Abstract
Activation of the lipid-regulated nuclear receptor peroxisome proliferator-activated receptor gamma (PPARγ) modifies the immunophenotype of monocyte-derived dendritic cells (DCs). However it has not been analyzed in a systematic manner how lipid metabolism and immune regulation are connected at the transcriptional level via this receptor. Here we present the genome-wide expression analyses of PPARγ-instructed human DCs. Receptor activation was achieved by exogenous, synthetic as well as endogenous, natural means. More than 1000 transcripts are regulated during DC development by activation of PPARγ; half of the changes are positive effects. These changes appear to enhance and modulate the robust gene expression alterations associated with monocyte to DC transition. Strikingly, only genes related to lipid metabolism are overrepresented among early induced genes. As a net consequence, lipid accumulation appears to be diminished in these cells. In contrast, genes related to immune response are regulated after 24 hours, implying the existence of indirect mechanisms of modulation. Receptor dependence was established by using DCs of patients harboring a dominant-negative mutation of PPARγ. Our data show that PPARγ acts as a mostly positive transcriptional regulator in human developing DCs, acting primarily through controlling genes involved in lipid metabolism and via this, indirectly modifying the immune phenotype.
Introduction
Dendritic cells (DCs) are professional antigen-presenting cells. They capture, process, and present antigen to T cells and thus initiate immune responses or under certain conditions promote immune tolerance.1 Although several transcription factors have been identified that participate in DC development, the transcriptional regulation of DC differentiation and subtype specification is poorly defined.2 In particular, the response to a changing milieu and tissue environment is not well understood. More than 30 studies have investigated the genome-wide transcription changes during human DC development and transition to the activated state.3-6 Thus, it is well documented that a very large number of genes are regulated during this cytokine-driven differentiation program. However beside cytokines, various hormones and lipids also profoundly affect the function and immunophenotype of DCs contributing to lineage, subtype, and functional specification. For example, DC phenotype is regulated by activation of nuclear hormone receptors such as the glucocorticoid receptor, vitamin D receptor, retinoic acid receptor (RAR), and also PPARs. We and others reported that the lipid-regulated nuclear receptor PPARγ is highly and acutely up-regulated during DC differentiation.7-9 PPARγ is a lipid-activated transcription factor that was originally identified by virtue of its role in adipocyte differentiation.10 This transcription factor directly regulates the expression of several genes participating in fatty acid uptake and lipid storage.11 The phenotypic effects of PPARγ ligand on DC development have been well characterized (ie, PPARγ-instructed DCs have an enhanced phagocytic activity and a modified cytokine-production profile, and these cells acquire an elevated natural killer T [NKT]–cell–activating capacity).7-9,12 However it is not clear whether these phenotypic changes are due to direct transcriptional effects on inflammatory gene expression or due to secondary changes induced in lipid metabolism. In order to gain insights into how PPARγ regulates different facets of DC differentiation, we sought to identify PPARγ-regulated genes and gene networks in monocyte-derived DCs using unbiased, global gene-expression profiling validated by quantitative reverse transcriptase–polymerase chain reaction (qRT-PCR) and complemented by a human genetic model. We employed an exogenous ligand activation approach using a highly selective synthetic ligand (rosiglitazone). In addition, we have defined culture conditions in which human serum induces PPARγ activation via a yet uncharacterized endogenous mechanism. We have found that more than 1000 transcripts (probes sets) were regulated by PPARγ during DC differentiation. We also compared the gene-expression profile of DCs obtained from patients13 harboring dominant-negative mutations of this receptor. Our results indicate that activation of PPARγ acutely up-regulated genes primarily involved in lipid metabolism and transport; and furthermore, changes in immune function appear to be secondary to these alterations in gene expression
Materials and methods
Approval was obtained from the institutional review boards of the University of Debrecen and the University of Cambridge for these studies. Informed consent was obtained in accordance with the Declaration of Helsinki.
Cell culture and ligand treatment
Monocytes (98% CD14+) were obtained from buffy coats by Ficoll gradient centrifugation and magnetic separation using anti-CD14–conjugated microbeads (VarioMACS; Miltenyi Biotec, Bergisch Gladbach, Germany). DCs were prepared as described.9 Cells were cultured in RPMI 1640 (Sigma, St Louis, MO) supplemented with 10% FBS (Invitrogen, Carlsbad, CA) or with 10% human AB serum (Sigma). To obtain macrophages, monocytes were cultured in the presence of 100 ng/mL M-CSF (Peprotech, Rocky Hill, NJ) for 5 days. Ligands or vehicle control (50% DMSO/ethanol) were added at day 0. Cells were treated with rosiglitazone (RSG), GW9662, or GW501516 (Alexis Biochemicals, San Diego, CA).
Microarray analysis
RNA isolation and labeling was performed as described.14 Hybridization was carried out at the Microarray Core Facility of European Molecular Biology Laboratory (EMBL; Heidelberg, Germany). Analyses were carried out using GeneSpring GX7.3.1 (Agilent, Santa Clara, CA) software. Raw data (cell files) were analyzed by the GC-RMA algorithm. Data were normalized using per-chip normalization (global scaling). First, genes (probe sets), which had low expression (raw expression < 20 in 90% of the experiments), were filtered; next, probe sets 2-fold up- or down-regulated with P values less than .01 were selected. Where indicated, hierarchical clustering was performed. To identify overrepresented gene ontology (GO) categories, Cytoscape2.1/BiNGO (Biological Networks Gene Ontological tool) software was used.15 All microarray data are available in the public Gene Expression Omnibus (GEO) database (accession no. GSE8658).
Real-time quantitative RT-PCR
Real-time quantitative RT-PCR was conducted as described.14 The comparative cycle threshold (Ct) method was used to quantify transcripts and normalize to cyclophilin A expression. In addition, Taqman low-density arrays (TLDAs) were used (Figures 4 and 7B) according to manufacturer's instructions. For the TLDA analyses, a high-capacity cDNA archive kit (Applied Biosystems, Foster City, CA) was used. Quantitative PCR was performed using real-time PCR (ABI PRISM 7900; Applied Biosystems). The sequences of the primers and probes are in Table S5 (available on the Blood website; see the Supplemental Materials link at the top of the online article).
Western-blot analysis
Twenty micrograms protein of whole-cell extracts was separated by electrophoresis on 12.5% polyacrylamide gel and transferred to PVDF membrane (Bio-Rad Laboratories, Hercules, CA). Membranes were probed with anti-FABP4 (Cayman Chemical, Ann Arbor, MI; rabbit polyclonal antibody [Ab]) and then stripped and reprobed with anti-GAPDH (ab8245–100; Abcam, Cambridge, United Kingdom) according to manufacturer's recommendations.
Transient transfections and reporter gene assays
MH100-TK-Luciferase reporter construct was transfected along with the GAL-PPARγ-LBD or GAL-PPARδ-LBD and CMX-β-gal into COS1 cells using jetPEI reagent (Qbiogene, Irvine, CA). Cells were lysed and assayed for reporter expression 24 hours after transfection. The luciferase and β-galactosidase assay activity was determined as described previously.16
Fluorescence-activated cell sorter analysis
Cell staining was performed using PE-conjugated monoclonal antibodies and isotype-matched controls (BD PharMingen, Heidelberg, Germany). The fluorescence of labeled cells was measured by using a FACSCalibur flow cytometer (Becton Dickinson, San Jose, CA).
Nile red staining
Nile red staining was carried out on unfixed cells as described.17 In brief, DCs (1 × 105/mL to 2 × 105/mL) were suspended in PBS, then Nile red (Sigma; concentration, 5 ng/mL) was added and the cells were incubated for 5 minutes and measured by a FACSCalibur flow cytometer (Becton Dickinson).
Lipid body staining
Osmium tetroxide staining of lipid bodies was performed as described previously.18 Coverslips were washed several times with PBS and mounted with Moviol (Sigma). The morphology of fixed cells was observed, and lipid droplets were enumerated with a 100×/1.3 NA oil-objective lens using an Axiovert 200 microscope (Zeiss, Oberkochen, Germany) connected to an AxioCam HR camera (Zeiss). Image analysis was carried out using AxioVision 4.5 software (Zeiss).
Results
Exogenous and endogenous activation of the PPARγ pathway during DC development
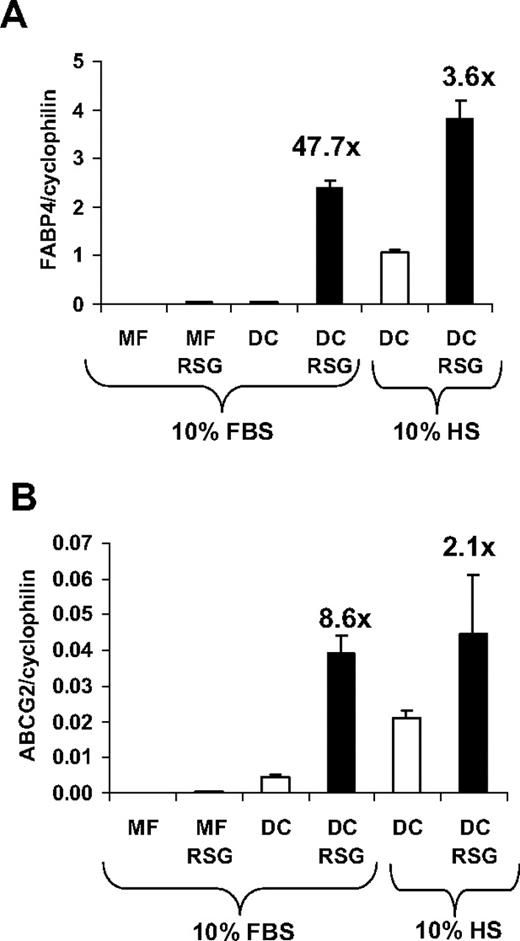
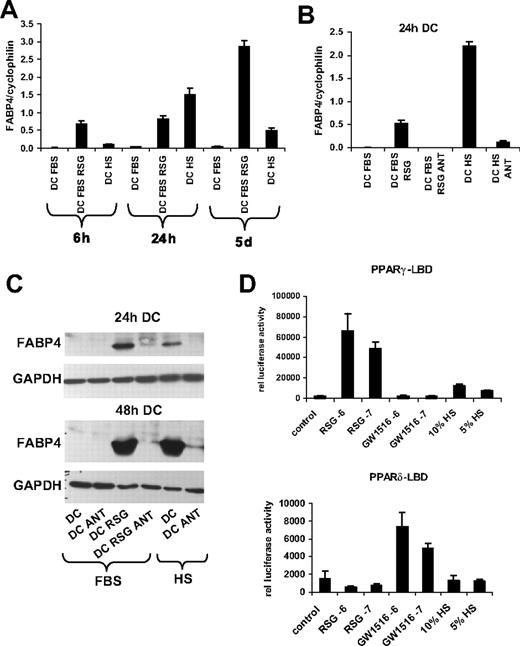
One of the early genome-wide expression studies indicated that PPARγ is highly up-regulated during monocyte-derived DC differentiation.19 We and others confirmed that the transcript level of PPARγ is immediately and acutely up-regulated during DC development7-9 ; and concordant with this effect, the bone fide PPARγ target gene FABP4 (adipose-specific fatty acid–binding protein; also known as aP2)20 was robustly induced by PPARγ-specific ligand treatment.9 In order to optimize the activation of PPARγ response during DC differentiation, we compare the induction of FABP4 using various conditions. When monocytes were cultured in the presence of M-CSF (macrophage differentiation), a weak induction of FABP4 was observed upon PPARγ-specific agonist (rosiglitazone; [RSG]) treatment (Figure 1A). In contrast, when cells were differentiated toward DCs in the presence of GM-CSF and IL-4, we obtained a marked up-regulation of FABP4 upon PPARγ ligand administration. Remarkably, an elevated expression of FABP4 was also detected in the absence of ligand if the cells were cultured in human serum. ABCG2, a recently described PPARγ target,16 also displayed a similar expression pattern (Figure 1B), suggesting that the activity of the receptor can be readily regulated by exogenous as well as endogenous means. Collectively, our data revealed that the optimal condition, providing the largest dynamic range, to study PPARγ response could be achieved if monocytes were cultured in the presence of GM-CSF and IL-4 in FBS-containing medium.
Induction of PPARγ target genes under different cell culture conditions. Transcript levels of FABP4 (A) and ABCG2 (B) were determined by qRT-PCR. RNA was obtained from monocyte-derived macrophages (MF) or monocyte-derived DCs in the presence or absence of 2.5 μM rosiglitazone (RSG). Cells were cultured in human AB serum (HS)– or fetal bovine serum (FBS)–containing cell culture medium as indicated. Error bars indicate the standard diviation (SD) of the relative expression.
Induction of PPARγ target genes under different cell culture conditions. Transcript levels of FABP4 (A) and ABCG2 (B) were determined by qRT-PCR. RNA was obtained from monocyte-derived macrophages (MF) or monocyte-derived DCs in the presence or absence of 2.5 μM rosiglitazone (RSG). Cells were cultured in human AB serum (HS)– or fetal bovine serum (FBS)–containing cell culture medium as indicated. Error bars indicate the standard diviation (SD) of the relative expression.
Next we sought to optimize the time course of ligand administration. Our early observations indicated that freshly isolated monocytes lack PPARγ expression, but the mRNA level of PPARγ was immediately and acutely up-regulated during DC development. It should be noted, however, that the largest PPARγ response was obtained when ligand was added at the beginning of differentiation.9 Given these considerations, we decided to add PPARγ agonist at the beginning of DC differentiation and cells were harvested 6 hours later in order to detect early responses. As 6 hours is obviously not sufficient for DC development, we also harvested the cells 24 hours and 5 days after to compare the genome-wide expression profile of control and PPARγ-activated DCs.
Global gene-expression profile of exogenous PPARγ ligand–treated cells
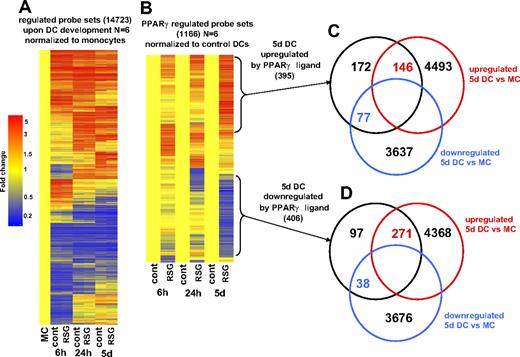
To monitor the whole-genome expression profile during DC development, we selected those transcripts (probes sets) for which expression was changed upon DC differentiation. Global gene-expression profiling was performed using Affymetrix GeneChips (HU133 Plus2; Santa Clara, CA) and data were obtained from 6 biologic replicates. Unexpectedly, a very high number of genes changed during DC development: 14 723 probe sets were up- or down-regulated if all of the transcripts regulated at any of the time points were added up (Figure 2A). It should be mentioned that numerous genes are covered by more than 1 probe set, thus the actual numbers of genes were lower. These results indicated that activation of PPARγ is not a general inhibitor of DC development because the vast majority of genes that are regulated upon DC differentiation showed an unaltered expression upon PPARγ ligand treatment (Figure 2A). Rather ligand activation appears to modify the differentiation in specific ways.
Gene-expression changes during DC development and upon PPARγ ligand treatment. (A) Probe sets (14 723) were regulated upon DC differentiation. Gene-expression data were normalized to freshly isolated monocytes. Dendograms were obtained from hierarchical clustering (standard correlation). Data were obtained using 6 biologic replicates. Cells were treated with 1 μM rosiglitazone (RSG; 6 hours, 24 hours DC) or with 2.5 μM RSG (5 days DC). RNA was isolated at the indicated time points. (B) Probe sets (1166) were regulated by PPARγ ligand during monocyte-derived DC differentiation. Microarray data were normalized to vehicle-treated cells. Venn diagram of the probe sets that were up-regulated (C) or down-regulated (D) after 5 days of PPARγ ligand treatment. Comparison of genes that were regulated in 5-day DCs versus monocytes is shown.
Gene-expression changes during DC development and upon PPARγ ligand treatment. (A) Probe sets (14 723) were regulated upon DC differentiation. Gene-expression data were normalized to freshly isolated monocytes. Dendograms were obtained from hierarchical clustering (standard correlation). Data were obtained using 6 biologic replicates. Cells were treated with 1 μM rosiglitazone (RSG; 6 hours, 24 hours DC) or with 2.5 μM RSG (5 days DC). RNA was isolated at the indicated time points. (B) Probe sets (1166) were regulated by PPARγ ligand during monocyte-derived DC differentiation. Microarray data were normalized to vehicle-treated cells. Venn diagram of the probe sets that were up-regulated (C) or down-regulated (D) after 5 days of PPARγ ligand treatment. Comparison of genes that were regulated in 5-day DCs versus monocytes is shown.
To select those transcripts that were induced or repressed by PPARγ agonist, we filtered those probe sets that exhibited changed expression after 6 hours, 24 hours, or 5 days of differentiation upon ligand treatment. Therefore we compared the gene expression of untreated versus RSG-treated cells at all time points and selected those probe sets that were at least 2-fold up- or down-regulated. As shown on the heat map in Figure 2B, the expression of 1166 probe sets was altered upon ligand treatment (all of the identified probe sets are presented in Table S1). It should be mentioned that a comparable number of probe sets were positively or negatively regulated by PPARγ ligand (633 up; 534 down). Interestingly, after 6 or 24 hours, more up-regulated genes were detected than down-regulated ones, but by day 5 this trend seemed to be reversed. Furthermore, we detected a large number of transcripts that were positively regulated across multiple time points by ligand treatment (40/633). We failed to detect any probe sets that were down-regulated at all time points. Next, we compared the genes regulated by activation of the nuclear receptor to the ones regulated during DC differentiation (Figure 2C,D). As shown in Figure 2D, in DCs harvested at day 5, 271 of 402 genes were induced; in contrast only 38 probe sets were down-regulated (DC versus isolated monocytes). These findings suggested that activation of PPARγ negatively regulates a small subset of genes that are induced during DC development. Interestingly, approximately 40% of the PPARγ ligand–up-regulated probe sets (146/395 in 5-day DCs) were also positively regulated upon DC development (Figure 2C). Several well-characterized PPARγ targets (ie, FABP4, ADFP) were included in this subgroup. Interestingly, some of these well-known PPAR targets were previously described to be up-regulated during DC development,19 indicating that a weak endogenous activation of PPARγ is likely to occur even if cells are cultured in FBS-containing medium. As a conclusion, PPARγ activation does not appear to grossly inhibit the DC differentiation program. Rather it seems to be an integral part of the differentiation, and additional activation leads to enhanced expression of a sector of genes.
Activation of PPARγ changes the lipid metabolism/storage of DCs
Next we systematically analyzed the PPARγ-regulated genes and investigated the potential role of these genes in DC development and function. To describe the dynamics of DC response upon PPARγ ligand treatment, we classified the regulated genes according to their kinetics of expression and known biologic functions. First we analyzed the gene lists containing the PPARγ-regulated genes with Cytoscape/BiNGO software15 to determine which gene ontology (GO) categories are overrepresented. Remarkably, we observed that only lipid/fatty acid metabolism–related categories were overrepresented among the genes up-regulated by PPARγ ligand after 6 hours (Table 1; see Table S2 for the full data set). Although several well-known PPAR targets were in this list (ABCG1, ANGPTL4, CPT1A, CD36), most of the uncovered lipid metabolism–related genes were not previously described as PPARγ-regulated genes and most likely several of them are directly and transcriptionally regulated by this receptor (Table 1). We also analyzed the biologic functions of those genes that were up-regulated after 24 hours and 5 days of ligand treatment. We found that after 24 hours or 5 days both lipid metabolism and immune response–related genes were highly overrepresented (Table 1). These data suggest it is very likely that activation of PPARγ indirectly modifies the immunophenotype of DCs via the activation of metabolic and signaling pathways. It has been shown that activation of PPARγ receptor modulates the cytokine production, immunogenicity, and antigen-presenting capacity of DCs.12 For instance, PPARγ-instructed DCs express lower levels of group I CD1s (CD1a, CD1b, CD1c, and CD1e), lipid-presenting molecules, and CD80 but were characterized by elevated expression of CD1d and CD86.8,9 Our gene-expression profiling confirmed these previously documented cell-surface marker expression changes. For example, group I CD1s (CD1a, CD1b, and CD1c) as well as CD80 were down-regulated following PPARγ ligand administration; in contrast, CD1d was positively regulated (Table 1), indicating that most of the previously described expression changes are regulated at the level of transcription or mRNA stability. In addition, after 24 hours, the expression of enzymes involved in collagen degradation (several matrix metalloproteinases: MMP1, MMP9, MMP10, and MMP12) was down-regulated, whereas adhesion-related molecules were induced by day 5 (Table S2). Genes associated with Th1 immune response were also down-modulated by day 5 (IRF4, TLR6, INHBA, CD80).
Comparison of the gene-expression pattern of lipid metabolism and immune response–related genes
| . | Immune response . | Lipid metabolism . | ||
|---|---|---|---|---|
| P . | Annotated genes . | P . | Annotated genes . | |
| 6 h, up-regulated by RSG; 203 probe sets, 80 annotated genes | NS | SPN, IL7R, BCL2, SEMA3C, TLR4, SAMHD1 | <.001 | LYPLA3, ACAA2, CD36, ECH1, FABP5, APOC1, SCARB1, ANGPTL4, CPT1A, MLYCD, SCD, PPAP2B, ABCG1, PLCL1, HADHSC |
| 24 h, up-regulated by RSG; 210 probe sets, 84 annotated genes | <.001 | MGLL, C1QB, C1QG, OAS1, OAS3, C1QA, TLR4, GBP2, TRIM22, OAS2, IL21R, GBP1, IL7R, ALOX5, CD1D, CCL15, MX2, NCK2, APOL3, SAMHD1 | .006 | MGLL, DDHD1, PLTP, ECH1, APOC1, PHYH, SCARB1, ACADVL, ALOX5, ACSL1, PPAP2B, SCD, HADHSC |
| 5 d, up-regulated by RSG; 395 probe sets, 157 annotated genes | .011 | MR1, MGLL, OAS1, SPG3A, OAS3, C3, TLR4, ADA, AOC3, CCL5, COL4A3BP, CCRL2, IFIT2, LTB4R, GPR65, IL7R, CD1D, S100A8, CD164, FOS, SPON2, ITGAL, C4A, S100A9 | .019 | MGLL, CD36, PCCA, PPP2R1B, ECH1, DHRS3, VLDLR, PECR, APOC1, ASAH1, PC, ACSL1, SCD, PPAP2B, HSD11B1, FDX1, HADHSC |
| 6 h, down-regulated by RSG; 33 probe sets, 12 annotated genes | NS | HAMP, CCL1 | ||
| 24 h, down-regulated by RSG; 144 probe sets, 72 annotated genes | NS | PPBP, BST1, HPSE, CD1B, FCAR, ENTPD1, CD1C, CD226, ALOX5AP, CD1E, IL1R2, CD1A, IL1RAP | NS | ALOX5AP |
| 5 d, down-regulatedby RSG; 406 probesets, 162 annotatedgenes | .036 | PPBP, CSF2RB, BCL6, CD1B, CCL4, CD1C, CCR7, TNFRSF4, RGS1, IRF4, INHBA, SERPINA1, TLR6, PTGER4, CD80, CD1E, IL1R2, IL1R1, MMP25, IFI16, CD1A, IL1RAP | NS | AGPAT1, ISYNA1, LTC4S, PLA2G4A, PPAP2A, OSBPL3 |
| . | Immune response . | Lipid metabolism . | ||
|---|---|---|---|---|
| P . | Annotated genes . | P . | Annotated genes . | |
| 6 h, up-regulated by RSG; 203 probe sets, 80 annotated genes | NS | SPN, IL7R, BCL2, SEMA3C, TLR4, SAMHD1 | <.001 | LYPLA3, ACAA2, CD36, ECH1, FABP5, APOC1, SCARB1, ANGPTL4, CPT1A, MLYCD, SCD, PPAP2B, ABCG1, PLCL1, HADHSC |
| 24 h, up-regulated by RSG; 210 probe sets, 84 annotated genes | <.001 | MGLL, C1QB, C1QG, OAS1, OAS3, C1QA, TLR4, GBP2, TRIM22, OAS2, IL21R, GBP1, IL7R, ALOX5, CD1D, CCL15, MX2, NCK2, APOL3, SAMHD1 | .006 | MGLL, DDHD1, PLTP, ECH1, APOC1, PHYH, SCARB1, ACADVL, ALOX5, ACSL1, PPAP2B, SCD, HADHSC |
| 5 d, up-regulated by RSG; 395 probe sets, 157 annotated genes | .011 | MR1, MGLL, OAS1, SPG3A, OAS3, C3, TLR4, ADA, AOC3, CCL5, COL4A3BP, CCRL2, IFIT2, LTB4R, GPR65, IL7R, CD1D, S100A8, CD164, FOS, SPON2, ITGAL, C4A, S100A9 | .019 | MGLL, CD36, PCCA, PPP2R1B, ECH1, DHRS3, VLDLR, PECR, APOC1, ASAH1, PC, ACSL1, SCD, PPAP2B, HSD11B1, FDX1, HADHSC |
| 6 h, down-regulated by RSG; 33 probe sets, 12 annotated genes | NS | HAMP, CCL1 | ||
| 24 h, down-regulated by RSG; 144 probe sets, 72 annotated genes | NS | PPBP, BST1, HPSE, CD1B, FCAR, ENTPD1, CD1C, CD226, ALOX5AP, CD1E, IL1R2, CD1A, IL1RAP | NS | ALOX5AP |
| 5 d, down-regulatedby RSG; 406 probesets, 162 annotatedgenes | .036 | PPBP, CSF2RB, BCL6, CD1B, CCL4, CD1C, CCR7, TNFRSF4, RGS1, IRF4, INHBA, SERPINA1, TLR6, PTGER4, CD80, CD1E, IL1R2, IL1R1, MMP25, IFI16, CD1A, IL1RAP | NS | AGPAT1, ISYNA1, LTC4S, PLA2G4A, PPAP2A, OSBPL3 |
Microarray gene lists were analyzed with Cytoscape/BiNGO software to assess Gene Ontology (GO) category enrichment. The corrected P values are also indicated. These were calculated by Cytoscape/BiNGO using a hypergeometrical test to assess whether the GO categories were overrepresented or not.
NS indicates nonsignificant.
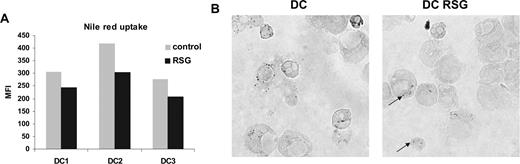
To explore the potential role of the up-regulated lipid metabolism–associated genes in PPARγ-activated DCs, we assessed the lipid content of the cells. We did not detect any changes in neutral lipid content using thin-layer chromatography (TLC; data not shown). However, Nile red–stained PPARγ ligand–treated cells exhibited less fluorescence intensity than control DCs (Figure 3A), suggesting that these cells contain fewer cytoplasmic lipid droplets. Consistent with this observation, we also detected fewer lipid droplets/body in PPARγ-instructed cells using osmium tetroxide staining (Figure 3B). These results altogether indicated that PPARγ-activated DCs have an elevated expression of numerous lipid metabolism–related genes; moreover, they have an enhanced capacity to metabolize/redistribute lipids, resulting in diminished cytoplasmic lipid content.
Modified lipid metabolism in PPARγ-instructed DCs. (A) Nile red uptake of control or PPARγ ligand (RSG)–treated DCs (5 days). Cells were stained with Nile red as described in “Nile red staining” for detection of lipid droplets. (B) Osmium tetroxide staining of DCs. DCs were cultured for 24 hours and control or RSG-treated cells were stained with osmium tetroxide. The arrows indicate the intracellular lipid droplets. See “Lipid body staining” for image acquisition information.
Modified lipid metabolism in PPARγ-instructed DCs. (A) Nile red uptake of control or PPARγ ligand (RSG)–treated DCs (5 days). Cells were stained with Nile red as described in “Nile red staining” for detection of lipid droplets. (B) Osmium tetroxide staining of DCs. DCs were cultured for 24 hours and control or RSG-treated cells were stained with osmium tetroxide. The arrows indicate the intracellular lipid droplets. See “Lipid body staining” for image acquisition information.
Validation of microarray expression data with TLDAs
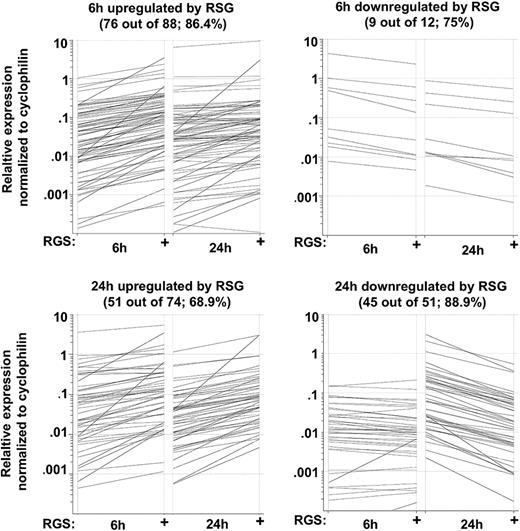
In order to validate the microarray (GeneChip) results by an independent method, we used real-time quantitative RT-PCR (qRT-PCR) using Taqman low-density arrays (TLDAs). We selected 174 genes that were up- or down-regulated by PPARγ activators (6 hours or 24 hours) from the gene lists. Altogether, 136 of 174 genes (78.2%) were validated by qRT-PCR (Figure 4). We used 3 biologic replicates and considered a gene validated if the fold change was at least 1.5 and P value was less than .05 (Table S3). These results prove that our microarray data are reliable and appropriate for further analyses.
Validation of microarray data using TLDA. RNA was isolated at the indicated time points and subjected to TLDA analyses. Cells were treated with 1 μM rosiglitazone (RSG). Data were obtained from 3 individuals. Black lines indicated those genes that were induced at least 1.5 fold and had a P value .05 or lower.
Validation of microarray data using TLDA. RNA was isolated at the indicated time points and subjected to TLDA analyses. Cells were treated with 1 μM rosiglitazone (RSG). Data were obtained from 3 individuals. Black lines indicated those genes that were induced at least 1.5 fold and had a P value .05 or lower.
Human serum–cultured DCs have enhanced PPARγ activity: evidence for endogenous ligand activation
As shown in Figure 1, we observed elevated expression of the PPARγ target genes (FABP4, ABCG2) if cells were cultured in human serum. This intriguing finding suggested that these cells possessed enhanced PPARγ activity. To take advantage of this finding and to further characterize our in vitro system to assess the gene-expression changes, a time course experiment was carried out. A moderate elevation of PPARγ targets (FABP4, ABCG2) was detected after 6 hours in the presence of human serum. Unexpectedly, after 24 hours, human serum–exposed cells exhibited a higher PPARγ target gene expression than those cultured in the presence of FBS and treated with a synthetic PPARγ ligand (Figure 5A). To prove the receptor dependence of this process, the cells were also treated with a PPARγ-specific antagonist (GW9662 [ANT]; Figure 5B). In this case we observed a substantially attenuated expression of PPARγ target genes, suggesting that enhanced expression of target genes upon human serum exposure is indeed PPARγ dependent (Figure 5B). Consistent with the enhanced mRNA expression, the protein level of FABP4 was also increased when cells were cultured in human serum (Figure 5C). Our results thus indicated that human serum instructs DCs to generate endogenous PPARγ ligand. The results of the time course experiments also support the notion that PPARγ ligand is produced endogenously, because after 6 hours a moderate induction of FABP4 was already detected, whereas after 24 hours a robust up-regulation of PPARγ targets was obtained, suggesting that at that stage PPARγ ligands would most likely have accumulated in the cells. It should be noted that the transcript level of PPARγ was about 2-fold higher in human serum–cultured cells compared with FBS-cultured cells (data not shown). To further characterize the source of PPARγ ligand, we performed transient transfection experiments using the Gal4-DBD (DNA-binding domain) fusion of the ligand-binding domain (LBD) of PPARγ or PPARδ receptor to detect PPAR ligands derived from human serum. A robust induction of reporter gene activity was obtained when COS cells were treated with synthetic PPARγ activator (RSG), and an elevated expression of reporter activity was measured upon 10% or 5% human serum exposure (Figure 5D). The induction was specific for PPARγ because using a PPARδ-LBD we failed to detect elevated reporter gene expression upon human serum administration. These data strongly suggest that human serum is likely to contain PPARγ activators or ligand precursors.
The effect of fetal bovine serum (FBS) and human AB serum (HS) on PPARγ response. (A,B) Transcript levels of FABP4 were determined by qRT-PCR. The indicated samples were cultured in fetal bovine serum (FBS)– or human AB serum (HS)–containing cell-cultured medium. (A) RNA was isolated at the indicated times; cells were treated with 1 μM rosiglitazone (RSG; 6 hours, 24 hours DC) or with 2.5 μM RSG (5 days DC). (B) Cells were cultured for 24 hours and treated with 1 μM rosiglitazone (RSG) and/or with 5 μM GW9662 (ANT). (C) Western-blot analysis of FABP4 protein expression in DCs. GAPDH was used as a loading control. Cells were cultured for 24 or 48 hours and treated with 1 μM rosiglitazone (RSG) and/or 5 μM GW9662 (ANT). Samples were cultured in fetal bovine serum (FBS)– or human AB serum (HS)–containing cell-cultured medium. (D) COS1 cells were cotransfected with MH100-TK-Luciferase reporter construct and GAL4-PPARγ–LBD or GAL4-PPARδ–LBD construct along with β-galactosidase construct. Cells were lysed after 24 hours, and the luciferase and β-galactosidase activity was assayed as described in “Transient transfections and reporter gene assays.” All experiments were done in triplicates; error bars represent SDs. Cell were treated with 1 μM (-6) or 100 nM (-7) PPARγ activator (RSG) or 1 μM (-6) or 100 nM (-7) PPARδ activator (GW501516 abbreviated as GW1516). The indicated cells were cultured in 10% or 5% human AB serum (HS)–containing medium.
The effect of fetal bovine serum (FBS) and human AB serum (HS) on PPARγ response. (A,B) Transcript levels of FABP4 were determined by qRT-PCR. The indicated samples were cultured in fetal bovine serum (FBS)– or human AB serum (HS)–containing cell-cultured medium. (A) RNA was isolated at the indicated times; cells were treated with 1 μM rosiglitazone (RSG; 6 hours, 24 hours DC) or with 2.5 μM RSG (5 days DC). (B) Cells were cultured for 24 hours and treated with 1 μM rosiglitazone (RSG) and/or with 5 μM GW9662 (ANT). (C) Western-blot analysis of FABP4 protein expression in DCs. GAPDH was used as a loading control. Cells were cultured for 24 or 48 hours and treated with 1 μM rosiglitazone (RSG) and/or 5 μM GW9662 (ANT). Samples were cultured in fetal bovine serum (FBS)– or human AB serum (HS)–containing cell-cultured medium. (D) COS1 cells were cotransfected with MH100-TK-Luciferase reporter construct and GAL4-PPARγ–LBD or GAL4-PPARδ–LBD construct along with β-galactosidase construct. Cells were lysed after 24 hours, and the luciferase and β-galactosidase activity was assayed as described in “Transient transfections and reporter gene assays.” All experiments were done in triplicates; error bars represent SDs. Cell were treated with 1 μM (-6) or 100 nM (-7) PPARγ activator (RSG) or 1 μM (-6) or 100 nM (-7) PPARδ activator (GW501516 abbreviated as GW1516). The indicated cells were cultured in 10% or 5% human AB serum (HS)–containing medium.
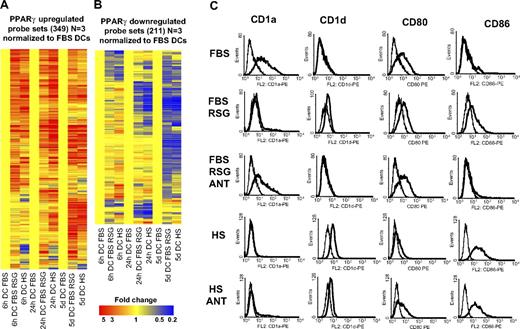
Next we wished to determine the global gene-expression profile of the human serum–cultured DCs to further characterize the effects of serum and compare it to RSG-induced gene-expression changes. A much higher number of genes were regulated by the change of serum if compared with the synthetic PPARγ ligand. For example, at 6 hours 789 probe sets showed a higher expression level in human serum– versus FBS-cultured cells. In contrast, only 154 probe sets showed higher expression in RSG-treated versus FBS-cultured cells (> 2 fold; P < .01, 3 biologic replicates; Figure S1; Table S4). Most importantly, the overlap was almost complete when we compared PPARγ ligand to serum-regulated gene sets (Figure 6A,B). It should be noted that in these experiments 3 biologic replicates were used and that is likely to be the reason for the lower numbers. Together, these findings suggest that human serum–exposed DCs have enhanced PPARγ signaling, and in these cells additional signaling pathways may also get activated, which then further modulate the phenotype and function of DCs. To further characterize the immunophenotype of human serum–cultured cells, we assessed the membrane expression pattern of several DC markers by flow cytometry. It is well established that PPARγ ligand–treated DCs cultured in FBS express less CD1a and CD80 but express more CD86 and CD1d.7-9 We confirmed these results; in addition, we proved that these changes were mostly receptor dependent, as administration of a PPARγ antagonist abolished the effect (Figure 6C). We found that human serum–cultured DCs displayed a similar cell-surface expression as PPARγ ligand–treated DCs (Figure 6C), but some of the markers (CD1d and CD86) showed a higher expression level in human serum–cultured cells. We also added PPARγ-specific antagonist (GW9662) to establish receptor dependency. In the case of CD80 and CD1d, we reverted almost completely the effects of human serum, but in the case of CD86 or CD1a, we could not revert the phenotype obtained in the presence of FBS, suggesting that besides PPARγ activation, other pathways are also active.
PPARγ and human serum–induced and repressed genes. (A) Three hundred forty-nine probe sets were up-regulated and 211 probe sets down-regulated (B) by PPARγ ligand (3 biologic replicates were used). Hierarchical cluster analysis was performed with GeneSpring software (standard correlation). Affymetrix microarray data were normalized to nontreated FBS-cultured cells. Monocytes were cultured for 6 or 24 hours or 5 days as described in “Cell culture and ligand treatment” and the indicated cells were cultured in human AB serum (HS) instead of FBS. Cells were treated with 1 μM rosiglitazone (RSG; 6 hours, 24 hours DC) or with 2.5 μM RSG (5 days DC). (C) Characterization of CD1a, CD1d, CD80, and CD86 cell-surface expression on DCs treated with ligands: 2.5 μM RSG alone or with 5 μM GW9662 (ANT). The indicated DCs were cultured in human AB serum (HS) instead of FBS. Data obtained with specific monoclonal antibody (mAb) indicated (—) versus isotype-matched control (–).
PPARγ and human serum–induced and repressed genes. (A) Three hundred forty-nine probe sets were up-regulated and 211 probe sets down-regulated (B) by PPARγ ligand (3 biologic replicates were used). Hierarchical cluster analysis was performed with GeneSpring software (standard correlation). Affymetrix microarray data were normalized to nontreated FBS-cultured cells. Monocytes were cultured for 6 or 24 hours or 5 days as described in “Cell culture and ligand treatment” and the indicated cells were cultured in human AB serum (HS) instead of FBS. Cells were treated with 1 μM rosiglitazone (RSG; 6 hours, 24 hours DC) or with 2.5 μM RSG (5 days DC). (C) Characterization of CD1a, CD1d, CD80, and CD86 cell-surface expression on DCs treated with ligands: 2.5 μM RSG alone or with 5 μM GW9662 (ANT). The indicated DCs were cultured in human AB serum (HS) instead of FBS. Data obtained with specific monoclonal antibody (mAb) indicated (—) versus isotype-matched control (–).
Impaired PPARγ response in DCs derived from patients harboring a mutated PPARG gene
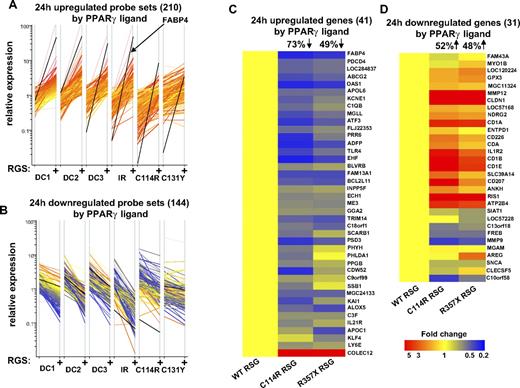
One of the main goals of this study was to identify PPARγ receptor–dependent gene-expression changes during DC development. One of our concerns was, however, that high concentration of PPARγ ligand might elicit a receptor-independent response as has been described in PPARγ knock-out macrophages.21 To assess the receptor dependency of gene-expression changes, we performed profiling experiments using control or ligand-treated DCs obtained from patients harboring various point mutations in the coding region of the PPARG gene. We recently reported that FABP4 induction is highly impaired in the cells having mutations in the DNA-binding domain or the ligand-binding domain of the PPARγ receptor (C114R, C131Y, R357X).13 Now we assessed the expression pattern of genes regulated upon ligand treatment (24 hours) in wild-type cells (210 probe sets up-regulated and 144 down-regulated by RSG). Our results indicated that PPARγ mutant cells (C114R, C131Y) exhibit a generally impaired PPARγ response (Figure 7A,B). As a control experiment we determined the PPARγ response using DCs obtained from a patient having insulin resistance but not carrying any PPARγ mutation. As expected, in this case we failed to detect any alternations. We also determined the expression of the previously validated genes with TLDAs (41 up- and 31 down-regulated validated genes by PPARγ ligand, 24 hours). The expression level was normalized to PPARγ-treated nonmutant cells and we determined the effects of the mutations (C131Y, R357X). We observed that approximately 50% to 70% of the genes showed impaired expression in the mutant cells (Figure 7C,D). Taken together, our results indicate that the observed PPARγ ligand–promoted transcription response was severely attenuated in primary cells with mutant PPARγ and, therefore, these effects appear to be receptor dependent.
Cells with PPARγ mutation have an impaired PPARγ response. (A-B) Microarray transcript profiles are shown for selected probe sets that were induced (A) or repressed (B) in normal DCs (24 hours cells) upon PPARγ ligand treatment. Data were normalized to the median expression. Data were obtained from normal (3 DC samples) and mutated cells (C114R, R357X). As a further control, microarray data were also obtained from a patient with insulin resistance (IR). Cells were cultured for 24 hours and treated with 1 μM rosiglitazone (RSG). (C,D) TLDA analyses of selected up- (C) or down-regulated (D) genes of normal and mutated cells (C114R, R357X). qRT-PCR data were normalized to PPARγ ligand–treated normal DCs (WT RSG) and dendograms were obtained from hierarchical clustering (standard correlation). On the top of the dendograms the percentage of genes significantly (changes were at least 1.5-fold and P < .05) up- or down-regulated is indicated compared with normal (WT RSG) DCs.
Cells with PPARγ mutation have an impaired PPARγ response. (A-B) Microarray transcript profiles are shown for selected probe sets that were induced (A) or repressed (B) in normal DCs (24 hours cells) upon PPARγ ligand treatment. Data were normalized to the median expression. Data were obtained from normal (3 DC samples) and mutated cells (C114R, R357X). As a further control, microarray data were also obtained from a patient with insulin resistance (IR). Cells were cultured for 24 hours and treated with 1 μM rosiglitazone (RSG). (C,D) TLDA analyses of selected up- (C) or down-regulated (D) genes of normal and mutated cells (C114R, R357X). qRT-PCR data were normalized to PPARγ ligand–treated normal DCs (WT RSG) and dendograms were obtained from hierarchical clustering (standard correlation). On the top of the dendograms the percentage of genes significantly (changes were at least 1.5-fold and P < .05) up- or down-regulated is indicated compared with normal (WT RSG) DCs.
Discussion
There are multiple reasons to be intrigued by the idea that a lipid-activated transcription factor such as PPARγ has a role in human immune cell regulation. One is that this receptor has been intimately linked to lipid metabolism and storage in fat cells and its role in human macrophages and dendritic cells is much less circumscribed and defined. Another is that studying this receptor provides an opportunity to find links between metabolism and immune response: 2 fields that are rarely linked at the molecular, transcriptional level. Finally the global effect of activation of PPARγ on the transcriptome in normal primary human cells has not been systematically and exhaustively studied and key questions such as how much activation and repression is mediated by the receptor remain to be answered. In order to address these issues, we have designed a robust (6 biologic replicates), global, and unbiased approach to assess the effect of exogenous synthetic ligand (rosiglitazone) activation and endogenous liganding (exposure to human serum) on gene-expression changes. The analyses were followed by 2 validation approaches: one is qRT-PCR and the other uses a human genetic model in order to establish receptor dependence. By carrying out this complex study, we could determine primary and secondary targets of PPARγ, assess the contribution of the receptor's activation to DC differentiation at the transcriptional level, and compare the effects of a high-affinity synthetic ligand to a yet unknown endogenous activator(s).
The global transcription effects of activating PPARγ
A very large amount of data are available about the global gene-expression profiles of DCs and the immature and activated DCs profiles were especially extensively compared.3-6 It is well established that PPARγ is indispensable for adipocyte formation and this transcription factor is up-regulated during adipocyte differentiation.20 Numerous reports characterized the global gene-expression profile of fat cell development using the murine 3T3 L1 preadipocyte model.22-24 In contrast to identifying PPARγ-regulated genes in myeloid cells, only macrophages were analyzed so far,21,25-27 and in that system overwhelmingly repressive events have been documented. Most of the available microarray data sets are using mouse cells or mouse tissues and merely a few reports investigated human cells.28,29 For this study we obtained expression data from primary human cells derived from healthy individuals. This approach provides us with a unique data set but also contributes to larger variability. In order to increase the robustness and reliability of the data, we used 6 biologic replicates. In addition we employed a very stringent filtering approach as described in “Microarray analysis.” We found that using this stringent filtering approach, 1166 probe sets were modulated by PPARγ ligand treatment. We could validate around 80% of the selected genes with qRT-PCR irrespective of the usage of any additional statistical tool such as multiple test corrections (Figure 4 and data not shown). Our results imply that numerous genes are likely to be directly regulated by PPARγ receptor, especially those induced after 6 hours of treatment. This is in contrast to previous reports on the gene-expression profile of mouse peritoneal macrophages activated by PPARγ activators and it is more likely to reflect differences in the cell types and cell states used rather than species-specific differences.27 This study also establishes that PPARγ acts as a predominantly positive transcription factor rather than a repressor in an immune cell type because more than half of the regulated genes are positively modulated. It is particularly intriguing that transcriptional repression brought about by the receptor's activation is not immediate but rather occurs over time, requiring at least 24 hours to develop (Figure 2B).
Genes regulated at early time points are associated with lipid metabolism
We observed that more than 200 probe sets were induced after 6 hours of PPARγ ligand treatment. Interestingly in this gene list the category of lipid/fatty acid metabolism was markedly overrepresented, consistent with the presumed role of this receptor in metabolism.11 Our understanding of the role of lipid metabolism in DCs remains limited. However our experimental analysis of global lipid changes indicated that PPARγ ligand–treated DCs accumulated fewer lipid droplets/body, suggesting that these cells have an enhanced capacity to metabolize or at least redistribute certain lipids. It is difficult to define which pathways of lipid metabolism are responsible for this regulation because several identified genes participate in antagonistic processes, such as fatty acid oxidation (ACOX1, HADHSC) or phospholipid and triacylglicerol synthesis (DGAT2, PPAP2B), and their relative contribution cannot be easily assessed. It should be noted that after 6 hours of cell differentiation a few genes related to lipid/fatty acid uptake and transport (CD36, FABP4) were also induced. These transporters might contribute to the distribution of intracellular lipids. Interestingly, LXRα (NR1H3), the previously described PPARγ target,30 was also up-regulated in PPARγ-activated DCs. LXRα is a key regulator of lipid and cholesterol metabolism in macrophages.31 Interestingly, administration of PPARγ ligand failed to modulate the expression of ABCA1 in human DCs, suggesting that PPARγ-dependent LXR response is very limited in this monocyte-derived DC model.16
PPARγ links lipid metabolism and immune function
The data presented here also document that many more genes were up- or down-regulated via PPARγ activation after 24 hours or 5 days than after 6 hours. Interestingly, a substantial fraction of these genes were related to immune response. Multiple pathways are probably responsible for this regulation. For example, we have recently described that PPARγ-activated DCs had an enhanced capacity to produce retinoic acid by inducing retinol- and retinal-metabolizing enzymes and approximately 30% of the PPARγ ligand–regulated genes (at 5 days) were RARα dependent.14 Similar pathways and mechanisms might exist where changes in metabolism and signaling could contribute direct regulation of immune function–associated genes.
The relationship between DC differentiation and PPARγ activation
It is a key issue how activation of PPARγ interferes with DC differentiation at the transcriptional level. Interestingly most of the identified genes, which were negatively regulated upon ligand treatment, were induced during DC development, suggesting that the receptor's activation inhibits some aspects of DC differentiation (group I CD1s, IL1R1, IL1R2, IRF4, CD80, DCNP1). Several models were proposed for this negative transcriptional, transrepression/anti-inflammatory effect of PPARγ.32,33 Additional mechanisms such as induction of signal transduction pathways and transcription factors including repressors should be also considered. On the flip side, PPARγ up-regulation of genes otherwise down-regulated include PDK4, CD1D, TLR4, CD36, the leukotriene B4 receptor, and transcription factor ATF3. These genes are likely to contribute to the development of the receptor-specific DC phenotype. Previous results suggested that the cell culture conditions have a dramatic effect on the phenotype and functional properties of DCs. For example, human serum–cultured monocyte-derived DCs express less CD1a and CD8034 ; in addition these cells express elevated levels of CD1d and FABP4, suggesting that human serum–cultured DCs have an enhanced PPARγ activation capacity.14 Here we confirmed and extended these studied by the comparison of the global gene-expression profile of FBS- and human serum–cultured cells. Our results indicated that most of the synthetic PPARγ ligand–induced or repressed genes showed a similar regulation if the cells were cultured in human serum without administration of any synthetic PPARγ agonist. We previously described that in human tonsils a significant number of PPARγ-positive DCs were detected,9 thus it is possible that in vivo at least a subset of myeloid DCs have an activated PPARγ transcription tone.
Impaired PPARγ response in DCs from patients harboring a mutated PPARγ
As part of our validation strategy we also obtained microarray and qRT-PCR data from patients harboring a mutated form of the PPARγ receptor. Fifty percent to 70% of the PPARγ-regulated genes exhibited an impaired regulation upon ligand treatment, indicating that the observed effects of PPARγ activator are receptor dependent. Our microarray and qRT-PCR data demonstrated that several established PPARγ targets (ie, FABP4, ABCG2) showed a consistently impaired expression in this cohort of patients. These mutations cause lipodystrophic insulin resistance.13 Determination of PPARγ responsiveness from diabetic or prediabetic patients might help to identify and further characterize this syndrome. Importantly, such an approach may also identify those individuals likely to be resistant to certain PPARγ activators and sensitive to others.
In summary, in this report we characterized the gene-expression pattern of PPARγ-activated DCs. Our results suggest that activation of this receptor has a limited but characteristic effect on DCs and probably contributes to the response to changing lipid environment and subtype specification.35 The receptor acts as a predominantly positive transcriptional regulator and controls primarily lipid metabolism–associated genes and immune function via indirect mechanisms; therefore, it provides a link between these 2 processes. In addition, several novel PPARγ-regulated genes were identified that are directly or indirectly modulated by this nuclear hormone receptor and can provide starting points for further investigations. The molecular details of the links between lipid metabolism and immune function remain to be determined, but clearly some of the players have been identified.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors are grateful for the excellent technical assistance of Ms Ibolya Furtos and for the bioinformatics support from the Hungarian Bioinformatics Institute. The authors are indebted to members of the Nagy laboratory and Dr Eva Rajnavolgyi for discussions and comments on the manuscript.
The work is supported by grants from the National Research and Technology Office (RET-06/2004, NKFP-1A/008/2004; L.N.). L.N. is an International Scholar of the Howard Hughes Medical Institute and holds a Wellcome Trust Senior Research Fellowship in Biomedical Sciences in Central Europe (no. 074021). M.A., M.G., and K.C. are supported by Wellcome Trust and National Institute for Health Research (NIHR) Biomedical Research Center funding.
National Institutes of Health
Wellcome Trust
Authorship
Contribution: I.S. performed research and wrote the paper; D.T. and M.A. performed research; T.N. and E.B. analyzed data; M.G. and K.C. designed research; and L.N. designed and directed research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Laszlo Nagy, Department of Biochemistry and Molecular Biology, University of Debrecen, Medical and Health Science Center, Nagyerdei krt. 98, Debrecen, Hungary, H-4012; e-mail: lnagy@indi.biochem.dote.hu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal