Abstract
A hallmark of various human malignancies is the expression of immunoinhibitory factors within the tumor microenvironment. There is indirect evidence based on in vitro experiments that tumor-infiltrating T cells in human malignancies are suppressed by such factors. Still, direct evidence of the influence of individual inhibitory factors on immune cells in human cancer in vivo is lacking. To address this question, we used Hodgkin lymphoma (HL) as a model because histopathological characteristics of HL are thought to be due mostly to the effects of a wide variety of cytokines, including TGFβ or membrane-bound receptors such as PD-1 that are suspected to contribute to immune evasion of tumor cells. Using a genome-wide transcriptional approach, we established specific RNA fingerprints of TGFβ and PD-1 signaling in human T cells in vitro. Applying these specific fingerprints, we directly demonstrate that CD4+ T cells in HL—but not in follicular lymphoma (FL)—are under the inhibitory influence of both TGFβ and PD-1 in vivo. This approach can be easily generalized to provide direct evidence of the impact of any given soluble or cell-bound factor on any cell type within diseased tissue.
Introduction
Tumors frequently develop evasion strategies that may influence every stage of the tumor-specific immune response, from the activation of antigen-presenting cells to T-cell recruitment, activation, and effector function.1-3 Hodgkin lymphoma (HL) seems to be a role model for an ineffective tumor immune response, because the minority of tumor cells, so called Reed-Sternberg (RS) cells, are surrounded mainly by a mixture of reactive cells including lymphocytes, plasma cells, eosinophils, and histiocytes.4,5 The majority of lymphocytes in close proximity to the tumor cells are CD4+ T cells with anergic phenotype, indicating poor immune surveillance of the tumor.6 In fact, several immune evasion strategies are described in HL, including down-regulation of immunodominant Epstein-Barr Virus (EBV) antigens, and secretion of soluble factors such as transforming growth factor beta (TGFβ), interleukin-10 (IL10), or prostaglandin E2 (PGE2), that have been shown to inhibit the activation of specific cytotoxic T lymphocytes (CTLs) and professional antigen-presenting cells in vitro and in murine models.7-10 Also cell-to-cell interactions via inhibitory receptors might lead to immune inhibition. For example, neoplastic CD20+ B cells in nodular lymphocyte-predominant HL express PDL-1, and the respective inhibitory receptor PD-1 is expressed by T cells in close proximity to the tumor cells.11
So far, a general limitation studying immune inhibition within the tumor microenvironment, particularly in humans, is the fact that we rely solely on indirect evidence. This is best exemplified for soluble factors such as TGFβ, which is found to be elevated within the tumor microenvironment of many human tumors including lymphomas.12,13 It is also well established that T cells isolated from tumor tissue exhibit an anergic phenotype14-17 and that such a phenotype can be induced in vitro in normal T cells exposed to inhibitory factors.14,17,18 However, no direct evidence exists that the anergic phenotype observed in tumor infiltrating T cells is indeed due to exposure to inhibitory cytokines such as TGFβ, IL10, or PGE2. Moreover, there is not even direct evidence that these factors lead to signaling events within tumor-infiltrating T cells in vivo.
HL is a very good example for the existing circumstantial evidence. Activated lymphocytes, binucleate RS cells, and mononuclear Hodgkin cells have been described to express activated TGFβ.7,13 The expression of TGFβ as well as the functional phenotype of T cells in HL suggested that this cytokine might be involved in immunosuppression in HL, however, direct evidence is lacking. Other possible mechanisms of immune escape in HL might involve ligands for inhibitory receptors such as PDL-1 on the surface of the neoplastic cells.11 PD-1 is a receptor of the Ig superfamily that negatively regulates T-cell antigen receptor signaling by interacting with the specific ligands (PDLs) and is suggested to play a role in the maintenance of self-tolerance.19-21 Ligands for PD-1 are expressed on the surface of different human cancers including HL. Again, this provides indirect evidence that PDL/PD-1 interactions might also lead to immune inhibition of tumor infiltrating T cells.22,23
If such inhibitory mechanisms are to be efficiently targeted therapeutically, it will be critical to better determine the overall impact of any given inhibitory factor on immune cells in vivo. A strategy to overcome the above-described limitations has been recently exemplified for the dissection of oncogenic pathways associated with malignant disease.24 While the potential role of particular oncogenes such as ras or myc has been clearly established in preclinical in vivo models, their individual role in human cancer is less well described and even less understood. Bild et al applied an elegant approach to shed light on this important issue.24 By transfecting normal human cells with single oncogenes followed by genome-wide transcriptional analysis they determined an oncogene-specific “RNA fingerprint.” Using descriptive and analytic bioinformatics, this fingerprint was then applied to genome-wide transcriptional profiles of human malignancies clearly demonstrating that these oncogene-specific RNA fingerprints can be recognized within the malignant cells. It was further shown that survival times of patients significantly differed when grouped by the activity of individual oncogenic RNA fingerprints.
We hypothesized that this approach should—in principle—be applicable to any other cell and factor that leads to transcriptional changes upon stimulation and signaling. Furthermore, this approach should—for the first time—provide direct evidence whether a particular cell is indeed under the control of a particular inhibitory factor in vivo. We established specific TGFβ and PD-1 RNA fingerprints in human CD4+ T cells and applied these RNA fingerprints to transcriptional profiles of CD4+ T cells isolated from HL lymph nodes. To determine whether the influence of TGFβ on CD4+ T cells is specific for HL or can also be detected in other lymphomas, we also applied the fingerprints to CD4+ T cells originated from follicular lymphoma (FL), thereby providing direct evidence that these inhibitory factors are clearly signaling in T cells infiltrating HL but not FL.
Patients, materials, and methods
Isolation of CD4+ T cells and stimulation
Blood samples were collected from healthy blood donors after informed written consent was obtained in accordance with the Declaration of Helsinki. CD4+ T cells were isolated by negative selection as described previously.9 Cells were stimulated by mixing with artificial antigen-presenting cells (aAPCs) at a ratio of 1:3 (cells-beads) composed of magnetic beads (Dynal Biotech, Oslo, Norway) coated with the following antibodies: anti-CD3 (OKT3), anti-CD28 (9.3),25 anti–PD-1–17,26 and anti–MHC-I (W6/32). For all experiments, these aAPCs were coated with suboptimal anti-CD3 Ab (5%), suboptimal levels of anti-CD28 Ab (14%), and either anti–MHC-I Ab (CD3/28/MHC-I) or anti–PD-1 Ab (CD3/28/PD-1), constituting the remaining 81% of protein added to the bead, as previously described.19 TGFβ was initially titrated at different concentrations ranging from 0 to 50 ng/mL to determine minimum concentration for maximum inhibitory effect in T-cell functions such as proliferation and cytokine production. For defining the TGFβ fingerprint, 30 ng/mL TGFβ was used. This concentration is also within the range of TGFβ described in serum derived from cancer patients of different origin.27,28
Patients
Lymph node specimens of 9 patients with classic Hodgkin lymphoma (HL), 9 patients with FL, and 9 patients with reactive lymph node reaction (RLN) of different causes were included. This study was performed within the framework of the German Hodgkin Study Group. When possible, samples were taken at primary diagnosis. Detailed information about sex and histologic diagnosis is given in Table 1. Also included were 3 samples with aberrant diagnosis: 1 patient with T-cell–rich B-cell lymphoma (B-NHL); 1 with lymphocyte-predominant HL (LPHL), but with tumor-free tissue in the removed lymph node; and 1 with HL, with histologically proven follicular lymphoma in prior medical history. CD4+ T cells from lymph node specimens were isolated by mechanical homogenization of the specimen and subsequently purified by positive selection on ice using magnetic cell sorting columns (Miltenyi Biotech, Bergisch Gladbach, Germany) according to the manufacturer's instructions. All samples were taken after informed consent following approval by the Ethik Kommission of the University of Cologne, Cologne, Germany.
Patient sex, diagnosis, and array platform used
| Patient no. . | Sex . | Diagnosis . | Primary diagnosis . | Array pPlatform . |
|---|---|---|---|---|
| 1 | Female | HL, NS | Yes | Affymetrix |
| 2 | Male | HL, MC | Yes | Affymetrix |
| 3 | Male | HL, NS | Yes | Affymetrix |
| 4 | Male | HL, not specified | No | Affymetrix |
| 5 | Male | HL, NS | Yes | Illumina |
| 6 | Female | HL, NS | Yes | Illumina |
| 7 | Male | HL, NS | Yes | Illumina |
| 8 | Female | HL, NS | Yes | Illumina |
| 9 | Female | HL, NS | No | Illumina |
| 10 | Male | Monocytoid B-cell reaction | Yes | Affymetrix |
| 11 | Female | Reactive changes | Yes | Affymetrix |
| 12 | Male | Lymphadenitis | Yes | Affymetrix |
| 13 | Female | Lymphadenitis | Yes | Affymetrix |
| 14 | Male | Reactive changes | Yes | Affymetrix |
| 15 | Female | Reactive changes | Yes | Illumina |
| 16 | Male | Lymphadenitis | Yes | Illumina |
| 17 | Female | Reactive changes | Yes | Illumina |
| 18 | Male | Reactive changes | Yes | Illumina |
| 19 | Male | FL, grade II | Yes | Affymetrix |
| 20 | Male | FL, grade I | No | Affymetrix |
| 21 | Male | FL, grade I | Yes | Affymetrix |
| 22 | Male | FL, grade I | Yes | Illumina |
| 23 | Male | FL, grade I | Yes | Illumina |
| 24 | Female | FL, grade II | Yes | Illumina |
| 25 | Male | FL, grade I | Yes | Illumina |
| 26 | Male | FL, grade II | Yes | Illumina |
| 27 | Female | FL, grade I | Yes | Illumina |
| 28 | Female | HL, MC; FL in premedical history | No | Illumina |
| 29 | Male | B-NHL | Yes | Illumina |
| 30 | Male | Reactive changes, proven LP-HD in different LN | No | Illumina |
| Patient no. . | Sex . | Diagnosis . | Primary diagnosis . | Array pPlatform . |
|---|---|---|---|---|
| 1 | Female | HL, NS | Yes | Affymetrix |
| 2 | Male | HL, MC | Yes | Affymetrix |
| 3 | Male | HL, NS | Yes | Affymetrix |
| 4 | Male | HL, not specified | No | Affymetrix |
| 5 | Male | HL, NS | Yes | Illumina |
| 6 | Female | HL, NS | Yes | Illumina |
| 7 | Male | HL, NS | Yes | Illumina |
| 8 | Female | HL, NS | Yes | Illumina |
| 9 | Female | HL, NS | No | Illumina |
| 10 | Male | Monocytoid B-cell reaction | Yes | Affymetrix |
| 11 | Female | Reactive changes | Yes | Affymetrix |
| 12 | Male | Lymphadenitis | Yes | Affymetrix |
| 13 | Female | Lymphadenitis | Yes | Affymetrix |
| 14 | Male | Reactive changes | Yes | Affymetrix |
| 15 | Female | Reactive changes | Yes | Illumina |
| 16 | Male | Lymphadenitis | Yes | Illumina |
| 17 | Female | Reactive changes | Yes | Illumina |
| 18 | Male | Reactive changes | Yes | Illumina |
| 19 | Male | FL, grade II | Yes | Affymetrix |
| 20 | Male | FL, grade I | No | Affymetrix |
| 21 | Male | FL, grade I | Yes | Affymetrix |
| 22 | Male | FL, grade I | Yes | Illumina |
| 23 | Male | FL, grade I | Yes | Illumina |
| 24 | Female | FL, grade II | Yes | Illumina |
| 25 | Male | FL, grade I | Yes | Illumina |
| 26 | Male | FL, grade II | Yes | Illumina |
| 27 | Female | FL, grade I | Yes | Illumina |
| 28 | Female | HL, MC; FL in premedical history | No | Illumina |
| 29 | Male | B-NHL | Yes | Illumina |
| 30 | Male | Reactive changes, proven LP-HD in different LN | No | Illumina |
NS indicates nodular sclerosis; MC, mixed cellularity; LP-HD, lymphocyte-predominant Hodgkin disease; and LN, lymph node.
Cytometric bead array for cytokines
The concentration of IFN-γ in cell culture supernatants was measured using the human Th1/Th2 Cytokine kit II (BD Pharmingen, San Diego, CA) as described previously.9
RNA preparation and microarray hybridization
CD4+ T cells of 4 different healthy donors were either left unstimulated or stimulated with magnetic beads coated with CD3/CD28/MHC-I or CD3/CD28/PD-1, respectively. TGFβ-treated cells were stimulated with CD3/CD28/MHC-I, and TGFβ was added at a concentration of 30 ng/mL. After 8 hours, magnetic beads were removed and cells were lysed in TRIzol reagent (Invitrogen Life Technologies, Frederick, MD). RNA isolation and quantification were performed as described previously.9 cRNA (1.5 μg) was hybridized to Sentrix whole genome bead chips 6 × 2 (Illumina, San Diego, CA) and scanned on Illumina BeadStation 500 ×. Microarrays of CD4+ T cells isolated from lymph node specimen were performed on the Illumina and the Affymetrix platform (Affymetrix, Santa Clara, CA). For samples performed on the Illumina platform RNA isolation, quantification and labeling were performed as described above. cRNA was hybridized to Sentrix whole genome bead chips 6 × 2. For samples performed on the Affymetrix platform, RNA isolation, quantification, and target preparation were performed according to standard protocols for small samples and cRNA was hybridized to HG-U133A arrays.
Statistical and bioinformatic data analysis
Raw data collection for Illumina BeadChip and Affymetrix HG-U133A arrays was performed using Illumina BeadStudio software or Affymetrix MAS5.0 software. Further statistical and bioinformatic analyses were performed using R language (http://www.r-project.org) and packages from the Bioconductor project (affy, multtest, hcluster, pamr, e1071, pcurve). For normalization of data from the 2 platforms, we used quantile and invariant set normalizations implemented in R. For comparison of the data derived from the 2 different platforms, we performed 2 distinct cross annotations. The first cross annotation was based on direct sequence comparison using the MAQC annotation information (http://www.switchtoi.com), and the second was a database annotation using Entrez gene IDs. In general, both annotation methods revealed similar results; shown here are results based on direct sequence comparison. When defining RNA fingerprints of TGFβ and PD-1, a 2-step analysis was performed. In the first step, resting CD4+ T cells were compared with CD4+ T cells stimulated with CD3/CD28/MHC-I (activated cells) in the absence or presence of TGFβ (TGFβ-treated cells), or to cells treated with CD3/CD28/PD-1 (PD-1–treated cells). Differentially expressed genes were selected using a fold change/P value filter with the following criteria: fold change of 2 or more, absolute difference in signal intensity between group means of 100 or more, and P value of .05 or less. In the second step, activated cells were compared with TGFβ-treated cells or PD-1–treated cells. Here, different fold change criteria (fold change of 1.5 or more for TGIβ and fold change of 3.5 or more for PD-1) were used. A gene was included in the RNA fingerprint only if its FCs derived from both comparisons (steps 1 and 2) showed the same direction. Four bioinformatics methods (hierarchic clustering, principal components analysis [PCA], and supervised classification using 2 different algorithms, the shrunken centroids method [the so-called prediction analysis for microarrays; PAM] and support vector machines [SVM]) were used to assess evidence for an in vivo influence of the RNA fingerprints. Hierarchic cluster analysis was performed using the hcluster method in R. Before clustering, the data were log2 transformed. Distances of the samples were calculated using a correlation coefficient (correlation similarity metric), and clusters were formed by taking the average of each cluster (average linkage). PCA analysis was performed using the pcurve package in R. Briefly, PCA is a technique for simplifying a dataset, by reducing multidimensional datasets to lower dimensions for analysis. PCA can be used for dimensionality reduction in a dataset while retaining those characteristics of the dataset that contribute most to its variance. When visualizing PCA results, the first 3 principal components (coordinates) were z-transformed (mean = 0, standard deviation = 1) and subsequently plotted in 3D. 3D plots show a 45° angle between x-coordinate and y-coordinate. When performing supervised classification, we first used the pamr package in R which uses the shrunken centroids method introduced by Tibshirani et al.29 Briefly, the method computes a standardized centroid for each class. Nearest centroid classification then takes the gene expression profile of a new sample, and compares it with each of these class centroids. The class whose centroid that it is closest to is the predicted class for that new sample. At last, we applied the e1071 package in R, which is used for support vector machine classification. A support vector machine is a classification method that separates a batch of objects into 2 classes, thereby maximizing the area around the class border that has no objects. A new sample is then predicted to belong to either side of the border. Cross-platform analysis was performed according to a method by Warnat et al.30 In short, the data were annotated across both platforms and were then transformed to derive numerically comparable measures of gene expression.
Results
Quantification of the inhibitory effect of TGFβ and PD-1 on human CD4+ T cells in vitro
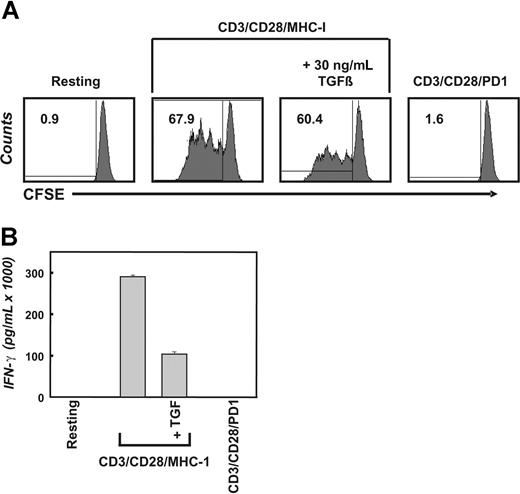
To directly determine the in vivo impact of inhibitory cytokines such as TGFβ or inhibitory surface receptors such as PD-1 on tumor-infiltrating T cells, we postulated that the factor-dependent transcriptional regulation assessed on a genome-wide scale should be comparable in T cells directly isolated from tumor tissue and T cells exposed to TGFβ or PD-1 in vitro. Prior to assessment of transcriptional changes as a consequence of stimulation with TGFβ or PD-1, we established the functional impact of both factors on highly purified CD4+ T cells derived from healthy donors. The impact of TGFβ and PD-1 was assessed in context of T-cell receptor–mediated activation because it has been previously shown that T cells in vivo would be exposed to inhibitory factors in the context of antigen recognition within the tumor microenvironment.6,7,31 The CD4+ T cells were labeled with 5,6-carboxyfluorescin-diacetat-succinimidyl-ester (CFSE) and subsequently stimulated with aAPCs (CD3/CD28/MHC-I) with or without TGFβ or aAPCs coated with CD3/CD28/PD-1 for up to 96 hours (Figure 1; shown here is the 96-hour time point). As expected, stimulation of primary CD4+ T cells with CD3/CD28/MHC-I resulted in robust T-cell expansion and cytokine secretion. Addition of TGFβ to the cultures reduced T-cell proliferation, albeit this effect was not as dramatic as that induced by PD-1 stimulation, which completely inhibited T-cell proliferation (Figure 1A). Additional experiments demonstrating the range of T-cell inhibition are shown in Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article. In contrast, IFN-γ secretion was clearly decreased by both inhibitory factors, TGFβ and PD-1 (Figure 1B).
Inhibition of T-cell proliferation and IFN-γ secretion by TGFβ and PD-1. (A) Freshly isolated primary human CD4+ T cells were labeled with CFSE and left unstimulated or were stimulated with the indicated magnetic beads (artificial antigen-presenting cells, or CD3/CD28/MHC-I or CD3/CD28/PD-1) in the absence or presence of 30 ng/mL TGFβ. After 4 days, CFSE dilution was analyzed by flow cytometry. The overall percentage of dividing cells is displayed inside the corresponding dot plot. (B) CD4+ T cells were stimulated as in panel A. After 4 days of incubation, the concentration of IFN-γ was determined using flow cytometric bead arrays. The presented data are representative for at least 3 independent experiments; error bars in panel B represent 1 representative experiment performed in triplicate.
Inhibition of T-cell proliferation and IFN-γ secretion by TGFβ and PD-1. (A) Freshly isolated primary human CD4+ T cells were labeled with CFSE and left unstimulated or were stimulated with the indicated magnetic beads (artificial antigen-presenting cells, or CD3/CD28/MHC-I or CD3/CD28/PD-1) in the absence or presence of 30 ng/mL TGFβ. After 4 days, CFSE dilution was analyzed by flow cytometry. The overall percentage of dividing cells is displayed inside the corresponding dot plot. (B) CD4+ T cells were stimulated as in panel A. After 4 days of incubation, the concentration of IFN-γ was determined using flow cytometric bead arrays. The presented data are representative for at least 3 independent experiments; error bars in panel B represent 1 representative experiment performed in triplicate.
TGFβ and PD-1 RNA fingerprints in CD4+ T cells from healthy donors
For establishing the TGFβ and PD-1 fingerprints, CD4+ T cells from 4 donors were either left unstimulated (resting cells) or stimulated with CD3/CD28/MHC-1 (activated cells) with or without addition of TGFβ (TGFβ-treated cells) or were stimulated with aAPCs coated with CD3/CD28/PD-1 (PD-1–treated cells). To filter genes regulated under direct influence of TGFβ or PD-1, we analyzed transcriptional changes in 2 different ways: In a first step (a) resting cells versus activated cells and (b) resting cells versus TGFβ or PD-1–treated cells were compared. Genes specifically regulated under the influence of TGFβ or PD-1 were determined using set theory supported by Venn diagrams as previously described.9 In a second step, we compared expression profiles of activated cells versus TGFβ or PD-1–treated cells. We defined the union of lists from steps 1 and 2 as direct impact of either factor on the CD4+ T-cell transcriptional profile and thus as the RNA fingerprints of TGFβ and PD-1 signaling in T cells (112 genes for TGFβ and 37 genes for PD-1; Tables S1–S2).
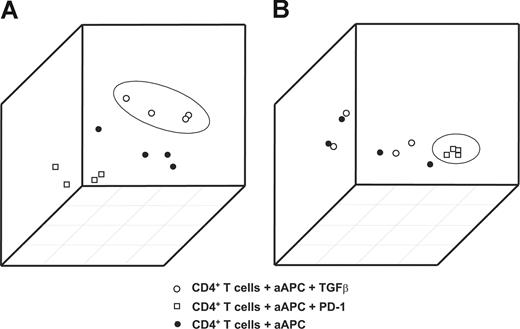
When plotting the first 3 principal components from the 2 signatures, samples treated with TGFβ (Figure 2A) or PD-1 (Figure 2B) were accurately distinguished from the other samples. This clear separation of samples treated with TGFβ or PD-1 from the remaining samples documents the particular impact of TGFβ and PD-1 on CD4+ T cells and therefore provides the rationale for defining these signatures as RNA fingerprints.
Generation of TGFβ and PD-1 genomic fingerprints in primary CD4+ T cells from healthy donors. Principal components analysis (PCA) using the RNA fingerprints of TGFβ or PD-1 was performed to distinguish samples treated with TGFβ or PD-1 from the respective control samples. The first 3 principal components derived from (A) the TGFβ and (B) the PD-1 fingerprints are plotted. Plots depict samples stimulated with magnetic beads coated with CD3/CD28/MHC-I (●); samples treated with CD3/CD28/PD-1 (□); and samples treated with CD3/CD28/MHC-I in the presence of TGFβ (○).
Generation of TGFβ and PD-1 genomic fingerprints in primary CD4+ T cells from healthy donors. Principal components analysis (PCA) using the RNA fingerprints of TGFβ or PD-1 was performed to distinguish samples treated with TGFβ or PD-1 from the respective control samples. The first 3 principal components derived from (A) the TGFβ and (B) the PD-1 fingerprints are plotted. Plots depict samples stimulated with magnetic beads coated with CD3/CD28/MHC-I (●); samples treated with CD3/CD28/PD-1 (□); and samples treated with CD3/CD28/MHC-I in the presence of TGFβ (○).
CD4+ T cells in Hodgkin lymphoma differ from T cells of reactive lymph nodes
To first assess overall differences between CD4+ T cells derived from HL and FL versus RLN, we performed descriptive biomathematic analysis. CD4+ T cells from RLN were used as a control, reflecting the characteristics of healthy T cells to the closest point possible. FL was used as a second malignancy to determine disease-specific differences. For this analysis, we used expression profiles of 5 samples from RLN patients, 4 samples from HL patients, and 3 samples from FL patients derived from the Affymetrix HG-U133A microarray. Explicit information about histologic diagnosis and type of array platform used is given in Table 1. Genes were defined as differentially regulated when FC was more than 2 or FC was less than − 2, with a P value less than .05 and difference in sample means more than 100. In total, we found 108 differentially expressed genes between CD4+ T cells derived from HL and RLN samples (42 up-regulated, 66 down-regulated) and 144 differentially expressed genes between CD4+ T cells derived from FL resp. RLN samples (144 down-regulated) (Table S3). Interestingly, when comparing for T-cell activation–induced genes, no significant differences between the patient groups were observed (data not shown). To link differential expression of genes to biologic processes, we postulated that it is possible to apply, for example, the RNA fingerprints we established for TGFβ and PD-1 in our in vitro system to answer the question of whether such inhibitory mechanisms play a role in HL in vivo.
CD4+ T cells in Hodgkin lymphoma harbor the TGFβ fingerprint
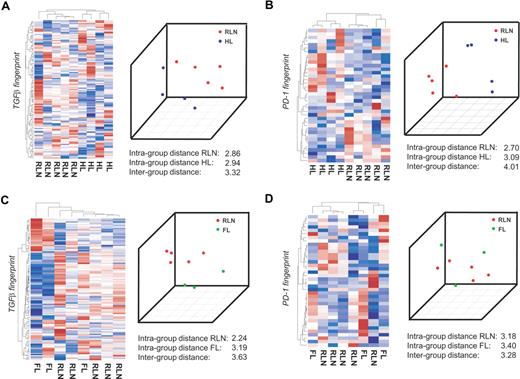
To separate distinct sample groups based on different biologies, several approaches including unsupervised as well as supervised approaches have been developed.32 If TGFβ indeed acts on CD4+ T cells in HL, it should be possible to correctly separate T cells isolated from HL from CD4+ T cells isolated from RLN within the gene space of the TGFβ fingerprint established in vitro. We therefore applied a total of 4 independent approaches, namely, (1) hierarchic clustering, (2) principal component analysis (PCA), (3) classification based on nearest shrunken centroids (PAM), and (4) support vector machines (SVMs). We first performed this analysis on the Affymetrix platform on a subgroup of patients, namely 5 samples from RLN patients, 4 samples from HL patients, and 3 samples from FL patients. By applying hierarchic clustering using the TGFβ fingerprint, HL and RLN were separated into 2 distinct clusters (Figure 3A left panel). The correct separation was still achieved when using less stringent filter criteria for generating the TGFβ fingerprint (data not shown). In contrast, when applying other gene sets established as biologically defined RNA fingerprints including those established by Bild et al,24 HL and RLN samples were not correctly separated. This analysis included fingerprints associated with transcriptional changes following activation of Ras, Myc, E2F3, Src, b-catenin, EGF, VEGF, or NFκB fingerprints associated with T-cell activation, cell cycle activity, apoptosis, inflammatory response, or chemokine activity, (data not shown). These findings further support the specificity of the TGFβ fingerprint within the HL samples.
CD4+ transcriptional profiles from patients with HL are separated from RLN on the basis of TGFβ-regulated genes. CD4+ T cells were isolated from lymph nodes of 4 different patients with HL, 3 patients with FL, and 5 patients with RLN. cRNA was hybridized to HG-U133A Affymetrix arrays. The RNA fingerprints of (A) TGFβ and (B) PD-1 were used to separate transcriptional profiles of HL from RLN. Left panel of A and B: Hierarchic cluster analysis using average linkage and correlation distance metric. Right panel of A and B: Result of PCA with the first 3 principal components is shown. RNA fingerprints of (C) TGFβ and (D) PD-1 were used to attempt to separate transcriptional profiles of FL from RLN. Again, hierarchic cluster analysis and PCA are shown.
CD4+ transcriptional profiles from patients with HL are separated from RLN on the basis of TGFβ-regulated genes. CD4+ T cells were isolated from lymph nodes of 4 different patients with HL, 3 patients with FL, and 5 patients with RLN. cRNA was hybridized to HG-U133A Affymetrix arrays. The RNA fingerprints of (A) TGFβ and (B) PD-1 were used to separate transcriptional profiles of HL from RLN. Left panel of A and B: Hierarchic cluster analysis using average linkage and correlation distance metric. Right panel of A and B: Result of PCA with the first 3 principal components is shown. RNA fingerprints of (C) TGFβ and (D) PD-1 were used to attempt to separate transcriptional profiles of FL from RLN. Again, hierarchic cluster analysis and PCA are shown.
As a second unsupervised approach we applied PCA. When plotting the first 3 principal components, HL and RLN samples were again separated using the TGFβ fingerprint. This was further supported by a larger intergroup distance (between HL and RLN) compared with the respective intragroup distances (Figure 3A right panel). To more formally assess the existence of a TGFβ fingerprint signature in HL, we applied leave-one-out cross validation based on PAM and SVMs. PAM analysis predicted HL respectively RLN cases with a 100% accuracy and posterior probability based on the genes within the TGFβ fingerprint. Using the SVM approach, again, a 100% accuracy was achieved (Figure S2A). So far, assessment of differential transcriptional regulation in CD4+ T cells from either HL or RLN based on specific RNA fingerprints indicated that TGFβ is an important component of the HL environment leading to signaling events in CD4+ T cells infiltrating the tumor site.
PD-1 signaling is also prominent in T cells derived from Hodgkin lymphoma
The same 4 bioinformatic approaches were used to determine whether genes of the PD-1 fingerprint were also harbored in HL-derived CD4+ T cells. As depicted in the left panel of Figure 3B, HL and RLN samples were correctly separated when applying hierarchic clustering based on the PD-1 fingerprint. Similarly, applying PCA led to a correct separation of HL and RLN samples, which was also supported by a larger intergroup distance. When applying PAM, one sample was always falsely predicted and the posterior probability never reached 100% for all samples. Using SVM though, the prediction accuracy was 100% based on the PD-1 fingerprint (Figure S2B). Taken together, the results indicate PD-1 is a further important factor in the HL environment.
RNA fingerprints reveal no impact of TGFβ or PD-1 on CD4+ T cells in follicular lymphoma
To determine whether the influence of TGFβ on CD4+ T cells is specific for HL or can also be detected in other lymphomas, we analyzed CD4+ T cells derived from patients with FL. We first assessed the influence of TGFβ by applying the TGFβ fingerprint. Hierarchic clustering never correctly separated FL from RLN samples (Figure 3C left panel). Similarly, the supervised approaches showed no correct prediction (Figure S2C). Only when applying PCA were FL and RLN samples correctly separated, and the intergroup variance was larger than the intragroup distances (Figure 3C right panel). Similarly, none of the fingerprints (eg, Ras, Myc) correctly separated FL from RLN samples, indicating that none of these pathways plays a major role in CD4+ T cells derived from FL tissue.
Validation of the method using additional patient samples and a different array platform
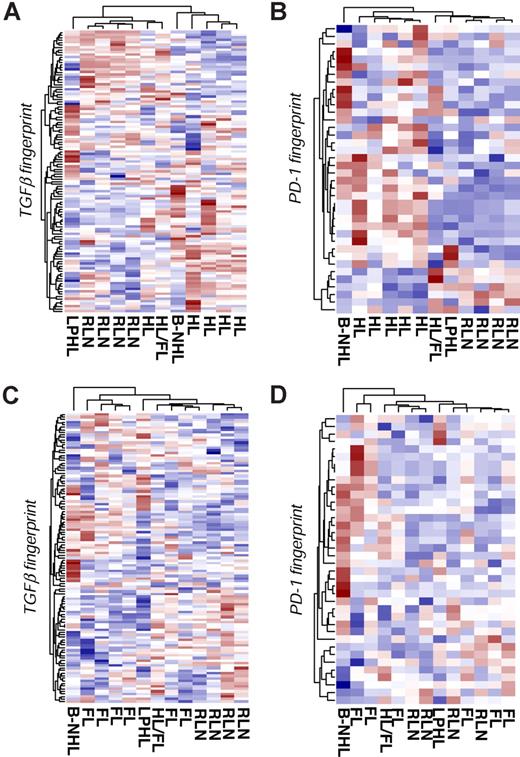
To validate our method and to show the independency of the results from the microarray platform, we used the Illumina BeadChip platform for further analysis. Here we analyzed 5 patients with HL, 6 patients with FL, and 4 patients with RLN. In addition, we included 3 samples with aberrant diagnosis to further specify our approach: 1 patient with T-cell–rich B-cell lymphoma (B-NHL); 1 patient with lymphocyte-predominant HL (LPHL), but with tumor-free tissue in the removed lymph node specimen; and 1 patient with HL, with histologically proven FL in the prior medical history. First, we tested the TGFβ fingerprint. As depicted in Figure 4A, the TGFβ fingerprint correctly separates the HL samples from the RLN samples, with only one HL sample falsely allocated to RLN. Interestingly, T cells derived from a tumor-free lymph node of a patient with LPHL clustered together with the RLN samples, suggesting that TGFβ-mediated signaling events are restricted to the tumor in HL. Similarly, T cells from the patient with prior history of FL were more closely related to T cells from RLN samples. The results of the PCA analysis mirrored the hierarchic clustering. Moreover, both supervised approaches resulted in a significant classification of the different samples, highlighting the impact of TGFβ on CD4+ T cells in HL (Figure S3A).
Validation using additional patient samples and a second array platform. CD4+ T cells were isolated from lymph nodes of 5 patients with HL, 4 patients with RLN, and 6 patients with FL. Three samples with aberrant diagnosis are labeled as follows: T-cell–rich B-cell lymphoma (B-NHL); lymphocyte-predominant Hodgkin lymphoma (LPHL); and Hodgkin lymphoma with premedical history of follicular lymphoma (HL/FL). cRNA was hybridized to Illumina Sentrix BeadChip Version 2. The RNA fingerprints of (A) TGFβ and (B) PD-1 were used to differentiate HL and RLN samples using hierarchic clustering. Hierarchic cluster analysis of FL and RLN samples based on the (C) TGFβ fingerprints and (D) PD-1 fingerprints.
Validation using additional patient samples and a second array platform. CD4+ T cells were isolated from lymph nodes of 5 patients with HL, 4 patients with RLN, and 6 patients with FL. Three samples with aberrant diagnosis are labeled as follows: T-cell–rich B-cell lymphoma (B-NHL); lymphocyte-predominant Hodgkin lymphoma (LPHL); and Hodgkin lymphoma with premedical history of follicular lymphoma (HL/FL). cRNA was hybridized to Illumina Sentrix BeadChip Version 2. The RNA fingerprints of (A) TGFβ and (B) PD-1 were used to differentiate HL and RLN samples using hierarchic clustering. Hierarchic cluster analysis of FL and RLN samples based on the (C) TGFβ fingerprints and (D) PD-1 fingerprints.
When using the PD-1 fingerprint, HL and RLN samples were correctly separated. Figure 4B displays the results of hierarchic clustering. The results of the PCA analysis and both supervised methods confirmed the separation and correct classification of the different samples (Figure S3B). Taken together, even when using a different array platform, both TGFβ and PD-1 fingerprints separate HL from RLN samples. This result gives further evidence for the impact of both TGFβ and PD-1 on CD4+ T cells in the tumor microenvironment of HL.
When analyzing CD4+ T cells from FL patients, FL and RLN samples were not separated by hierarchic clustering using either the TGFβ (Figure 4C) or the PD-1 (Figure 4D) fingerprint. In addition, PCA and both supervised methods failed to classify the samples accordingly (Figure S4A,B), thereby supporting the specificity of both factors toward HL.
Cross-platform analysis further supports the impact of TGFβ and PD-1 on CD4+ T cells in HL but not in FL
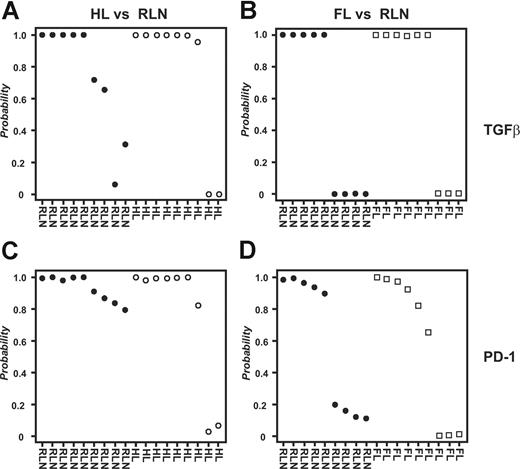
To analyze all samples together irrespective of array platform used, we applied an approach for cross-platform analysis introduced by Warnat et al.30 Due to the quantitative nature of the method, hierarchic clustering was not a useful tool for analyzing data derived from different array platforms because it regularly separates samples based on technology used rather than biology (D.E., unpublished results, November 2006). In contrast, supervised approaches can be performed on data derived by cross-platform analysis. As shown in Figure 5A, PAM analysis predicted HL respectively RLN samples with a 79% accuracy and high posterior probabilities based on the genes within the TGFβ fingerprint. Using the PD-1 fingerprint, we derived a total accuracy of 89% (Figure 5C). In contrast, when classifying FL and RLN samples, the overall prediction accuracy was only 53% for the gene spaces of both the TGFβ and the PD-1 fingerprints (Figure 5B,D). Again, we verified the specificity of the fingerprints, this time by analyzing 335 biologically defined gene spaces (terms defined by Gene Ontology; GO Terms) chosen based on size of the respective GO Terms (including 50-100 genes). Less than 9% of these gene spaces derived a correct classification of HL versus RLN samples respectively FL versus RLN (data not shown). These data further strengthen the hypothesis for both TGFβ and PD-1 to play a role in inhibiting CD4+ T cells specifically in HL but not FL.
Combined analysis of all samples irrespective of array platform used. Supervised classification (PAM) using the data derived from cross-platform analysis; 9 samples from HL patients, 9 samples from FL patients, and 9 samples from patients with RLN were used for analysis. For each sample the posterior probability, that is, the percentage of certainty of a correct class prediction, is plotted. The TGFβ fingerprint was used to classify (A) HL and RLN samples or (B) FL and RLN samples, respectively. The PD-1 fingerprint was used to classify (C) HL and RLN samples or (D) FL and RLN samples.
Combined analysis of all samples irrespective of array platform used. Supervised classification (PAM) using the data derived from cross-platform analysis; 9 samples from HL patients, 9 samples from FL patients, and 9 samples from patients with RLN were used for analysis. For each sample the posterior probability, that is, the percentage of certainty of a correct class prediction, is plotted. The TGFβ fingerprint was used to classify (A) HL and RLN samples or (B) FL and RLN samples, respectively. The PD-1 fingerprint was used to classify (C) HL and RLN samples or (D) FL and RLN samples.
Discussion
Clear evidence has been accumulated that anergic T cells can be isolated from various types of human cancers.1,15,33 Almost exclusively, T-cell function in human cancer is defined by in vitro experiments assessing T-cell activation, thereby providing only indirect evidence for the role of particular inhibitory factors in vivo. Both soluble factors and cell-to-cell–mediated mechanisms have been shown to induce anergy in T cells from healthy donors in vitro. So far, however, there is no direct evidence that inhibitory factors such as TGFβ, PD-1, CTLA-4, or IL10 signal efficiently in tumor-infiltrating T cells in human cancer.
To overcome these limitations, we have adapted a recently introduced functional genomics approach.24 Bild et al established gene expression signatures in vitro, reflecting the activation status of particular oncogenic pathways. Using these RNA fingerprints, human tumors could be clustered clearly defining prognosis in respective patient subsets.24 Here, we provide clear evidence that RNA fingerprints of T cells derived from healthy individuals activated in the presence of inhibitory cytokines such as TGFβ or inhibitory receptors such as PD-1 can be used to directly determine whether T cells isolated from diseased tissue are indeed under the influence of these inhibitory factors in vivo. We show that TGFβ has distinct impact on CD4+ T cells in HL but not in FL. Hierarchic clustering, principal component analysis, and supervised classification based on the TGFβ-induced RNA fingerprint all demonstrated that this inhibitory factor indeed induces signaling in CD4+ T cells in HL. Besides a direct inhibitory effect on CD4+ CD25− conventional T cells, TGFβ might be involved in the conversion of CD4+ CD25− T cells to CD4+ CD25+ T cells with regulatory function.34 Such cells are described to be abundant within the HL lymph node.35
Because only PCA indicated a separation of FL from RLN samples, we conclude that TGFβ has a minor impact on CD4+ T cells in FL. These data are most likely reflecting that—in contrast to RS cells—most non-HL lymphoma (NHL) cells do not produce TGFβ in excess amounts.36,37
Because ligands for PD-1 are expressed on the surface of various tumors including neoplastic B cells in LPHL11 and PDL/PD-1 interactions are discussed to provide a potent mechanism of immune escape,22,23,38 we therefore assessed the impact of PD-1 on CD4+ T cells in HL. Similar to TGFβ, different independent biomathematic approaches indicated an influence of PD-1 on CD4+ T cells in classical HL. These data are insofar surprising, because PD-1 expression is described only in T cells from LPHL. PDL1/PD-1 interactions are also described in NHL of B-cell origin.39 In B-NHL, PD-1 was found to be constitutively expressed on a subset of infiltrating CD4+ CD25− T cells and PDL-1 could be induced on intratumoral CD4+ CD25+ T cells. In vitro, anti–PDL-1 antibody or PD-1 fusion protein partly restored the proliferation of infiltrating CD4+ CD25− T cells when cocultured with intratumoral regulatory T (Treg) cells.39 We analyzed the possible in vivo influence of PD-1 on CD4+ T cells in FL, a subset of NHL. Because none of the 4 applied statistical tests gave evidence of an impact of PD-1 on CD4+ T cells in FL, we conclude that PD-1 has minor influence on infiltrating T cells in this particular disease.
To further demonstrate that these results are not tributary to the array technology used, we applied Illumina as a second platform. Moreover, in a cross-platform analysis, we performed a combined analysis of all samples together irrespective of array platform. These data clearly demonstrated that the effect we observed was independent of sample size and array technology used. The specificity of the inhibitory influence of TGFβ and PD-1 especially in HL was further underscored by analyzing 3 samples with aberrant diagnosis. Especially when analyzing a tumor-free specimen from a patient with LPHL, we found that this sample showed more similar expression patterns to RLN samples than to HL samples based on the TGFβ and PD-1 fingerprints, thereby demonstrating the specificity of the fingerprints toward the analyzed sample type.
We have previously shown that PGE2 might also be an inhibitory factor leading to signaling in T cells in HL,9 however, at that time we applied only hierarchic clustering analysis on HL samples using an in vitro established PGE2 signature. Applying the more differentiated biostatistical analysis introduced here, all 4 tests resulted in a separation and prediction of HL and RLN samples (data not shown). These results substantiate our recent findings on PGE2, however identify TGFβ and PD-1 as further major inhibitory factors leading to specific transcriptional changes in T cells derived from HL samples.
In conclusion, we have successfully applied the concept of RNA fingerprints to study the individual role of inhibitory factors acting on CD4+ T cells within the tumor microenvironment. Using this approach, we demonstrated a significant impact of TGFβ and PD-1 on CD4+ T cells in HL but not FL. Because we had access only to lymph node material of the patients, it would be interesting in future studies to analyze peripheral blood CD4+ T cells of HL patients for existing TGFβ and PD-1 signatures. These studies would be of particular interest in light of our findings from the tumor-free specimen of a patient with LPHL suggesting that the TGFβ and PD-1 signatures in T cells might be restricted to the tumor microenvironment. Following Bild et al's suggestions for the direct assessment of individual oncogenic pathways for tumor cells,24 we propose the combination of controlled in vitro experiments for the generation of specific RNA fingerprints with the analysis of tumor-infiltrating immune cells as a generally applicable and efficient method to quickly assess upstream events—here, cytokine signaling and cell surface receptor signaling—of transcriptional changes observed in diseased tissue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Alexander von Humboldt Foundation via a Sofia-Kovalevskaja Award (J.L.S.). J.M.C. was supported by the Köln Fortune Program of the Faculty of Medicine of the University of Cologne; T.Z. was supported by the Frauke-Weiskam-Christel Ruranski Foundation. J.L.S. is a member of the National Genome Research Network.
We thank our colleagues at the Division of Transfusion Medicine for their technical support.
Authorship
Contribution: J.M.C. designed research, performed research, and wrote the paper; D.E. performed research and wrote the paper; J.D., S.C., S.D.-P., M.B., and A.P. performed research; J.L.R. designed research and contributed new reagents; T.Z. designed research and performed research; J.L.S. designed research and wrote the paper. J.M.C. and D.E. contributed equally to this work.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Joachim L. Schultze, Molecular Tumor Biology and Tumor Immunology, Department of Internal Medicine I, University of Cologne, Kerpener Str. 62, 50924 Cologne, Germany; e-mail: joachim.schultze@uk-koeln.de.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal