impaired engraftment relative to wild-type controls in secondary transplantation assays. These findings suggest a unique role for Id1 in LT-HSC maintenance and hematopoietic development.
Introduction
Current models of hematopoiesis often list short-term (ST) hematopoietic stem cells (HSCs) as the immediate progeny of long-term (LT) HSCs.1,–3 Both progenitors can repopulate all blood cell lineages; however, ST-HSCs are limited in their renewal capacity,4,5 while LT-HSCs can be serially transplanted and self-renew beyond the life of the organism.6,–8 Technologies developed in the past 2 decades have enabled the identification and separation of LT- and ST-HSCs from other bone marrow populations and from each other. Validated with functional assays of in vivo repopulating potential, detailed phenotypes of several hematopoietic progenitor populations have been established
Studies in mice have shown that both LT- and ST-HSCs lack an array of lineage markers (Lin−) while simultaneously expressing stem-cell antigen-1 (Ly6A/E; Sca1) and high levels of the receptor tyrosine kinase, c-kit (CD117).9,10 This phenotype has been abbreviated Lin−Sca1+c-kitHi, or LSK. It constitutes only about 0.5% of whole bone marrow (WBM), but includes a heterogeneous population of HSCs and oligopotent progenitors. In mice expressing the Thy1.1 allele of CD90, HSCs are confined to the LSK subset expressing low levels of this marker (LSK-Thy1.1Lo).11 This represents 0.15% of WBM, and 1 in 22 LSK-Thy1.1Lo cells transplanted into radiation-conditioned hosts can concurrently repopulate B-cell, T-cell, and myeloid lineages.12 However, ST multilineage repopulating cells within the LSK-Thy1.1Lo compartment outnumber LT repopulating cells by 10 to 1, illustrating the paucity of LT-HSCs. Surface markers such as CD27, CD48, CD150, and Flt3/Flk2 (CD135) have been used in independent studies to further define and enrich LT-HSCs, as will be discussed in our results.
The ability to identify distinct yet primitive hematopoietic progenitors has made it possible to question the molecular mechanisms governing differentiation between these compartments. The factors governing LT- to ST-HSC differentiation and how these differ from stimuli directing LT-HSC renewal are of special interest due to their potential therapeutic promises. However, characterizing molecular events in HSCs is not trivial due to the scarcity of these bone marrow progenitors and the labor-intensive protocols required to isolate them at high purities. Nevertheless, studies using genetic approaches are beginning to implicate several gene families as influential regulators of these processes.13,,,,–18 Among these are the cell-cycle regulators p21cip1/waf1 and p16INK4a, which have been shown to influence LT-HSC maintenance by governing depletion of this compartment.19,–21
Helix-loop-helix (HLH) transcriptional regulators have also been shown to play a role in HSC maintenance.22,–24 These proteins can function as either transcriptional activators or repressors. E-proteins, encoded by E2A, HEB, and E2–2 genes, are a family of activators that may influence HSC maintenance by influencing transcription of p21cip1/waf1 and p16INK4a genes.25,,–28 E-proteins are almost ubiquitously present; hence, their transcriptional activity is controlled by expression of one or more inhibitory proteins. One prominent class of E-protein inhibitors is the Id family, which includes Id1 through Id4. Id proteins have HLH domains similar to E-proteins, which allows avid binding to E-proteins to form inactive heterodimers.29,30 Through this mechanism, Id proteins regulate a wide range of differentiation programs. For example, ectopic expression of Id1 from lymphoid-specific promoters abolishes B- or T-cell development,31,32 and overexpression in bone marrow progenitors affects other hematopoietic lineages23
Several studies have suggested that mRNA for Id1 and Id3 are present in HSCs from mice and humans.33,34 This prompted us to evaluate the physiologic effect of these important regulators on HSC maintenance and differentiation. We used a knock-in mouse model to evaluate Id1 expression on an individual-cell basis in rare bone marrow subsets including LT- and ST-HSCs. This was done by inserting the green fluorescent protein (GFP) sequence into the Id1 locus such that its expression was driven by the Id1 promoter. Since mice homozygous for this insertion did not produce Id1 protein, it was also possible to evaluate how Id1 ablation affected the frequency of phenotypic HSCs in bone marrow, as well as the function of Id1−/− marrow in transplant assays. These data indicate a specific role for Id1 in modulating LT-HSC but not ST-HSC renewal and differentiation. In contrast, similar experiments with Id3−/− marrow indicate that Id3 does not contribute to LT-HSC activity.
Materials and methods
All animal protocols were first approved by the Internal Animal Care and Use Committee of the Oklahoma Medical Research Foundation.
Mice
C57BL/6J (B6), B6.PL-Thy1a/CyJ (B6-CD90.1), and B6.SJL-PtprcaPepcb/BoyJ (B6-CD45.1) mice were originally obtained from Jackson Laboratories (Bar Harbor, ME). B6-Id3−/− mice were kind gifts from Dr Yuan Zhuang (Duke University School of Medicine, Durham, NC).35 Id1/GFP knock-in mice were generated and backcrossed as described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article) and used at 4 to 8 weeks of age. B6-CD45.1 transplant hosts were 6 to 18 weeks old.
Bone marrow analyses and cell sorting
For all experiments, cell collection and staining were conducted in Hanks balanced salt solution supplemented with 5% fetal calf serum (H5F). Analyses of WBM composition were conducted with phycoerythrin (PE)– or allophycocyanin (APC)–conjugated antibodies against CD2 (clone RM2–5), CD3 (145–2C11), CD5 (53–7.3), CD11b (Mac-1; M1/70), CD19 (1D3), B220 (RA3–6B2), IgM (R6–60.2), Ly6G (1A8), NK1.1 (PK136), and the TER-119 antigen. Unless noted otherwise, antibodies were purchased from BD Pharmingen (San Diego, CA). After antibody staining, dead cells were excluded by propidium iodide incorporation (Molecular Probes, Eugene, OR). Relative fluorescence parameters were collected with a FACSCalibur flow cytometer and analyzed via CellQuest software (BD Immunocytometry, San Jose, CA).
Lin− marrow was prepared as previously described through immunomagnetic depletion.36 Briefly, WBM was first labeled with rat Ig against CD2, CD3 (17A2), CD5, CD8 (53–6.7), CD11b, CD19, B220, Ly6G and Ly6C (RB6–8C5), and the TER-119 antigen. Labeled cells were then incubated with BioMag anti-Rat Ig conjugated magnetic particles (Qiagen, Valencia, CA). Subsequent staining with PE anti–rat Ig and APC anti–c-kit (CD117; clone 2B8) and flow cytometric analysis showed that more than 99% of c-kit+ cells were Lin−/Lo.
Initial fractionation of Lin− marrow was done with APC anti-kit and PE-Cy5 anti-Sca1 (Ly6A/E; clone D7; eBiosciences, San Diego, CA). Lin−Sca1+ cells were analyzed with APC anti–c-kit and PE-conjugated antibodies against CD27 (LG.7F9; eBiosciences), Flt3/Flk2 (A2F10.1), or CD150 (SLAM; 9D1; eBiosciences). Samples examined for CD150 expression were first stained with biotin-conjugated anti-CD48 (HM48–1; eBiosciences), followed by subsequent incubation with APC-Cy7–conjugated streptavidin. Likewise, biotin-conjugated anti-Thy1.1 (CD90.1; OX-7) was used with APC-Cy7 streptavidin for expression analysis and for sorting Thy1.1LoLSK-enriched HSCs. Lin− marrow analyses and cell sorting were conducted using a FACSAria flow cytometer with FACSDiva software (BD Immunocytometry).
Transplant assays
Transplantation experiments were conducted as previously described.37 Briefly, B6-CD45.1 recipient mice were preconditioned with 6.5 Gy in a Mark I gamma irradiator using a 137Cs source (J. L. Shepard & Associates, Glendale, CA). A sublethal conditioning model was used in these assays to ensure stringent competition from the host marrow with the transplanted cells.36,–38 Cells to be transplanted from CD45.2 donors were suspended at the desired dose in 200 μL H5F and injected intravenously. At the indicated times after transplantation, marrow and thymus samples from matched cohorts of hosts receiving either knock-out or control grafts were evaluated as described for bone marrow analyses. Donor cells were identified by staining with FITC anti-CD45.2 (clone 104).
Results
An Id1/GFP knock-in mouse model
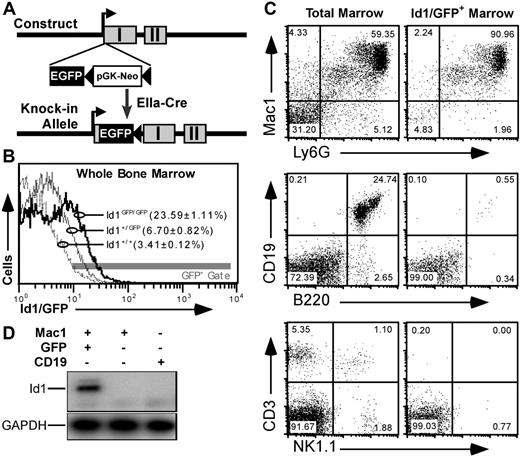
To better understand Id1 expression in primitive bone marrow, we constructed a mouse model where the GFPgene was inserted in the 5′ untranslated sequence of the Id1 gene. This was accomplished through homologous recombination with the targeting vector shown in Figure 1A and described in Document S1. GFP expression from the Id1 locus generated a distinct fluorescence shift in Id1GFP/GFP marrow relative to wild-type control (Figure 1B). Further analysis showed that GFP expression was specific to bone marrow populations expected to express Id1 based on data from reverse transcription–polymerase chain reaction (RT-PCR) assays.23 For example, most (> 90%) GFP+ bone marrow cells were of myeloid lineages expressing both Mac1 and Ly6G markers (Figure 1C). Furthermore, GFP fluorescence was rarely observed in bone marrow cells expressing B220 and/or CD19, as has been reported for Id1 expression.34 Likewise, neither CD3+ nor NK1.1+ cells expressed GFP.
GFP knock-in as a model for tracking Id1 expression. (A) Schematic diagrams showing the knock-in construct and knock-in allele. EGFP and pGK-Neo were inserted in the 5′ untranslated region in exon I (▩) of the Id1 gene. (B) Relative GFP fluorescence in WBM from Id1+/+ (- - -), Id1+/GFP (–), and Id1GFP/GFP (–) mice. Parentheses indicate the average fraction of marrow ± SEM from 9 samples from each genotype falling within the indicated fluorescence gate (▩). (C) Immunofluorescence showing Id1/GFP expression in specific bone marrow lineages. Left panels give total marrow profiles for the indicated markers. Right panels show those cells falling within the GFP+ gate indicated in panel B. Numbers are percentages of events in each quadrant. (D) Correlation between Id1 and GFP expression. Id1+/GFP marrow was labeled with antibodies against Mac1 (PE) and CD19 (APC), and sorted for the indicated subsets. RNA was extracted from the sorted samples and probed by RT-PCR for Id1 and GAPDH transcripts.
GFP knock-in as a model for tracking Id1 expression. (A) Schematic diagrams showing the knock-in construct and knock-in allele. EGFP and pGK-Neo were inserted in the 5′ untranslated region in exon I (▩) of the Id1 gene. (B) Relative GFP fluorescence in WBM from Id1+/+ (- - -), Id1+/GFP (–), and Id1GFP/GFP (–) mice. Parentheses indicate the average fraction of marrow ± SEM from 9 samples from each genotype falling within the indicated fluorescence gate (▩). (C) Immunofluorescence showing Id1/GFP expression in specific bone marrow lineages. Left panels give total marrow profiles for the indicated markers. Right panels show those cells falling within the GFP+ gate indicated in panel B. Numbers are percentages of events in each quadrant. (D) Correlation between Id1 and GFP expression. Id1+/GFP marrow was labeled with antibodies against Mac1 (PE) and CD19 (APC), and sorted for the indicated subsets. RNA was extracted from the sorted samples and probed by RT-PCR for Id1 and GAPDH transcripts.
To verify that our gating method accurately selected Id1-expressing cells, we sorted GFP+Mac1+ or GFP−Mac1+ bone marrow cells from Id1+/GFP mice. GFP−Mac1−CD19+ B cells were collected as a control. Id1 expression in these sorted populations was determined by RT-PCR using primers selectively amplifying Id1 transcripts derived from the wild-type allele. As shown in Figure 1D, Id1 expression was only detectable in the GFP+ population, indicating that GFP in this mouse model faithfully marks cells that are actively transcribing the Id1 gene. Further analysis with cultured WBM showed that Id1/GFP expression was induced by myeloid cytokines as has been noted previously23 (Figure S1). Immunoblotting showed that cytokine-stimulated bone marrow cells from wild-type but not Id1GFP/GFP mice expressed Id1 protein (Figure S2). Id1-deficient mice have been previously shown to have normal gross hematopoietic profiles.39 Consistent with these findings, detailed comparisons between Id1GFP/GFP, Id1+/GFP, and Id1+/+ littermates showed no significant variation in different hematopoietic lineages (data not shown).
Id1 expression is enriched in phenotypic LT-HSCs
Figure 1C suggested that approximately 5% of total Id1/GFP+ bone marrow cells did not express any of the lineage markers tested. Accordingly, we next analyzed Id1/GFP expression in bone marrow immunomagnetically depleted of cells expressing a panel of lineage markers (Lin−). We found that this primitive population had clearly more GFP fluorescence relative to wild-type samples (Figure 2A). Both LT- and ST-HSCs are contained within the LSK population.12,40,41 Interestingly, we found that a distinct subset of LSK expressed Id1/GFP (Figure 2B).
Id1/GFP expression in primitive bone marrow. (A) Id1/GFP expression in Lin− marrow. WBM of the indicated genotypes was immunomagnetically depleted of cells expressing an array of lineage markers (“Bone marrow analyses and cell sorting”), then evaluated by flow cytometry for relative GFP expression. Parentheses show average and SEM of Lin− cell fractions included in the indicated gate (n ≥ 12). (B) Id1/GFP expression in LSK marrow from wild-type (top) and Id1GFP/GFP mice (bottom). Lin− marrow was stained for Sca1 and c-kit expression. Left plots show staining of total Lin− marrow for these markers. Right panels show cells included in the GFP+ gate given in panel A. Boxes outline the LSK fraction. (C) Id1/GFP expression in HSC-enriched fractions of Id1GFP/GFP marrow. Lin−Sca1+ marrow was subdivided according to immunofluorescent intensity from additional HSC-related markers as indicated. Left plots show total Lin−Sca1+ marrow, center plots show Lin−Sca1+Id1/GFP− cells, and right plots show the GFP+ subset. Letters above gates correlate with populations listed in panel E. (D) Id1/GFP expression relative to CD150 and CD48 in WBM (top) and LSK marrow (bottom) as labeled. Box outlines the CD150+CD48− HSC-inclusive gate. (E) Relative Id1/GFP expression among several LSK marrow subsets. Each bar represents the average percentage of Id1/GFP+ cells of the named population. Phenotype definitions correlate to gated regions with the indicated letters in panels C and D. Averages were calculated from at least 3 independent samples. Error bar represents SEM. Populations are arranged in an ascending order for LT-HSC enrichment.
Id1/GFP expression in primitive bone marrow. (A) Id1/GFP expression in Lin− marrow. WBM of the indicated genotypes was immunomagnetically depleted of cells expressing an array of lineage markers (“Bone marrow analyses and cell sorting”), then evaluated by flow cytometry for relative GFP expression. Parentheses show average and SEM of Lin− cell fractions included in the indicated gate (n ≥ 12). (B) Id1/GFP expression in LSK marrow from wild-type (top) and Id1GFP/GFP mice (bottom). Lin− marrow was stained for Sca1 and c-kit expression. Left plots show staining of total Lin− marrow for these markers. Right panels show cells included in the GFP+ gate given in panel A. Boxes outline the LSK fraction. (C) Id1/GFP expression in HSC-enriched fractions of Id1GFP/GFP marrow. Lin−Sca1+ marrow was subdivided according to immunofluorescent intensity from additional HSC-related markers as indicated. Left plots show total Lin−Sca1+ marrow, center plots show Lin−Sca1+Id1/GFP− cells, and right plots show the GFP+ subset. Letters above gates correlate with populations listed in panel E. (D) Id1/GFP expression relative to CD150 and CD48 in WBM (top) and LSK marrow (bottom) as labeled. Box outlines the CD150+CD48− HSC-inclusive gate. (E) Relative Id1/GFP expression among several LSK marrow subsets. Each bar represents the average percentage of Id1/GFP+ cells of the named population. Phenotype definitions correlate to gated regions with the indicated letters in panels C and D. Averages were calculated from at least 3 independent samples. Error bar represents SEM. Populations are arranged in an ascending order for LT-HSC enrichment.
Since LSK is a heterogeneous population, of which LT-HSCs constitute only 1%,12 we next resolved GFP expression within the LSK compartment for other markers characteristic of HSCs. Flt3/Flk2 is not expressed by LT-HSCs, but is expressed by lymphoid-primed multipotent progenitors (MPPs).2,40,41 Flt3 expression evenly split the LSK compartment (Figure 2C top left panel); however, most Id1/GFP+LSK cells did not express Flt3. Specifically, Id1/GFP was expressed by 10.1% of Flt3−LSK cells and 3.1% of Flt3+LSK cells. Most Flt3+LSK cells that did express Id1/GFP had intermediate levels of Flt3, and comprised a cluster spanning the positive and negative populations.
CD27 is more widely expressed in LSK marrow (Figure 2C middle left panel), and the rare CD27−LSK fraction has been shown to be enriched for LT repopulating cells.34,42 Id1/GFP+ cells were also enriched in this population, representing 18.2% (± 2.4%) of total CD27−LSK cells. In contrast, the much larger CD27+LSK population contains fewer LT repopulating progenitors, and also fewer Id1/GFP+ cells (5.3% ± 0.9% of total CD27+LSK cells; Figure 2C middle right panel). Thy1.1 also marks a large subset of LSK marrow, and is expressed at low levels by both LT- and ST-HSCs.11,12,43 Id1/GFP+ cells represent 5.2% (± 1.1%) of Thy1.1LoLSK cells (Figure 2C bottom panels).
Recently, Kiel and colleagues have shown that the SLAM-family members CD48 and CD150 are relatively specific identifiers of HSCs.44 Approximately 1 in 5 CD48−CD150+ WBM cells, and 1 in 2 CD48−CD150+LSK cells had a LT repopulating potential characteristic of LT-HSCs. Interestingly, almost 1 in 6 CD48−CD150+ WBM cells express Id1/GFP (Figure 2D top panels). This compares to about 1 Id1/GFP+ per 2 CD48−CD150+LSK cells (Figure 2D bottom panels). Figure 2E shows the mean Id1/GFP+ cell frequency in each of the bone marrow compartments described. These data suggest that Id1 expression in bone marrow fractions increases in direct correlation to the enrichment of LT-HSCs in each fraction. A similar pattern of Id1/GFP expression was observed in the marrow of Id1+/GFP mice; however, the fluorescence levels were typically 2-fold lower, and the resulting overlap with wild-type autofluorescence prevented a reliable estimate of GFP+ cells (data not shown).
Id1/GFP+LSK cells are enriched for LT multilineage-repopulating progenitors
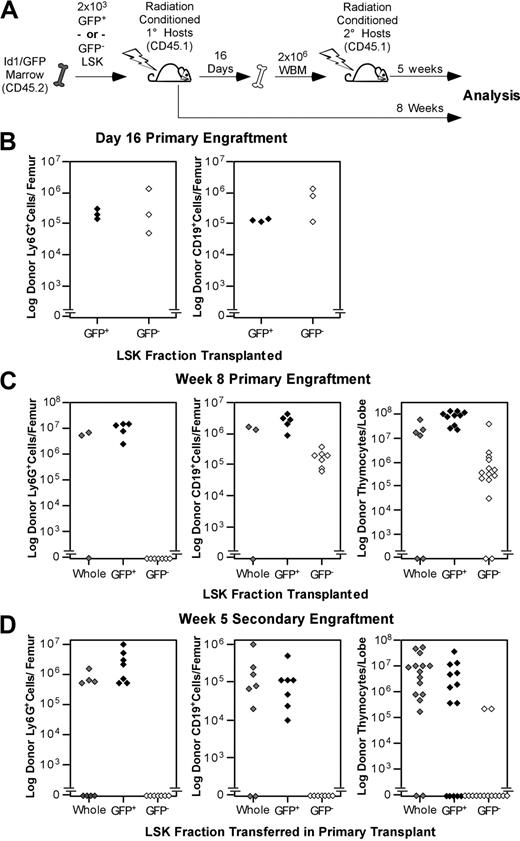
The abundance of Id1/GFP+ cells in phenotypic LT-HSC populations implied that Id1/GFP might be specifically expressed by LT-HSCs. To test this hypothesis, we sorted LSK from Id1GFP/GFP bone marrow into GFP+ and GFP− fractions and assayed these subsets for LT multilineage repopulating abilities. LSK subsets or total LSK were collected from CD45.2 donors and transferred into parallel cohorts of CD45.1 hosts as depicted in Figure 3A. Immunostaining of bone marrow from 3 primary hosts 16 days after transplantation showed that both Id1/GFP+ and Id1/GFP−LSK subsets produced comparable numbers of myeloid and B-cell progeny (Figure 3B). Thymic engraftment was not assessed at day 16 because thymic repopulation by stem cells is typically sparse at this time.36 Marrow was pooled from these primary hosts and transferred into fresh cohorts of hosts at 2 × 106 WBM cells per mouse.
Functional evaluation of Id1/GFP+LSK. (A) Schematic representation of the transplantation protocol used to determine the presence of LT-HSCs in Id1/GFP-expressing LSK cells. Subsets were sorted from CD45.2 Id1GFP/GFP donors. GFP+ preparations transplanted were at least 10-fold enriched for Id1/GFP+ cells compared with unseparated LSK cells. GFP− preparations included no more than 0.8% GFP+ cells. Primary and secondary hosts were treated prior to transplantation with sublethal doses of radiation. Separate cohorts of CD45.1 host mice each received 2 × 103 sorted Id1/GFP+, Id1/GFP−, or whole LSK cells. After 16 days, WBM was collected from a subset of primary hosts. Marrow was pooled from hosts receiving the same LSK subsets. A total of 2 × 106 cells were then transferred to each mouse in fresh cohorts of CD45.1 hosts. The remaining primary hosts were evaluated for marrow and thymic engraftment 8 weeks following transplantation. Secondary hosts were assayed for engraftment 5 weeks after secondary transplantatoin. (B) Day-16 analyses of primary engraftment. One femur was saved from each mouse for analysis, while marrow from the remaining femur and both tibia was used for secondary transplantations. For analysis, marrow was immunofluorescently labeled with antibodies against the donor CD45 isoform, Ly6G, and CD19. Donor fractions observed from flow cytometry were multiplied by cell counts to determine total donor-derived cells per femur in either the B-cell or myeloid lineages. (C) Long-term primary engraftment from Id1/GFP LSK subsets. Primary hosts were evaluated for engraftment 8 weeks after transplantation. Bone marrow was analyzed as described in panel B. In addition, thymic lobes from each host were evaluated separately for donor progeny. (D) Secondary engraftment analyses. Marrow and thymus from secondary hosts were analyzed as in panels B and C 5 weeks following secondary marrow transfer. For all panels, progeny from unseparated (whole) LSK, Id1/GFP+LSK, and Id1/GFP−LSK cells are represented by gray, black, and white symbols, respectively. Each symbol represents the total donor-derived progeny of the indicated lineage for 1 femur or thymic lobe, shown in log scale.
Functional evaluation of Id1/GFP+LSK. (A) Schematic representation of the transplantation protocol used to determine the presence of LT-HSCs in Id1/GFP-expressing LSK cells. Subsets were sorted from CD45.2 Id1GFP/GFP donors. GFP+ preparations transplanted were at least 10-fold enriched for Id1/GFP+ cells compared with unseparated LSK cells. GFP− preparations included no more than 0.8% GFP+ cells. Primary and secondary hosts were treated prior to transplantation with sublethal doses of radiation. Separate cohorts of CD45.1 host mice each received 2 × 103 sorted Id1/GFP+, Id1/GFP−, or whole LSK cells. After 16 days, WBM was collected from a subset of primary hosts. Marrow was pooled from hosts receiving the same LSK subsets. A total of 2 × 106 cells were then transferred to each mouse in fresh cohorts of CD45.1 hosts. The remaining primary hosts were evaluated for marrow and thymic engraftment 8 weeks following transplantation. Secondary hosts were assayed for engraftment 5 weeks after secondary transplantatoin. (B) Day-16 analyses of primary engraftment. One femur was saved from each mouse for analysis, while marrow from the remaining femur and both tibia was used for secondary transplantations. For analysis, marrow was immunofluorescently labeled with antibodies against the donor CD45 isoform, Ly6G, and CD19. Donor fractions observed from flow cytometry were multiplied by cell counts to determine total donor-derived cells per femur in either the B-cell or myeloid lineages. (C) Long-term primary engraftment from Id1/GFP LSK subsets. Primary hosts were evaluated for engraftment 8 weeks after transplantation. Bone marrow was analyzed as described in panel B. In addition, thymic lobes from each host were evaluated separately for donor progeny. (D) Secondary engraftment analyses. Marrow and thymus from secondary hosts were analyzed as in panels B and C 5 weeks following secondary marrow transfer. For all panels, progeny from unseparated (whole) LSK, Id1/GFP+LSK, and Id1/GFP−LSK cells are represented by gray, black, and white symbols, respectively. Each symbol represents the total donor-derived progeny of the indicated lineage for 1 femur or thymic lobe, shown in log scale.
The remaining primary hosts were evaluated for donor-derived B-cell, myeloid, and thymocyte development at 8 weeks after transplantation. Remarkably, donor-derived Ly6G+ cells were not detectable in the marrow of hosts receiving Id1/GFP−LSK (Figure 3C). In contrast, Id1/GFP+LSK uniformly produced between 106 and 108 Ly6G+ cells per femur at this time. As Ly6G is expressed by short-lived myeloid cell types, donor-derived cells with this phenotype are a sensitive indicator of persisting stem cell engraftment.45,,–48 Id1/GFP+LSK also generated robust B-cell and thymocyte engraftment in LT primary hosts. Conversely, the number of B cells produced by GFP−LSK did not increase between day 16 and week 8, and thymocyte production typically lagged 2 logs behind that of GFP+LSK.
Hosts receiving secondary grafts showed a similar pattern of engraftment (Figure 3D). At 5 weeks after secondary transplantation, GFP−LSK consistently failed to produce detectable myeloid progeny. Notably, CD19+ progeny were also absent in secondary grafts from Id1/GFP−LSK, and thymocyte engraftment was very rare (2 of 14 lobes examined). On the other hand, secondary GFP+LSK grafts produced myeloid and B cells in all hosts examined, as well as thymocyte engraftment in 9 of 14 thymic lobes. Together, these observations suggest that GFP+LSK cells are specifically enriched for serially transplantable multilineage progenitors reminiscent of LT-HSCs, which is consistent with the phenotypic data presented in Figure 2. Since Id1-deficient mice exhibit normal steady-state hematopoiesis,39 it is not surprising that LT-HSCs from Id1GFP/GFP mice showed secondary transplantation potential.
Id1−/− marrow contains numerically fewer cells with LT-HSC phenotypes
The correlation between Id1/GFP expression and LT-HSC phenotypes suggested that Id1 may play a specific role in LT-HSC function. Accordingly, we next compared the frequency of phenotypic HSCs in wild-type and Id1−/− marrow. A principal characteristic of LT-HSCs is their ability to generate multilineage engraftment after serial transplantation in irradiated hosts. Typically, HSCs with this capacity constitute approximately 1 to 3 cells per 105 WBM.4 This statistic correlated well with the frequency we observed for CD48−CD150+LSK in wild-type marrow (1.73 ± 0.2/105 WBM). In contrast, parallel samples of Id1−/− marrow harbored 0.95 (± 0.1) cells with this phenotype (45% reduction; Figure 4).
Relative frequencies of phenotypic LT-HSCs between wild-type and Id1−/− marrow. Numbers of cells meeting the given LT-HSC phenotypes per 100 000 WBM cells were calculated. Bars show the average of at least 3 samples per phenotype. Error bars indicate SEM. Statistically significant differences between wild-type and Id1−/− samples were determined by means of the Student t test (P values).
Relative frequencies of phenotypic LT-HSCs between wild-type and Id1−/− marrow. Numbers of cells meeting the given LT-HSC phenotypes per 100 000 WBM cells were calculated. Bars show the average of at least 3 samples per phenotype. Error bars indicate SEM. Statistically significant differences between wild-type and Id1−/− samples were determined by means of the Student t test (P values).
The difference in phenotypic LT-HSC frequency in wild-type and Id1−/− marrow was less obvious in fractions not as enriched for Id1-expressing cells. The CD27−LSK population is about 10-fold larger than the CD48−CD150+LSK fraction, and represents 23.2 ± 0.8/105 WBM cells. Further, CD27−LSK cells do not include all LT repopulating cells42 ; therefore, this population is proportionately less enriched for LT-HSCs. Reflecting this difference, the CD27−LSK population is only reduced by 37% in Id1−/− marrow (Figure 4).
LSK marrow fractions with lower frequencies of Id1/GFP+ cells did not differ in absolute numbers between wild-type and Id1−/− marrow. Flt3−LSK cells include all LT-HSCs as well as a substantial fraction of ST-HSCs and MPPs.2,40,41 This population was unchanged between wild-type and Id1−/− mice (Figure 4), as were other populations predominated by ST-HSCs and MPPs, including the Thy1.1Lo, Flt3+, and CD27+ subsets of LSK (data not shown). In contrast with the findings by others,49 we found no difference in the frequency of total LSK.
Loss of Id1 does not affect primary bone marrow engraftment
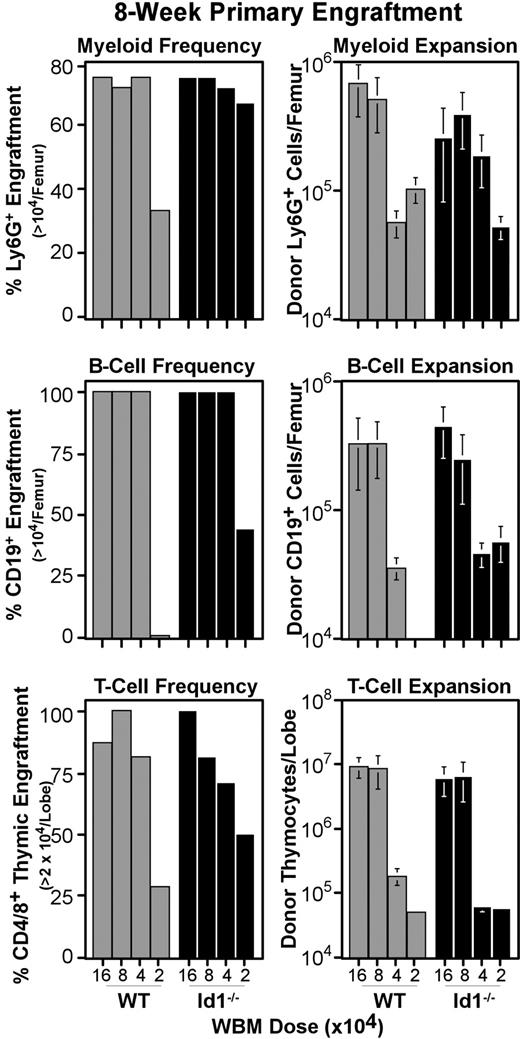
The reduced number of phenotypic LT-HSCs prompted us to test the function of Id1−/− marrow. Transplantation assays of Id1−/− and wild-type WBM were conducted as described for Figure 3, except graft doses ranged in 2-fold dilutions from 160 000 to 20 000 cells per host (Figure 5). At 8 weeks after transplantation, marrow and thymic samples were collected and analyzed by immunofluorescent staining for specific lineage engraftment. Control mice that were irradiated but did not receive transplants were included in each experiment, and their tissues were used to establish the background for detecting donor CD45.2 levels. Accordingly, mice were considered positive for donor myeloid or B cells if more than 104 donor Ly6G+ or CD19+ cells were generated per femur, respectively. Likewise, the thymic engraftment threshold was set at 2 × 104 donor thymocytes per lobe. Figure 5 (left panels) shows the fraction of host mice that met these thresholds for each WBM dose transplanted. Most mice were successfully engrafted at all but the lowest WBM dose tested; however, we did not see consistent differences in absolute hematopoietic output between wild-type and Id1−/− marrow (Figure 5 right panels). This result is consistent with the low frequency of Id1/GFP+ cells with ST-HSC phenotypes (Figure 2), and suggests that Id1 plays a minimal role in differentiating ST-HSCs.
Primary engraftment from Id1−/− versus wild-type marrow. WBM was collected from Id1−/− and wild-type donors, and transferred intravenously to parallel cohorts of radiation-conditioned hosts, as described for primary transplantations in Figure 3. Doses transferred are indicated on the x-axes. Bone marrow and thymic samples were collected 8 weeks after transplantation and evaluated for specific lineage engraftment as described for Figure 3. Left panels give the fraction of hosts engrafted for each lineage by transplantation dose. Right panels give average numbers of cells produced by grafts in each lineage. Error bars indicate SEM. At least 7 hosts pooled from 2 independent transplantation experiments were evaluated per WBM dose for each genotype. Mice were considered engrafted in B-cell or myeloid lineage if their femora harbored at least 104 donor-type cells expressing CD19 or Ly6G, respectively. For the T lineage, the engraftment threshold was set at 2 × 104 donor cells per thymus, as noted on the y-axes. For all 3 lineages, engraftment thresholds were set based on background levels in controls that were irradiated but did not receive transplants included in each cohort of hosts.
Primary engraftment from Id1−/− versus wild-type marrow. WBM was collected from Id1−/− and wild-type donors, and transferred intravenously to parallel cohorts of radiation-conditioned hosts, as described for primary transplantations in Figure 3. Doses transferred are indicated on the x-axes. Bone marrow and thymic samples were collected 8 weeks after transplantation and evaluated for specific lineage engraftment as described for Figure 3. Left panels give the fraction of hosts engrafted for each lineage by transplantation dose. Right panels give average numbers of cells produced by grafts in each lineage. Error bars indicate SEM. At least 7 hosts pooled from 2 independent transplantation experiments were evaluated per WBM dose for each genotype. Mice were considered engrafted in B-cell or myeloid lineage if their femora harbored at least 104 donor-type cells expressing CD19 or Ly6G, respectively. For the T lineage, the engraftment threshold was set at 2 × 104 donor cells per thymus, as noted on the y-axes. For all 3 lineages, engraftment thresholds were set based on background levels in controls that were irradiated but did not receive transplants included in each cohort of hosts.
Id1−/− bone marrow has impaired secondary engraftment potential
The association of Id1 expression with phenotypic LT-HSCs and their reduced frequency in Id1−/− marrow led us to suspect that Id1 may function specifically in these cells. Therefore, we tested the dependence of LT-HSC function on Id1 by comparing the serial transplantation potential of Id1−/− and wild-type bone marrow. For these experiments, primary hosts each received 500 000 WBM cells and were killed 16 days later to provide secondary marrow grafts. Chimerism of secondary marrow was evaluated prior to transplantation, and was found to be uniformly 35% to 50% donor derived. As outlined in Figure 3A, secondary grafts were each 2 × 106 WBM, and engraftment was evaluated after 5 weeks.
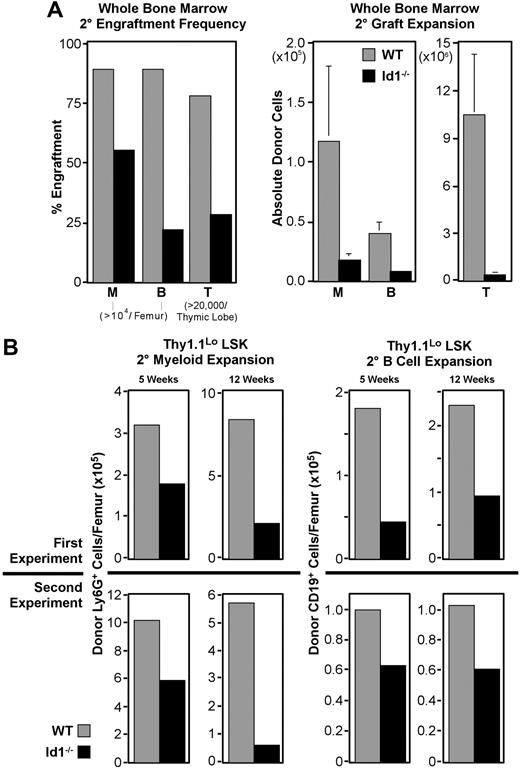
In 5 replicate experiments, 22 of the 25 mice receiving wild-type secondary marrow developed more than 104 Ly6G+ myeloid cells from the primary graft, while only 14 of 25 mice receiving Id1−/− marrow met this threshold (Figure 6A). Engraftment frequencies from Id1−/− secondary marrow in other lineages were also reduced. Absolute numbers of cells produced in these secondary grafts followed a similar pattern, with Id1−/− marrow narrowly exceeding thresholds in most cases. Collectively, these data indicate that LT-HSC function is impaired in the absence of Id1.
Secondary engraftment potential of Id1−/− bone marrow. For all panels, ▩ indicates wild-type and ■ indicates Id1−/− graft contributions. Secondary transplantations were conducted as described for Figure 3, except parallel cohorts of mice received either wild-type or Id1−/− marrow. Cells for each secondary transplantation experiment were pooled from at least 3 primary donors per genotype. Primary and secondary hosts were treated prior to transplantation with 6.5 Gy radiation. Evaluation of bone marrow chimeras 16 days after transplantation by immunofluorescent staining and flow cytometry showed that primary bone marrow was more than 40% donor derived. Marrow was pooled from at least 4 primary hosts before transplanting 2 × 106 cells per each secondary host. Secondary engraftment was evaluated 5 to 12 weeks later, as noted for each dataset. Engraftment analysis was also conducted as described for Figure 3. (A) Secondary engraftment from WBM transplantations. Left panel shows frequencies of host meeting myeloid (“M”), B-cell (“B”), or thymocyte (“T”) engraftment thresholds for each graft genotype 5 weeks after secondary marrow transplantation. Right panels give average donor-derived Ly6G+ (“M”) and CD19+ (“B”) cell numbers per femur, and donor-derived CD4+ and/or CD8+ cells per thymus (“T”). Error bars indicate SEM. Data were pooled from 5 replicate experiments where 25 mice received transplants per secondary marrow genotype. (B) Secondary engraftment from LSK-Thy1.1Lo enriched HSCs. In 2 replicate experiments, 103 LSK-Thy1.1Lo cells from either wild-type or Id1−/− donors were transferred to primary hosts, which served as donors for secondary grafts 16 days later. Top panels show average donor cells per femur observed from experiment 1, while bottom panels give results from experiment 2. Left panels show Ly6G+ myeloid engraftment at both 5 and 12 weeks after transplantation. Right panels show donor CD19+ B cells for the same time points. A minimum of 14 secondary hosts were evaluated at each time point per genotype.
Secondary engraftment potential of Id1−/− bone marrow. For all panels, ▩ indicates wild-type and ■ indicates Id1−/− graft contributions. Secondary transplantations were conducted as described for Figure 3, except parallel cohorts of mice received either wild-type or Id1−/− marrow. Cells for each secondary transplantation experiment were pooled from at least 3 primary donors per genotype. Primary and secondary hosts were treated prior to transplantation with 6.5 Gy radiation. Evaluation of bone marrow chimeras 16 days after transplantation by immunofluorescent staining and flow cytometry showed that primary bone marrow was more than 40% donor derived. Marrow was pooled from at least 4 primary hosts before transplanting 2 × 106 cells per each secondary host. Secondary engraftment was evaluated 5 to 12 weeks later, as noted for each dataset. Engraftment analysis was also conducted as described for Figure 3. (A) Secondary engraftment from WBM transplantations. Left panel shows frequencies of host meeting myeloid (“M”), B-cell (“B”), or thymocyte (“T”) engraftment thresholds for each graft genotype 5 weeks after secondary marrow transplantation. Right panels give average donor-derived Ly6G+ (“M”) and CD19+ (“B”) cell numbers per femur, and donor-derived CD4+ and/or CD8+ cells per thymus (“T”). Error bars indicate SEM. Data were pooled from 5 replicate experiments where 25 mice received transplants per secondary marrow genotype. (B) Secondary engraftment from LSK-Thy1.1Lo enriched HSCs. In 2 replicate experiments, 103 LSK-Thy1.1Lo cells from either wild-type or Id1−/− donors were transferred to primary hosts, which served as donors for secondary grafts 16 days later. Top panels show average donor cells per femur observed from experiment 1, while bottom panels give results from experiment 2. Left panels show Ly6G+ myeloid engraftment at both 5 and 12 weeks after transplantation. Right panels show donor CD19+ B cells for the same time points. A minimum of 14 secondary hosts were evaluated at each time point per genotype.
HSC-enriched Id1−/− marrow has impaired secondary engraftment potential
Thy1.1LoLSK cells are enriched for LT repopulating potential and include all LT-HSCs.12,41,50 A graft of 103 Thy1.1LoLSK cells is more than sufficient to rescue lethally irradiated hosts.37 This population is present in wild-type and Id1−/− marrow at approximately equal frequencies, where 600 000 cell WBM transplants deliver roughly 103 Thy1.1LoLSK cells for both genotypes. Observations summarized in Figure 4 suggested that the secondary engraftment deficiencies of Id1−/− WBM might result from fewer LT-HSCs present in WBM grafts relative to wild-type. If so, this deficit should be reflected in Thy1.1LoLSK transplants as well, given the constant frequency of this population in both genotypes.
We conducted serial transplantation experiments with 103 sorted Thy1.1LoLSK cells as the primary graft using the protocol described in Figure 3A, except cohorts of mice were analyzed at both 5 and 12 weeks after secondary transplantation. As with WBM, Id1−/− Thy1.1LoLSK cells consistently produced fewer secondary progeny, regardless of the lineage, relative to wild-type (Figure 6B). In 2 independent experiments, the difference in secondary myeloid engraftment at week 5 was almost 2-fold between wild-type and Id1−/− grafts, which is consistent with the difference between these genotypes in phenotypic LT-HSC frequency. Also consistent between both experiments was an increase in the difference in myeloid engraftment between genotypes by week 12, suggesting that the competitive ability of Id1−/− LT-HSCs may be decreasing with time.
B cells are longer lived than myeloid-lineage cells, and unlike mature granulocytes, can divide extensively in response to appropriate stimulation. Consequently, they are not as sensitive at indicating graft persistence as are myeloid-lineage cells. Nonetheless, we monitored B-cell engraftment as an indicator of multilineage stem-cell activity. This showed that absolute numbers of CD19+ B cells produced by secondary Id1−/− Thy1.1LoLSK grafts were also depressed relative to wild-type at both time points for both experiments (Figure 6B), ranging in difference from 1.5- to 2.5-fold.
The high dose of Thy1.1Lo LSK used for primary grafts in these experiments was designed to ensure a clear readout. We estimate that 103 Thy1.1Lo LSK cells include about 20% more HSCs than the 5 × 105 cell doses used in the WBM transplants described here. One result of this increased progenitor dose was a correlative increase in engraftment frequencies. At week 5, all hosts receiving wild-type grafts had at least 104 donor Ly6G+ myeloid cells per femur; however, only 10 of 14 Id1−/− graft recipients met this threshold. This trend was inverted for B-cell engraftment, with secondary grafts generating 104 CD19+ B cells in 9 of 15 mice receiving wild-type cells, compared with 12 of 14 mice receiving Id1−/− cells. However, by week 12, both myeloid and B-cell lineage engraftment frequencies were depressed in Id1−/− secondary graft recipients relative to wild-type (Table 1).
LSK-Thy1.1Lo secondary engraftment frequency
| Time after transplantation, wk . | Primary-graft genotype . | Hosts analyzed, no. . | Myeloid-engrafted hosts, no. (%) . | B-cell–engrafted hosts, no. (%) . |
|---|---|---|---|---|
| 5 | WT | 15 | 15 (100) | 9 (60.0) |
| 5 | Id1−/− | 14 | 10 (71.4) | 12 (85.7) |
| 12 | WT | 16 | 13 (81.3) | 13 (81.3) |
| 12 | Id1−/− | 14 | 10 (71.4) | 11 (78.6) |
| Time after transplantation, wk . | Primary-graft genotype . | Hosts analyzed, no. . | Myeloid-engrafted hosts, no. (%) . | B-cell–engrafted hosts, no. (%) . |
|---|---|---|---|---|
| 5 | WT | 15 | 15 (100) | 9 (60.0) |
| 5 | Id1−/− | 14 | 10 (71.4) | 12 (85.7) |
| 12 | WT | 16 | 13 (81.3) | 13 (81.3) |
| 12 | Id1−/− | 14 | 10 (71.4) | 11 (78.6) |
WT indicates wild-type.
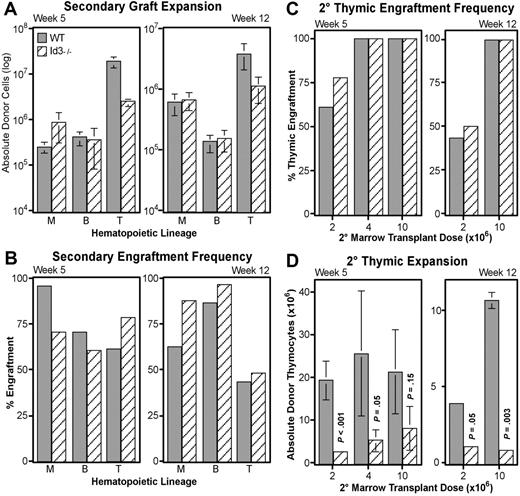
Loss of Id3 specifically diminishes thymocyte expansion, but does not impair LT-HSC function
Id3 is another HLH transcriptional inhibitor,51 and acts redundantly with Id1 in important developmental systems.52 Interestingly, other laboratories have indicated that Id3 mRNA may be present in human HSCs.33 This information suggests that Id3 may play a role similar to Id1 in LT-HSC maintenance. To test this hypothesis, we conducted additional transplantation experiments with WBM obtained from B6-Id3−/− donors and matched wild-type mice. Serial transplantations followed the protocol described in Figure 3A, with each primary host receiving 5 × 105 WBM. In contrast to Id1−/− transplants, secondary engraftment from Id3−/− WBM did not significantly differ from wild-type in absolute numbers of myeloid or B cells produced (Figure 7A). Likewise, the frequency of secondary hosts engrafted in these lineages fluctuated, but did not consistently favor either genotype (Figure 7B). These data suggest that loss of Id3 is not sufficient to alter the function of LT-HSCs.
Secondary transplantation potential of Id3−/− marrow. Graft expansion (A) and frequency (B) were evaluated for matched wild-type (▩) and Id3−/− (▧) WBM by following the secondary engraftment assay protocol described in Figure 3. Total donor-type cells engrafting in myeloid (“M”), B-cell (“B”), and thymocyte (“T”) lineages at 5 (left) or 12 (right) weeks after secondary transplantation are given in panel A. Numbers are averaged from 3 independent transplantation experiments where a total of 32 hosts received wild-type and 34 received Id3−/− secondary grafts. Error bars indicate SEM. (C) Thymic engraftment frequency as a function of increasing secondary marrow transplantation doses (x-axes) at either 5 or 12 weeks after secondary transplantation. (D) Thymocyte expansion differences between Id3−/− and wild-type secondary grafts as a function of secondary marrow doses at 5 or 12 weeks after secondary transplantation. y-axes give numbers of CD4+ and/or CD8+ donor-derived thymocytes for each of the indicated secondary marrow transplantation doses. Error bars indicate SEM. Each bar represents the average of at least 3 secondary hosts. Significance was determined using the Student t test (P).
Secondary transplantation potential of Id3−/− marrow. Graft expansion (A) and frequency (B) were evaluated for matched wild-type (▩) and Id3−/− (▧) WBM by following the secondary engraftment assay protocol described in Figure 3. Total donor-type cells engrafting in myeloid (“M”), B-cell (“B”), and thymocyte (“T”) lineages at 5 (left) or 12 (right) weeks after secondary transplantation are given in panel A. Numbers are averaged from 3 independent transplantation experiments where a total of 32 hosts received wild-type and 34 received Id3−/− secondary grafts. Error bars indicate SEM. (C) Thymic engraftment frequency as a function of increasing secondary marrow transplantation doses (x-axes) at either 5 or 12 weeks after secondary transplantation. (D) Thymocyte expansion differences between Id3−/− and wild-type secondary grafts as a function of secondary marrow doses at 5 or 12 weeks after secondary transplantation. y-axes give numbers of CD4+ and/or CD8+ donor-derived thymocytes for each of the indicated secondary marrow transplantation doses. Error bars indicate SEM. Each bar represents the average of at least 3 secondary hosts. Significance was determined using the Student t test (P).
However, Id3 did appear to play a role in the expansion of thymocyte progenitors. At both 5 and 12 weeks after transplantation, wild-type grafts consistently generated significantly more donor thymocytes (Figure 7A). Secondary wild-type grafts produced an average of 8-fold more thymocytes than did Id3−/− grafts at week 5, and more than 3-fold by week 12. To verify this difference in thymocyte production, we conducted further transplantations with higher doses of secondary bone marrow. As shown in Figure 7C, we found that 4 × 106 secondary WBM was sufficient to generate 100% thymic engraftment in this assay from either genotype by week 5. Nevertheless, fewer thymocytes continued to develop from Id3−/− grafts unless the secondary WBM dose was increased to 107 cells (Figure 7D). However, at 12 weeks after transplantation, a significant defect in Id3−/− thymocyte expansion persisted even at the 107 cell transplant dose. High thymic engraftment frequencies in the absence of Id3 indicate that loss of Id3 does not affect the production of thymic-seeding progenitors from HSCs; rather, the deficiencies in thymocyte production suggest a role for Id3 at a downstream expansion point during T-cell development.
Discussion
The data reported in this study illustrate the importance of E-protein inhibitors in maintaining the earliest forms of HSCs. Our investigation focused on possible roles for Id1 and Id3 in LT-HSCs because previous work has suggested that these E-protein regulators may be expressed in this primitive population.33 Although other studies have relied on RT-PCR data to chart Id1 expression in less-defined primitive bone marrow populations,23,33 the ability to track individual cells in Id1/GFP knock-in mice allowed us to examine rare populations and detect differences in Id1 expression between LT- and ST-HSCs. This revealed that Id1-expressing cells are more abundant in LT-HSCs than in ST-HSCs, and that LT-HSC activity resides in the Id1/GFP-expressing fraction of LSK (Figures 2–3).
Consistent with this expression pattern, Id1-deficient mice harbor fewer LT-HSCs as determined by phenotypic characterization (Figure 4). It is important to note that LT-HSC function is impaired but not abolished in Id1−/− marrow, as shown by the competitive repopulation studies summarized in Figure 6. This is in contrast with the apparently normal steady-state hematopoiesis found in Id1−/− mice; however, it is known from previous studies that very few stem cells are required to sustain hematopoiesis.10,12,43,44 While the 50% depletion of phenotypic LT-HSCs we observed in the absence of Id1 demonstrates the potential influence of E-protein activity on LT-HSC maintenance, the remaining LT-HSCs are nonetheless sufficient to sustain blood formation.
SCL/TAL1 is another HLH protein that functions as an E-protein inhibitor that has been shown to influence hematopoiesis. Ablation of SCL/TAL1 causes embryonic lethality due to abrogation of fetal blood formation.24,53,54 In contrast, conditional mutations that limit SCL/TAL1 deletion to adult HSCs have a mild effect on primary bone marrow engraftment, and no further effect on secondary engraftment.22,55 These observations suggest that SCL/TAL1 mainly affects ST-HSC activity during adult hematopoiesis. RT-PCR studies with human bone marrow progenitors support this conclusion in that SCL/TAL1 levels appear to be greater in ST-HSC subsets relative to LT-HSCs.56
Our studies indicate that Id1 plays a role distinct from SCL/TAL1 in adult hematopoiesis. Id1-expressing cells are associated with LT-HSC phenotypes, but are reduced in ST-HSC populations (Figure 2). Further, loss of Id1 does not appear to diminish primary engraftment (Figure 5), yet secondary Id1−/− grafts poorly reconstitute bone marrow and peripheral blood (Figure 6; data not shown). Considering that Id1 and SCL/TAL1 both function primarily through inhibiting E-protein activity,29,57,58 it is curious that one mainly influences LT-HSCs, and the other influences ST-HSCs. One possibility may be that Id1 functions to prevent LT-HSC differentiation into ST-HSCs, and directs daughter cells from LT-HSC divisions to remain in this primitive state. Signals that trigger down-regulation of Id1 in LT-HSCs may allow E-proteins to drive differentiation until expression of SCL/TAL1 at the ST-HSC stage checks further differentiation and facilitates ST-HSC expansion. In this paradigm, loss of Id1 in LT-HSCs would promote differentiation into ST-HSCs, resulting in depletion of LT-HSCs (Figure 4). Conversely, loss of SCL/TAL1 allows extended E-protein activity, promoting differentiation of ST-HSCs into more mature progenitors and resulting in loss of expansion potential.22
Alternatively, Id1 may affect the maintenance of LT-HSCs through its influence on cell cycle. Cell-cycle regulators, including p21cip1/waf1 and p16INK4a, are important for LT-HSCs to maintain quiescence.19,20,27,59 E-proteins have been shown to promote transcription of these cell-cycle inhibitors,25,60 which may increase in the absence of Id1. In turn, increased p21cip1/waf1 activity could contribute to the reduced LT-HSC numbers and engraftment potential we observed by inhibiting expansion of the LT-HSC pool. Others have evaluated cell cycle in Id1−/− LSK by bromodeoxyuridine incorporation and found that loss of Id1 is associated with increased cell cycle.49 We suspect this is due to enriched ST-HSCs and MPPs in Id1−/− LSK, since rare LT-HSCs were not identified in those studies. Further, Hoechst/Pyronin-Y analysis of HSC-enriched Thy1.1Lo LSK did not show increased cell cycling in Id1−/− HSC (Figure S3), but it should be considered that LT-HSC constitute less than 5% of Thy1.1Lo LSK; hence, even these data are insufficient to draw conclusions regarding the effect of Id1 on cell cycle in LT-HSCs.
As with p21cip1/waf1, Id1 may influence LT-HSC maintenance by blocking transcription of the p16INK4a gene.27 This cell-cycle inhibitor is normally only present in aged LT-HSCs or during times of stress, such as in posttransplantation recovery. These observations have lead to the hypothesis that p16INK4a transcription is a possible cause of age-associated reduction in LT-HSC numbers, and may be important in returning LT-HSCs to quiescence after cycling.20 Given the putative role of Id1 in affecting p16INK4a expression,26,–28 excess p16INK4a in Id1−/− LT-HSCs may account for the reduced LT-HSC number as we observed at steady state.
It is noteworthy that loss of Id3 did not impair LT-HSC activity (Figure 7A,B). Although Id1 and Id3 might have distinct functions, different patterns of Id1 and Id3 expression in LT-HSCs could be an explanation. For example, RT-PCR analysis of human bone marrow suggests that Id1 expression is higher in LT- than ST-HSCs, while Id3 expression is constant.33 However, it is difficult to compare the relative levels of Id1 and Id3 by RT-PCR. It is thus possible that Id1 expression is sufficient to compensate for loss of Id3 expression in LT-HSCs. Regardless of the contribution of Id3 to HSC function, the competitive transplantation experiments revealed a role for Id3 in thymocyte expansion (Figure 7C,D), which is not obvious during steady-state thymopoiesis in Id3−/− mice.61 This function appears to be after the point of thymic seeding progenitors, since the frequency of engraftment from Id3−/− grafts was not impaired. The greatest surge of expansion during thymocyte development is triggered by pre–T-cell receptor signaling, and correlates with up-regulation of Id3 expression.62 It has been suggested that Id3 contributes to this expansion by suppressing E-protein function.63 Our findings concur with this hypothesis, and illustrate the competitive advantage Id3 renders to thymopoiesis.
Taken together, this study has identified a novel function of Id1, namely regulating hematopoietic stem-cell renewal or maintenance. Such a role for Id1 or Id proteins is unlikely to be restricted to hematopoietic lineage cells. In fact, Id proteins have also been detected in other stem or progenitor cells, such as epithelial stem cells and neuronal progenitors.64,65 Whether Id proteins function in stem cells through inhibition of E-protein functions remains to be determined. If so, this opens a new area of investigation regarding the role of E-proteins in stem-cell renewal and differentiation. Considering the prominent effects of basic HLH proteins on the differentiation of diverse cell types, knowledge about how they operate in stem cells would be of great importance in various aspects of developmental biology and tissue regeneration.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Drs Paul Kincade and Darryl Dudley for critical reading of the manuscript. We are grateful to Amanda Crosbie and Erin Beeston for technical support and Dr Chuxia Deng (the National Institute of Diabetes, Digestive and Kidney Diseases) for providing the ploxP-neo vector and mouse embryonic stem cells. We are indebted to the transgenic and flow cytometry facilities at the Oklahoma Medical Research Foundation for technical support.
This work was supported by grants from the National Institutes of Health to X.-H. S. (AI33597, AI56129). X.-H.S. holds the Eli Lilly Distinguished Chair in Biomedical Research. S.S.P. was supported by training grant T32-AI07633 from the National Institute of Allergy and Infectious Diseases.
National Institutes of Health
Authorship
Contribution: S.S.P. designed and performed research, analyzed data, and wrote the paper; Y.Z., L.N., S.W.C., and Z.H. contributed to parts of the research; and X.-H.S. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Xiao-Hong Sun, Program in Immunobiology and Cancer Research, Oklahoma Medical Research Foundation, 825 NE 13th St, Oklahoma City, OK 73104; e-mail:sunx@omrf.ouhsc.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal