Abstract
The proteasome has emerged as an important target for cancer therapy with the approval of bortezomib, a first-in-class, reversible proteasome inhibitor, for relapsed/refractory multiple myeloma (MM). However, many patients have disease that does not respond to bortezomib, whereas others develop resistance, suggesting the need for other inhibitors with enhanced activity. We therefore evaluated a novel, irreversible, epoxomicin-related proteasome inhibitor, carfilzomib. In models of MM, this agent potently bound and specifically inhibited the chymotrypsin-like proteasome and immunoproteasome activities, resulting in accumulation of ubiquitinated substrates. Carfilzomib induced a dose- and time-dependent inhibition of proliferation, ultimately leading to apoptosis. Programmed cell death was associated with activation of c-Jun-N-terminal kinase, mitochondrial membrane depolarization, release of cytochrome c, and activation of both intrinsic and extrinsic caspase pathways. This agent also inhibited proliferation and activated apoptosis in patient-derived MM cells and neoplastic cells from patients with other hematologic malignancies. Importantly, carfilzomib showed increased efficacy compared with bortezomib and was active against bortezomib-resistant MM cell lines and samples from patients with clinical bortezomib resistance. Carfilzomib also overcame resistance to other conventional agents and acted synergistically with dexamethasone to enhance cell death. Taken together, these data provide a rationale for the clinical evaluation of carfilzomib in MM.
Introduction
The 26S proteasome plays a critical role in cellular homeostasis through its function in ubiquitin-dependent protein turnover, including targets involved in cell-cycle progression, apoptosis, DNA repair, stress response, and misfolded and obsolete proteins.1 Major catalytic activities of the 20S core of the proteasome include a chymotrypsin-like (ChT-L) activity found in the β5 subunit, a trypsin-like (T-L) activity in subunit β2, and a postglutamyl peptide hydrolyzing (PGPH) or caspase-like activity in the β1 subunit.2 Of these, the ChT-L activity has been shown to be the rate-limiting step of proteolysis in vitro and in vivo.3 Two major isoforms of the proteasome have been described, including the constitutive proteasome, which is present in most cells, and the immunoproteasome, which in place of the above subunits incorporates 3 related proteins, β1i, β2i, and β5i. The immunoproteasome is predominantly expressed in cells of lymphoid origin and plays a role in major histocompatibility complex class I antigen presentation and other constitutive proteolytic activities.3-5
Bortezomib (VELCADE)6 is the first proteasome inhibitor to enter clinical practice for the treatment of relapsed/refractory multiple myeloma (MM), based in part on pioneering preclinical studies.7 Phase I-III trials showed bortezomib had impressive antimyeloma activity,8-10 and additional studies are ongoing to define its role in that disease and other hematologic malignancies such as non-Hodgkin lymphoma (NHL).11,12 However, the overall response rate from the phase III experience was 43%,10 and bortezomib resistance is emerging as well, underscoring the need for a next generation of proteasome inhibitors with greater efficacy. One such inhibitor is NPI-0052, or salinosporamide A, a compound related to lactacystin that showed antitumor activity predominantly through caspase-8 activation.13 Because lactacystin binds to several proteasome subunits, however, and may inhibit other cellular proteases,14 this class of agents may be less specific. In contrast, epoxomicin is a natural product isolated from an Actinomycetes15 that forms an irreversible, selective, and highly specific morpholino adduct only with the N-terminal threonine of the β5 subunit.16 Because the activity of this class of inhibitors against MM has not been well studied, it was of interest to evaluate the effects of carfilzomib (formerly PR-171; Proteolix), an epoxyketone related to epoxomicin (Figure 1A).
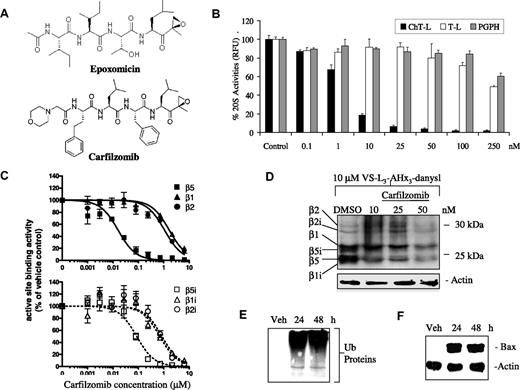
Inhibition of the proteasome by carfilzomib. (A) Structures of epoxomicin (top) and carfilzomib (bottom) are shown. (B) Quantitative representation of the in vitro inhibition of the 20S proteasome catalytic activities in ANBL-6 cellular lysates (10 μg per reaction) in the absence or presence of carfilzomib with fluorogenic peptide substrates for the proteasomal ChT-L, PGPH, and T-L activity, as indicated, followed by the measurement of free AMC groups is shown. (C) In cellulo measurement using ELISA techniques of the 20S proteasome subunit targets of carfilzomib in cell extracts from ANBL-6 cells pulse treated for 1 hour with carfilzomib is shown. Error bars in panels B and C are SD. (D) Competitive binding experiment in ANBL-6 cells between carfilzomib (5-hour pretreatment) followed by VS-L3-AHx3-danysl (2 hours) determined the in cellulo specificity of carfilzomib to individual proteasome catalytic subunits. (E,F) Shown is Western blot analysis of the accumulation of ubiquitinated (Ub) substrates (E) and proapoptotic Bax (F) after 1-hour pulse exposure to carfilzomib (100 nM) in RPMI 8226 cells, followed by 24- and 48-hour recovery times. Actin was used as a loading control. Veh indicates vehicle.
Inhibition of the proteasome by carfilzomib. (A) Structures of epoxomicin (top) and carfilzomib (bottom) are shown. (B) Quantitative representation of the in vitro inhibition of the 20S proteasome catalytic activities in ANBL-6 cellular lysates (10 μg per reaction) in the absence or presence of carfilzomib with fluorogenic peptide substrates for the proteasomal ChT-L, PGPH, and T-L activity, as indicated, followed by the measurement of free AMC groups is shown. (C) In cellulo measurement using ELISA techniques of the 20S proteasome subunit targets of carfilzomib in cell extracts from ANBL-6 cells pulse treated for 1 hour with carfilzomib is shown. Error bars in panels B and C are SD. (D) Competitive binding experiment in ANBL-6 cells between carfilzomib (5-hour pretreatment) followed by VS-L3-AHx3-danysl (2 hours) determined the in cellulo specificity of carfilzomib to individual proteasome catalytic subunits. (E,F) Shown is Western blot analysis of the accumulation of ubiquitinated (Ub) substrates (E) and proapoptotic Bax (F) after 1-hour pulse exposure to carfilzomib (100 nM) in RPMI 8226 cells, followed by 24- and 48-hour recovery times. Actin was used as a loading control. Veh indicates vehicle.
In the current work, we demonstrate that carfilzomib inhibited the ChT-L activity of the proteasome both in vitro and in cellulo in models of MM. Experiments modeling the anticipated in vivo pharmacokinetics of drug exposure showed that carfilzomib inhibited proliferation in a variety of cell lines and patient-derived neoplastic cells, including MM, and induced intrinsic and extrinsic apoptotic signaling pathways and activation of c-Jun-N-terminal kinase (JNK). Furthermore, carfilzomib showed enhanced anti-MM activity compared with bortezomib, overcame resistance to bortezomib and other agents, and acted synergistically with dexamethasone (Dex). Taken together, these data indicate that carfilzomib is a promising proteasome inhibitor with activity against MM, providing a rational basis for its translation into the clinic.
Materials and methods
The proteasome inhibitor bortezomib was obtained from the University of North Carolina at Chapel Hill pharmacy, and carfilzomib was provided by Proteolix, Inc. Stock solutions were prepared in dimethyl sulfoxide (DMSO) and used as indicated, with a final vehicle concentration that did not exceed 0.5% (vol/vol). All chemicals, unless otherwise indicated, were from Fisher Scientific (Pittsburgh, PA).
Cell models, cell culture, and experimental conditions
RPMI 8226, U266, ANBL-6, KAS-6/1, H-929, 8226.Dox40, 8226.LR5, MM1.S, MM1.R, bortezomib-resistant (BR) cells, and patient-derived cells were cultured in RPMI 1640 (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (Sigma; St Louis, MO), 100 U/mL penicillin, and 100 μg/mL streptomycin (Tissue Culture Facility, University of North Carolina), and maintained in 5% CO2 at 37°C. Interleukin (IL)-6-dependent cells and purified plasma cells from patients were supplemented with an additional 1 ng/mL of IL-6 (R&D Systems, Minneapolis, MN).
Patient samples were collected under a protocol approved by the Institutional Review Board of the University of North Carolina. Informed consent was obtained in accordance with the Declaration of Helsinki. Mononuclear cells from bone marrow aspirates or peripheral blood samples were isolated by density gradient centrifugation over Ficoll-Paque Plus (Amersham Biosciences, Piscataway, NJ). Malignant cells were then isolated by immunomagnetic bead positive selection in a Midi MACS LS column following the manufacturer's protocol (Miltenyi Biotec, Auburn, CA). The purity of MM cells was confirmed by flow cytometric analysis using phycoerythrin-conjugated anti-CD138 antibody (Miltenyi Biotec).
Cells in culture were exposed to proteasome inhibitors in either a continuous fashion or a discontinuous one designed to mimic the known pharmacokinetics of bortezomib in vivo, which is rapidly cleared from the circulation with a t1/2α of 0.22 to 0.46 hour.17 In the latter, proteasome inhibitors were added for 1 hour, after which cultures were washed once with phosphate-buffered saline (PBS) and resuspended in drug-free media.
Measurement of 20S proteasome activity
In vitro measurements of catalytic 20S proteasome activities were performed as described.18
Enzyme-linked immunosorbent assay for subunit profiling of carfilzomib
ANBL-6 cells (2 × 106/well) were plated in 96-well plates and treated with carfilzomib doses from 0.001 to 10 μM for 1 hour. Cells were then lysed (20 mM Tris-HCl, 0.5 mM EDTA), and cleared lysates were transferred to polymerase chain reaction (PCR) plates. A standard curve was generated using untreated ANBL-6 cell lysates starting at a concentration of 6 μg protein/μL. The active site probe [biotin-(CH2)4-Leu-Leu-Leu-epoxyketone; 20 μM] was added and incubated at room temperature for 1 hour. Cell lysates were then denatured by adding 1% sodium dodecyl sulfate (SDS) and heating to 100°C, followed by mixing with 20 μL per well streptavidin-sepharose high-performance beads (GE Healthcare, Chalfont St. Giles, United Kingdom) in a 96-well multiscreen DV plate (Millipore, Billerica, MA) and incubated for 1 hour. These beads were then washed with enzyme-linked immunosorbent assay (ELISA) buffer (PBS, 1% bovine serum albumin, and 0.1% Tween-20), and incubated overnight at 4°C on a plate shaker with antibodies to proteasome subunits. Antibodies used included mouse monoclonal anti-β1, anti-β2, anti-β1i, and anti-β5i (Biomol, Plymouth Meeting, PA), goat polyclonal anti-β2i (Santa Cruz Biotechnology, Santa Cruz, CA), and rabbit polyclonal anti-β5 (affinity-purified antiserum against KLH-CWIRVSSDNVADLHDKYS peptide). The beads were washed and incubated for 2 hours with horseradish peroxidase-conjugated secondary goat antirabbit, goat antimouse (BioSource, Camarillo, CA), or rabbit antigoat (Invitrogen) antibodies. After washing, the beads were developed using the supersignal ELISA picochemiluminescence substrate (Pierce, Rockford, IL). Luminescent detection was performed on the Tecan Genius Pro (Mannedorf, Zurich, Switzerland). Raw luminescence was converted to μg/mL by comparison with the standard curve and expressed as the % inhibition relative to vehicle control. Curve fits were generated using the following nonsigmoidal dose-response equation: Y = Bottom + (Top-Bottom)/(1 + 10̂((LogEC50 − X) × HillSlope)), where X is the logarithm of concentration, Y is the % inhibition, and EC50 is the dose showing 50% effect.
Competitive binding for subunit profiling of carfilzomib
The protocol used to determine carfilzomib subunit specificity via competitive binding was adapted from Berkers et al.19 Briefly, ANBL-6 cells were preincubated with increasing carfilzomib doses at 37°C, followed by addition of the hapten-labeled cell-permeant vinyl sulfone (VS) proteasome inhibitor VS-L3-AHx3-danysl. Western blots were then prepared as detailed in the next section and probed with polyclonal antidansyl antibodies (Invitrogen).
Cell extract preparation and Western blotting
Whole-cell extracts (WCEs) were prepared and separated by denaturing gel electrophoresis as described.20 The antibodies used for immunoblotting included anti–Actin, anti-JNK, anti-ubiquitin, and rat secondary antibodies (Santa Cruz Biotechnology), anti-Bax, anti-p-JNK (Thr138/Tyr185), and anti–poly(adenosine diphosphate [ADP]-ribose) polymerase (PARP) (EMD Biosciences, San Diego, CA), anti-HSC-70 and anti–second mitochondrial activator of caspases (Smac) antibodies (Stressgen, San Diego, CA), anti–mouse secondary (Amersham Biosciences), anti-rabbit secondary (Bio-Rad, Hercules, CA), anti–cytochrome c (BioVision, Mountain View, CA), and anti-Cox II (Invitrogen).
Cell proliferation assay
WST-1 (Roche Diagnostics, Indianapolis, IN) was used to determine the effects of proteasome inhibitors on cell proliferation according to the manufacturer's protocol. The inhibition of proliferation was calculated in relation to parallel control cells that received vehicle alone and tabulated in KaleidaGraph 3.0.1 (Synergy Software, Reading, PA) or Excel 2000 (Microsoft, Redmond, WA). A linear spline function was used to interpolate the median inhibitory concentration (IC50) using XLfit 4 software (ID Business Solutions, Guildford, United Kingdom). The degree of resistance (DOR) was calculated using the formula: DOR = IC50(resistant cells)/IC50(sensitive cells).
Apoptotic DNA fragmentation assay
For apoptosis experiments, cells were seeded onto 96-well plates, treated with a 1-hour pulse of 300 nM (RPMI 8226, ANBL-6) or 100 nM carfilzomib (KAS-6/1, U266), and allowed to recover for 24 hours before analysis with the Cell Death Detection ELISAPLUS kit (Roche Diagnostics) according to the manufacturer's specifications. The fold increase in DNA fragmentation is presented as the mean relative to vehicle-treated control cells.
Mitochondrial membrane potential (ΔΨm)
ANBL-6 cells pulsed with 100 nM carfilzomib were washed and suspended in PBS containing 5 μg/mL of JC-1 (Invitrogen), which exhibits potential-dependent accumulation in mitochondria. Analysis of the mitochondrial membrane potential-dependent color shift from 525 to 590 nm was carried out on a FacScan (Becton Dickinson, Franklin Lakes, NJ), and the data were analyzed with CellQuest software (Becton Dickinson).
Caspase activation assay
Cellular lysates were incubated in Tris-HCl with 40-μM fluorogenic substrates (Biomol) specific for caspase-3, caspase-8, or caspase-9. Measurement of free 7-amido-4-methylcoumarin (AMC) groups was performed at 380/460 nm on a multilabel FLUOstar Optima (BMG Labtech, Durham, NC). Results were expressed as fold activation over the vehicle control from experiments performed in triplicate.
Infection with recombinant adenovirus
Recombinant adenoviruses carrying the dominant-negative (DN) c-Jun (TAM67 mutant) were from Vector Biolabs (Philadelphia, PA). ANBL-6 cells were incubated in medium containing the adenovirus carrying DN-c-Jun or an empty cytomegalovirus (CMV) promoter at a multiplicity of infection of 1000 for 24 hours at 37°C.
Apoptosis detection with Annexin V and TO-PRO-3
Staining of cells by Annexin V was performed according to the manufacturer's specifications (BioVision), and TO-PRO-3 was used to identify necrotic or late-stage apoptosis. Data were collected using CellQuest software on a FacsCalibur (Becton Dickinson), and 10,000 events were analyzed with FlowJo version 6.3.3 (Tree Star, Ashland, OR). Results are expressed as percentage specific apoptosis and were calculated using the following formula: [(% apoptotic cells in experimental − % apoptotic cells in control)/(100 − % apoptotic cells in control) × 100].
Results
Impact of carfilzomib on the constitutive and immunoproteasome
Carfilzomib is a tetrapeptide epoxyketone related to epoxomicin (Figure 1A), the latter of which shows high specificity in vitro for the ChT-L proteasome activity. To evaluate the proteasomal inhibitory potential of carfilzomib in MM, extracts from ANBL-6 cells were exposed to increasing concentrations of carfilzomib (Figure 1B) and assayed for 20S catalytic activities. Carfilzomib displayed preferential in vitro inhibitory potency against the ChT-L activity in the β5 subunit, with over 80% inhibition at doses of 10 nM and above (Figure 1B) and little or no effect on the PGPH and T-L activities at doses up to 100 nM. It has been reported that binding of proteasome inhibitors can differ in vitro and in live cells (in cellulo),19 so it was of interest to determine whether carfilzomib interacted with other catalytic subunits in cellulo using an ELISA designed to identify subunit binding. This assay demonstrated that short exposure to low-dose carfilzomib led to preferential binding specificity for the β5 constitutive 20S proteasome and the β5i immunoproteasome subunits (Figure 1C). However, at very high concentrations over 1 μM, carfilzomib did interact with the β1 and β2 subunits as well as β1i and β2i. To confirm the subunit specificity of carfilzomib in another assay, a competitive binding technique was performed with a dansyl-linked peptide VS inhibitor (VS-L3-AHx3-dansyl), which binds to free catalytic subunits and can be detected by Western blotting.19 Extended exposure to carfilzomib for 5 hours saturated the β5 and β5i active sites in a dose-dependent manner and also led to increased binding to the β1, β1i, β2, and β2i subunits, with maximal binding observed at 50 nM (Figure 1D). To mimic the in vivo pharmacokinetics of proteasome inhibitor treatment, RPMI 8226 cells were pulsed with carfilzomib for 1 hour, and then allowed to recover in drug-free media. Accumulation of ubiquitin-conjugated proteins was observed up to 48 hours after recovery from the pulse (Figure 1E). In addition, the accumulation of proapoptotic Bax, a proteasome target,21 was also observed at 24 and 48 hours after treatment, signifying a potent and durable proteasome inhibition by carfilzomib (Figure 1F).
Antiproliferative and proapoptotic impact of carfilzomib
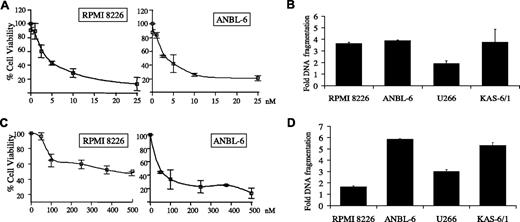
To determine whether carfilzomib-mediated proteasome inhibition impacted proliferation, its effect in IL-6–independent (RPMI 8226, U266) and IL-6–dependent (ANBL-6, KAS-6/1) myeloma cell lines was tested. A dose-dependent decrease in cell viability was found in ANBL-6 and RPMI 8226 cells (Figure 2A), and U266 and KAS-6/1 cells (data not shown), treated continuously with carfilzomib for 24 hours, with an IC50 less than 5 nM in both after 24 hours. This effect was further enhanced at 48 and 72 hours (data not shown). To determine whether the observed viability changes were caused by induction of apoptosis, MM cell lines were treated continuously with carfilzomib at 5 nM (U266, KAS-6/1) or 10 nM (RPMI 8226, ANBL-6). An apoptosis-specific DNA fragmentation ELISA revealed that low-dose carfilzomib induced apoptosis in all cell lines to varying degrees (Figure 2B).
Inhibition of proliferation and induction of apoptosis by carfilzomib. (A,C) IL-6–independent RPMI 8226 and IL-6–dependent ANBL-6 MM cells (2 × 104) were treated continuously (A) or pulsed for 1 hour (C) with increasing concentrations of carfilzomib. Cellular viability was determined at 24 hours using the water-soluble tetrazolium salt WST-1. (B,D) IL-6–dependent (ANBL-6, KAS-6/1) and IL-6–independent (RPMI 8226, U266) myeloma cells were continuously exposed (B) or pulsed (D) with carfilzomib for 1 hour and allowed to recover for 24 hours. Programmed cell death was then evaluated using a DNA fragmentation ELISA. Results are expressed as a fold-increase of DNA fragmentation over DMSO control, and error bars are SD.
Inhibition of proliferation and induction of apoptosis by carfilzomib. (A,C) IL-6–independent RPMI 8226 and IL-6–dependent ANBL-6 MM cells (2 × 104) were treated continuously (A) or pulsed for 1 hour (C) with increasing concentrations of carfilzomib. Cellular viability was determined at 24 hours using the water-soluble tetrazolium salt WST-1. (B,D) IL-6–dependent (ANBL-6, KAS-6/1) and IL-6–independent (RPMI 8226, U266) myeloma cells were continuously exposed (B) or pulsed (D) with carfilzomib for 1 hour and allowed to recover for 24 hours. Programmed cell death was then evaluated using a DNA fragmentation ELISA. Results are expressed as a fold-increase of DNA fragmentation over DMSO control, and error bars are SD.
Pulse treatment for 1 hour with carfilzomib followed by exposure to drug-free medium also induced a dose-dependent inhibition of proliferation, though higher drug concentrations were necessary (Figure 2C). This activity was associated with DNA fragmentation (Figure 2D), and enhanced levels of programmed cell death were also detected by staining with Annexin V, which showed a 2.5-fold increase after a 1-hour pulse with 100 nM carfilzomib in ANBL-6 cells (data not shown).
Mechanisms of carfilzomib-induced apoptosis
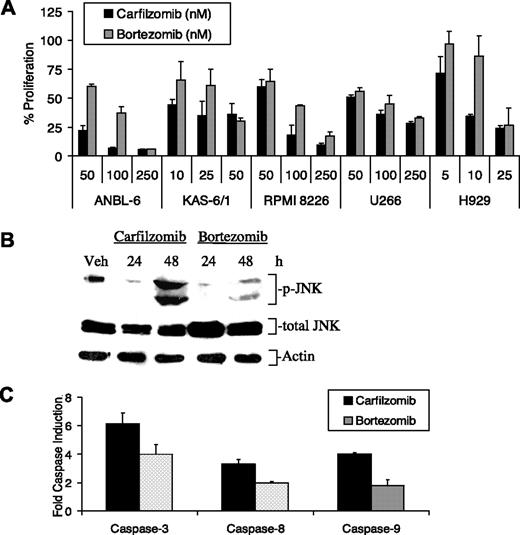
Initiation of apoptosis can occur through an intrinsic pathway involving cytochrome c release and caspase-9 activation or an extrinsic pathway mediated by activation of Fas/caspase-8-dependent signaling, which then converge on a common effector, caspase-3.22 Bortezomib-induced apoptosis activates both pathways,23 and we therefore examined whether carfilzomib had similar proapoptotic effects. Measurement of caspase activity in ANBL-6 cells pulsed with carfilzomib revealed substantial increases in caspase-8, caspase-9, and caspase-3 activity after 8 hours, giving a 3.2-, 3.9- and 6.9-fold increase, respectively, over control cells after 8 hours (Figure 3A). Caspase-9 and caspase-3 activities continued to increase up to 24 hours later, though no discernible increase in caspase-8 activity was found beyond 8 hours. To confirm that caspase activation was necessary for carfilzomib-induced cell death, ANBL-6 cells were treated with caspase inhibitors, followed by a brief pulse of carfilzomib (Figure 3B). Inhibition of caspase-3, caspase-8, and caspase-9 rescued 95%, 76%, and 94% of ANBL-6 cells, respectively (Figure 3B) from carfilzomib-mediated growth inhibition. These data indicated that carfilzomib-induced apoptosis occurred through both intrinsic and extrinsic caspase pathways.
Molecular events associated with exposure to carfilzomib. (A) ANBL-6 cells were exposed to a pulse of 100 nM carfilzomib, bortezomib, or vehicle (Veh) control and allowed to recover for 8 or 24 hours. Cellular lysates (30 μg per reaction) were incubated with 40-μM fluorogenic substrates specific for caspase-3, caspase-8, and caspase-9. Results are expressed as fold relative fluorescence units over DMSO control. (B) ANBL-6 (2 × 104 cells per reaction) were pretreated for 20 hours with caspase-3 (C-3)-, caspase-8 (C-8)-, and caspase-9 (C-9)-specific inhibitors, a negative control (Neg), a pan-caspase inhibitor (Pan), or no inhibitor (−), followed by a 1-hour pulse with 100 nM carfilzomib. Fresh media containing caspase inhibitors were then added, and cellular proliferation was determined after a 24-hour recovery period. Data were expressed as percent inhibition compared with vehicle (DMSO) controls. (C) Carfilzomib induces depolarization of mitochondria. The fluorescent shift of the JC-1 cationic dye from cytosol (green fluorescence) to mitochondria (red fluorescence) in live cells was analyzed by flow cytometry in ANBL-6 cells pulsed with 100 nM carfilzomib (Q1: red fluorescence; Q2: red and green fluorescence; Q3: green fluorescence). (D) ANBL-6 cells treated for 1 hour with 100 nM carfilzomib were subjected to centrifugal cellular fractionation into the cytosolic fraction and HMF that included mitochondria. Release of cytochrome c and Smac from the mitochondria was assessed by Western blot. Cox II, an intramitochondrial protein, was used as a control. WCEs were monitored as a control for protein expression. (E) Western blot analysis for the phosphorylation of JNK (ie, activated JNK) and cleavage of PARP in RPMI 8226 cells, which were pulsed with 100 nM carfilzomib and allowed to recover for the indicated time periods. HSC-70 was used as a loading control. To determine whether abrogation of JNK signaling through c-Jun affects carfilzomib's antiproliferative and proapoptotic action, ANBL-6 cells (2 × 104 per well) were infected with DN-c-Jun adenovirus for 24 hours, followed by addition of 100 nM pulse with carfilzomib. (F) Cellular growth was assessed using the WST-1 reagent. (G) Apoptosis was measured by DNA fragment production. Error bars for panels A, B, F, and G are SD.
Molecular events associated with exposure to carfilzomib. (A) ANBL-6 cells were exposed to a pulse of 100 nM carfilzomib, bortezomib, or vehicle (Veh) control and allowed to recover for 8 or 24 hours. Cellular lysates (30 μg per reaction) were incubated with 40-μM fluorogenic substrates specific for caspase-3, caspase-8, and caspase-9. Results are expressed as fold relative fluorescence units over DMSO control. (B) ANBL-6 (2 × 104 cells per reaction) were pretreated for 20 hours with caspase-3 (C-3)-, caspase-8 (C-8)-, and caspase-9 (C-9)-specific inhibitors, a negative control (Neg), a pan-caspase inhibitor (Pan), or no inhibitor (−), followed by a 1-hour pulse with 100 nM carfilzomib. Fresh media containing caspase inhibitors were then added, and cellular proliferation was determined after a 24-hour recovery period. Data were expressed as percent inhibition compared with vehicle (DMSO) controls. (C) Carfilzomib induces depolarization of mitochondria. The fluorescent shift of the JC-1 cationic dye from cytosol (green fluorescence) to mitochondria (red fluorescence) in live cells was analyzed by flow cytometry in ANBL-6 cells pulsed with 100 nM carfilzomib (Q1: red fluorescence; Q2: red and green fluorescence; Q3: green fluorescence). (D) ANBL-6 cells treated for 1 hour with 100 nM carfilzomib were subjected to centrifugal cellular fractionation into the cytosolic fraction and HMF that included mitochondria. Release of cytochrome c and Smac from the mitochondria was assessed by Western blot. Cox II, an intramitochondrial protein, was used as a control. WCEs were monitored as a control for protein expression. (E) Western blot analysis for the phosphorylation of JNK (ie, activated JNK) and cleavage of PARP in RPMI 8226 cells, which were pulsed with 100 nM carfilzomib and allowed to recover for the indicated time periods. HSC-70 was used as a loading control. To determine whether abrogation of JNK signaling through c-Jun affects carfilzomib's antiproliferative and proapoptotic action, ANBL-6 cells (2 × 104 per well) were infected with DN-c-Jun adenovirus for 24 hours, followed by addition of 100 nM pulse with carfilzomib. (F) Cellular growth was assessed using the WST-1 reagent. (G) Apoptosis was measured by DNA fragment production. Error bars for panels A, B, F, and G are SD.
A major characteristic of intrinsic caspase-stimulated apoptosis is the loss of mitochondrial transmembrane potential (ΔΨm) and release of proapoptotic proteins, including cytochrome c and Smac/DIABLO.24,25 To examine whether carfilzomib-mediated apoptosis led to mitochondrial membrane depolarization, ANBL-6 cells were pulsed with 100 nM carfilzomib and stained with the cationic dye, JC-1, and the polarity of the mitochondrial membrane was assessed by flow cytometry. JC-1 undergoes a shift from green to red fluorescence in nonapoptotic cells. In carfilzomib pulse-treated cells, the mitochondrial membrane integrity was decreased to 41% (Q1 + Q2), compared with 75% in vehicle-treated control cells (Figure 3C). Similar results were obtained in RPMI 8226 cells analyzed by fluorescent microscopy for ΔΨm (data not shown). Formation of the apoptosome, a large multisubunit molecule, requires the release of cytochrome c from mitochondria and the inclusion of both cytochrome c and caspase-9 to signal to the effector caspase-3.26 Cellular fractionation for the cytosolic component and heavy membrane fraction (HMF), including intact and disrupted mitochondria, revealed that a brief pulse with carfilzomib led to cytosolic accumulation of cytochrome c and Smac and a concomitant decrease of these proteins in the HMF (Figure 3D).
Proapoptotic signaling through JNK and its downstream targets, including c-Jun, plays an important role in cell death due to other proteasome inhibitors.27-29 To determine whether JNK activation is important in mediating carfilzomib-induced apoptosis, RPMI 8226 cells were exposed to a pulse of carfilzomib, and JNK activation status was determined. Indeed, brief pulses with carfilzomib led to a significant increase in activated JNK, lasting up to 48 hours (Figure 3E). In addition, cleavage of PARP, a late-stage apoptotic event, correlated well with JNK activity levels. To study whether c-Jun phosphorylation by activated JNK is important in the apoptotic signaling cascade induced by carfilzomib, ANBL-6 cells were infected with a DN-c-Jun adenovirus, after which the antiproliferative function of carfilzomib was assessed. Disruption of c-Jun activation led to nearly complete inhibition of the antiproliferative activity of carfilzomib (Figure 3F). Similarly, DN-c-Jun suppressed carfilzomib-induced apoptosis in pulse-treated ANBL-6 cells, as measured by a 2-fold decrease in caspase-3 activity (Figure 3G).
Effects of carfilzomib and bortezomib in MM cell lines
Bortezomib is approved for the treatment of relapsed/refractory MM patients, where it has been shown to improve survival, even in patients with poor-risk disease, such as those with chromosome 13 deletion.10,30 The efficacy of pulse carfilzomib and bortezomib treatment was therefore compared in cell line models of MM (Figure 4). First, we evaluated the viability of both IL-6–dependent and –independent cell lines pulse-treated with increasing doses of proteasome inhibitor and allowed to recover for 24 hours. Statistically significant (P < .05) decreases in the viable cell population treated with carfilzomib were observed in ANBL-6, KAS-6/1, H929, and RPMI 8226 cells compared with their bortezomib-treated counterparts, indicating an overall trend toward increased sensitivity in myeloma cell lines to carfilzomib (Figure 4A). Furthermore, JNK phosphorylation was enhanced with carfilzomib treatment compared with bortezomib (Figure 4B). Comparisons of caspase activity levels also indicated that carfilzomib was more potent than bortezomib in increasing caspase-3, caspase-8, and caspase-9 activity and did so by 1.5-, 1.8-, and 2.0-fold more, respectively, compared with bortezomib (Figure 4C).
Activity of carfilzomib and bortezomib against myeloma models. (A) Several MM cell lines were treated with a 1-hour pulse of increasing concentrations of carfilzomib or bortezomib. After 24 hours, the number of live cells was determined with a WST-1 assay. (B) Activation of the stress response in RPMI 8226 cells pulse treated for 1 hour with proteasome inhibitors and allowed to recover for the indicated time points is shown. Protein expression levels of activated JNK were examined after treatment with carfilzomib or bortezomib. (C) ANBL-6 cells were exposed to a 1-hour pulse of 100 nM carfilzomib or bortezomib and allowed to recover for 8 hours. Cellular lysates (30 μg per reaction) were then incubated with 40-μM fluorogenic substrates specific for caspase-3, caspase-8, and caspase-9 activity. Results are expressed as fold relative fluorescence units over DMSO control and determined as described in Figure 1. Error bars for panels A and C are SD.
Activity of carfilzomib and bortezomib against myeloma models. (A) Several MM cell lines were treated with a 1-hour pulse of increasing concentrations of carfilzomib or bortezomib. After 24 hours, the number of live cells was determined with a WST-1 assay. (B) Activation of the stress response in RPMI 8226 cells pulse treated for 1 hour with proteasome inhibitors and allowed to recover for the indicated time points is shown. Protein expression levels of activated JNK were examined after treatment with carfilzomib or bortezomib. (C) ANBL-6 cells were exposed to a 1-hour pulse of 100 nM carfilzomib or bortezomib and allowed to recover for 8 hours. Cellular lysates (30 μg per reaction) were then incubated with 40-μM fluorogenic substrates specific for caspase-3, caspase-8, and caspase-9 activity. Results are expressed as fold relative fluorescence units over DMSO control and determined as described in Figure 1. Error bars for panels A and C are SD.
Carfilzomib effects in patient-derived samples
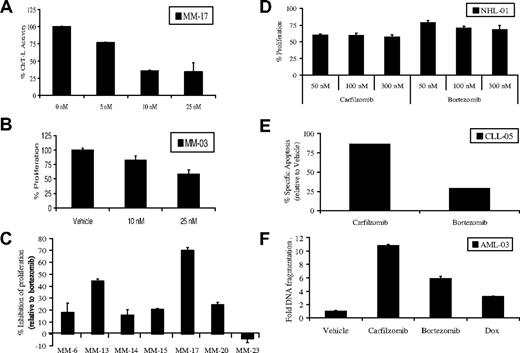
Based on the results obtained from MM cell culture models, it was of interest to evaluate the activity of carfilzomib in purified CD138+ plasma cells derived from patients. Exposure of such cells to escalating doses of carfilzomib for 24 hours continuously, followed by measurement of the ChT-L activity, revealed that carfilzomib induced a dose-dependent proteasome inhibition (Figure 5A). Similarly, cellular viability decreased significantly in such plasma cells treated continuously with carfilzomib (Figure 5B). We then examined whether a brief pulse of carfilzomib had greater antiproliferative effects compared with a similar pulse of bortezomib. Plasma cells from either bone marrow aspirates or peripheral blood of patients, some with chromosome 13 deletions, were pulsed with 100 nM carfilzomib or bortezomib. Carfilzomib inhibited proliferation to a greater extent than bortezomib by 11% to 70% in 8 of 9 samples from patients who were clinically bortezomib-naive (Figure 5C). However, bortezomib did display increased antiproliferative efficacy in one sample compared with carfilzomib (Figure 5C). It is important to note that all experiments were performed in purified CD138+ plasma cells, and the effect of carfilzomib on other clonal subsets of cells is currently unknown.
Activity of carfilzomib and bortezomib in patient samples. (A) Purified plasma cells were continuously treated with increasing doses of carfilzomib for 24 hours. Cells were then lysed, and the ChT-L activity was determined (10 μg per reaction). (B) CD138+ cells were treated with continuous exposure to the indicated concentrations of carfilzomib, followed by a WST-1 cell viability assay. (C) Purified plasma cells were pulse-treated with 100 nM carfilzomib or bortezomib, followed by recovery in drug-free media for 24 hours. WST-1 was used to assess proliferation. Several of the samples are from patients with chromosome 13 deletions (MM-13, MM-15, MM-17, MM-20, and MM-23). Results are expressed as the percentage (%) inhibition of proliferation of carfilzomib-treated cells relative to bortezomib-treated cells, which were set at 0, with a positive result indicating the amount of enhanced antiproliferative activity of carfilzomib over that of bortezomib. (D) Pulse carfilzomib and bortezomib exposure in an NHL patient sample and determination of antiproliferation activity by WST-1 assay is shown. (E) Flow cytometric analysis of carfilzomib-induced versus bortezomib-induced specific apoptosis in patient-derived CD19+ CLL B-cells is shown. Patient cells were pulsed (100 nM) for 1 hour with the indicated drug and allowed to recover for 24 hours. Apoptosis was assessed in cells stained with Annexin V/TO-PRO-3/anti-CD19. Specific apoptosis is shown in the CD19+ gated population relative to vehicle controls. (F) AML cells from a patient with progressive disease after multiple chemotherapeutic treatments were pulsed for 1 hour with 100 nM carfilzomib or bortezomib or continuously treated with 1 μM Dox. Apoptosis was measured by DNA fragmentation ELISA and expressed as fold induction over DMSO control in CD33+cells purified from peripheral blood mononuclear cells (PBMCs). Error bars in panels A,B,C,D, and F are SD.
Activity of carfilzomib and bortezomib in patient samples. (A) Purified plasma cells were continuously treated with increasing doses of carfilzomib for 24 hours. Cells were then lysed, and the ChT-L activity was determined (10 μg per reaction). (B) CD138+ cells were treated with continuous exposure to the indicated concentrations of carfilzomib, followed by a WST-1 cell viability assay. (C) Purified plasma cells were pulse-treated with 100 nM carfilzomib or bortezomib, followed by recovery in drug-free media for 24 hours. WST-1 was used to assess proliferation. Several of the samples are from patients with chromosome 13 deletions (MM-13, MM-15, MM-17, MM-20, and MM-23). Results are expressed as the percentage (%) inhibition of proliferation of carfilzomib-treated cells relative to bortezomib-treated cells, which were set at 0, with a positive result indicating the amount of enhanced antiproliferative activity of carfilzomib over that of bortezomib. (D) Pulse carfilzomib and bortezomib exposure in an NHL patient sample and determination of antiproliferation activity by WST-1 assay is shown. (E) Flow cytometric analysis of carfilzomib-induced versus bortezomib-induced specific apoptosis in patient-derived CD19+ CLL B-cells is shown. Patient cells were pulsed (100 nM) for 1 hour with the indicated drug and allowed to recover for 24 hours. Apoptosis was assessed in cells stained with Annexin V/TO-PRO-3/anti-CD19. Specific apoptosis is shown in the CD19+ gated population relative to vehicle controls. (F) AML cells from a patient with progressive disease after multiple chemotherapeutic treatments were pulsed for 1 hour with 100 nM carfilzomib or bortezomib or continuously treated with 1 μM Dox. Apoptosis was measured by DNA fragmentation ELISA and expressed as fold induction over DMSO control in CD33+cells purified from peripheral blood mononuclear cells (PBMCs). Error bars in panels A,B,C,D, and F are SD.
In addition to activity against MM, bortezomib is clinically active against other hematologic malignancies, so we also sought to examine the impact of carfilzomib in other models. Carfilzomib inhibited proliferation and induced apoptosis in purified samples from patients with diffuse large B-cell NHL (Figure 5D), chronic lymphocytic leukemia (CLL; Figure 5E), and acute myeloid leukemia (AML; Figure 5F). Inhibition of the ChT-L activity was confirmed in purified CD19+ cells from a patient with B-cell CLL (data not shown), and this ChT-L inhibition correlated well with induction of apoptosis in carfilzomib pulse-treated cells (Figure 5E). Relative to bortezomib, carfilzomib appeared to show enhanced activity against all of these samples (Figure 5D-F).
Carfilzomib and drug resistance
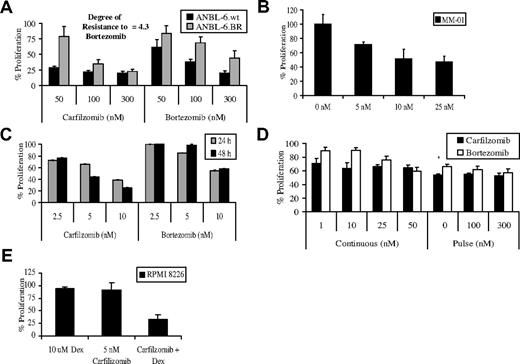
The potent activity of carfilzomib led us to evaluate the possibility that it could overcome bortezomib resistance. Resistant (BR) cell lines were prepared by exposing wild-type (wt) cells to increasing dosages of bortezomib over several months until discernible changes in bortezomib sensitivity were observed. Resistant cell lines displayed significant decreases in their sensitivity to the antiproliferative effects of bortezomib treatment. For example, ANBL-6.wt cells exposed continuously to 5 nM bortezomib for 24 hours experienced a 78% decrease in the viable cell population, compared with ANBL-6.BR cells, where only a 7% loss of viability was seen, indicating a 4-fold loss of bortezomib sensitivity (additional results are shown in Figure S1A, available on the Blood website; see the Supplemental Materials link at the top of the online article). Similar results were found in comparisons of RPMI 8226.BR and OPM-2.BR cells with their wt parentals. ANBL-6.wt and BR cells were then pulse-treated with comparable concentrations of carfilzomib or bortezomib, and the antiproliferative effects were assessed after 24 hours. Resistance to bortezomib did result in some cross-resistance with carfilzomib, because BR cells were less sensitive than wt cells to this irreversible inhibitor (Figure 6A). However, carfilzomib was more potent at inhibiting proliferation in BR cell lines than bortezomib, with ANBL-6.BR (Figure 6A), RPMI 8226.BR (Figure S1B), and OPM-2.BR (Figure S1C) cells exhibiting a 2.0-, 1.5-, and 2.1-fold increased sensitivity to carfilzomib, respectively. Similarly, in CD138+ plasma cells from a patient who had no clinical benefit after bortezomib therapy, carfilzomib retained the ability to inhibit proliferation (Figure 6B). Exposure of plasma cells from 2 patients who both clinically progressed while on bortezomib to carfilzomib showed enhanced cytotoxic effects compared with treatment with bortezomib (Figure 6C,D).
Carfilzomib and chemotherapeutic resistance. (A) ANBL-6.BR cells, and their wt counterparts, were pulsed with carfilzomib for 1 hour, and proliferation was assessed using the WST-1 reagent after a 24-hour recovery period. The DOR to bortezomib was computed by comparing the IC50 of bortezomib-sensitive and BR cells. (B) CD138+ cells from a MM patient with a chromosome 13 deletion who did not have a clinical response to bortezomib were treated with continuous exposure to the indicated concentrations of carfilzomib or bortezomib for 24 hours in triplicate, followed by measurement of proliferation with the WST-1 assay. (C) CD138+ cells from a MM patient who progressed while on bortezomib treatment were exposed to increasing concentrations of carfilzomib or bortezomib for 24 hours before assessment for cellular proliferation. (D) CD138+ plasma cells from a patient who progressed on bortezomib were exposed to continuous and pulse treatments with equivalent concentrations of carfilzomib and bortezomib, followed by a WST-1 cellular proliferation assay. (E) RPMI 8226 cells were treated continuously with 5 nM carfilzomib and 10 μM Dex for 48 hours to determine the effect of this combined therapy against cellular proliferation. Error bars are SD.
Carfilzomib and chemotherapeutic resistance. (A) ANBL-6.BR cells, and their wt counterparts, were pulsed with carfilzomib for 1 hour, and proliferation was assessed using the WST-1 reagent after a 24-hour recovery period. The DOR to bortezomib was computed by comparing the IC50 of bortezomib-sensitive and BR cells. (B) CD138+ cells from a MM patient with a chromosome 13 deletion who did not have a clinical response to bortezomib were treated with continuous exposure to the indicated concentrations of carfilzomib or bortezomib for 24 hours in triplicate, followed by measurement of proliferation with the WST-1 assay. (C) CD138+ cells from a MM patient who progressed while on bortezomib treatment were exposed to increasing concentrations of carfilzomib or bortezomib for 24 hours before assessment for cellular proliferation. (D) CD138+ plasma cells from a patient who progressed on bortezomib were exposed to continuous and pulse treatments with equivalent concentrations of carfilzomib and bortezomib, followed by a WST-1 cellular proliferation assay. (E) RPMI 8226 cells were treated continuously with 5 nM carfilzomib and 10 μM Dex for 48 hours to determine the effect of this combined therapy against cellular proliferation. Error bars are SD.
The ability of carfilzomib to overcome resistance to other antimyeloma agents was examined in melphalan-, Dex-, and doxorubicin (Dox)-resistant cell lines (Table 1). Carfilzomib overcame Dex resistance, in that MM1.R cells revealed an IC50 of 15.2 nM, which, interestingly, was less than the value of 29.3 nM for parental MM1.S cells. Resistance to melphalan also did not impact carfilzomib, because the IC50 in RPMI 8226.wt cells was comparable with that in melphalan-resistant 8226.LR5 cells. In contrast, Dox-resistant 8226.Dox40 cells did not respond to carfilzomib, as there was minimal proliferation inhibition at concentrations up to 1000 nM (Figure S1D). However, pretreatment with the p-glycoprotein inhibitor verapamil partially overcame this resistance (Table 1), supporting the possibility that carfilzomib may be subject to p-glycoprotein–mediated drug resistance.
Carfilzomib and resistance to antimyeloma agents
| Cell line . | IC50 . |
|---|---|
| MM1.S | 29.3 nM |
| MM1.R | 15.2 nM |
| RPMI 8226.wt | 89.9 nM |
| 8226.LR5 | 83.3 nM |
| 8226.Dox40 | >1000 nM |
| 8226.Dox40 and Verapamil | 829 nM |
| Cell line . | IC50 . |
|---|---|
| MM1.S | 29.3 nM |
| MM1.R | 15.2 nM |
| RPMI 8226.wt | 89.9 nM |
| 8226.LR5 | 83.3 nM |
| 8226.Dox40 | >1000 nM |
| 8226.Dox40 and Verapamil | 829 nM |
Carfilzomib activity was assessed in wt (MM1.S and RPMI 8226.wt), melphalan-resistant (8226.LR5), Dex-resistant (MM1.R), and Dox-resistant (8226.Dox40) myeloma cells. 8226.Dox40 cells were also pretreated for 24 hours with the p-glycoprotein inhibitor verapamil (15 μM). Cells were pulsed for 1 hour with increasing concentrations of carfilzomib, and the IC50 of proliferation was assessed after a 24-hour recovery.
Antimyeloma agents are often used in combinations to enhance their antitumor activity, and addition of Dex to bortezomib is one strategy that has shown improved efficacy.31 To determine whether carfilzomib works in concert with Dex, KAS-6/1 and RPMI 8226 cells were treated simultaneously with both drugs for 24 or 48 hours, respectively. Compared with Dex or carfilzomib alone, the combination demonstrated a greater antiproliferative effect (Figure 6F). Moreover, statistical analysis indicated that a high degree of synergy was present between the 2 drugs at all concentrations tested in both cell lines (Tables 2,3).
Combination indices of carfilzomib and Dex for RPMI 8226 cells
| Carfilzomib (nM) . | Dex (μM) . | ||
|---|---|---|---|
| 1 . | 5 . | 10 . | |
| 1 | 0.1 | 0.2 | 0.1 |
| 2.5 | 0.3 | 0.3 | 0.3 |
| 5 | 0.5 | 0.5 | 0.5 |
| 10 | 0.8 | 0.9 | 0.9 |
| Carfilzomib (nM) . | Dex (μM) . | ||
|---|---|---|---|
| 1 . | 5 . | 10 . | |
| 1 | 0.1 | 0.2 | 0.1 |
| 2.5 | 0.3 | 0.3 | 0.3 |
| 5 | 0.5 | 0.5 | 0.5 |
| 10 | 0.8 | 0.9 | 0.9 |
Combination indices of RPMI 8226 cells treated for 48 hours with the indicated concentrations of carfilzomib and Dex are shown. Data were analyzed using Calcusyn (version 2) software (BioSoft, Milltown, NJ). The combination index (CI) is a quantitative measure of the degree of drug interaction, with a CI less than 1 indicating synergy, a CI of 1 indicating additive effects, and a CI over 1 indicating antagonist.
Combination indices of carfilzomib and Dex for KAS-6/1 cells
| Carfilzomib (nM) . | Dex (μM) . | ||
|---|---|---|---|
| 1 . | 5 . | 10 . | |
| 1 | 0.1 | 0.3 | 0.6 |
| 2.5 | 0.2 | 0.3 | 0.7 |
| 5 | 0.9 | 0.5 | 0.6 |
| 10 | 0.1 | 0.2 | 0.5 |
| Carfilzomib (nM) . | Dex (μM) . | ||
|---|---|---|---|
| 1 . | 5 . | 10 . | |
| 1 | 0.1 | 0.3 | 0.6 |
| 2.5 | 0.2 | 0.3 | 0.7 |
| 5 | 0.9 | 0.5 | 0.6 |
| 10 | 0.1 | 0.2 | 0.5 |
Combination indices of KAS-6/1 cells treated for 24 hours with the indicated concentrations of carfilzomib and Dex are shown. Details as in footnote to Table 2.
Discussion
The reversible proteasome inhibitor bortezomib represents an important advance in the treatment of MM, where it has become one standard of care for relapsed/refractory disease and is under active investigation as a front-line agent. Combination regimens based on bortezomib may have further enhanced activity, including regimens such as bortezomib with Dox.32-35 Such findings have validated the ubiquitin-proteasome pathway as both a target by itself for cancer therapy and a rational target to induce chemosensitization and overcome chemoresistance to conventional agents. Despite these encouraging data, some patients have disease that shows primary resistance and does not respond to bortezomib therapy. Others develop secondary resistance, progress after an initial response, or do not respond when they are rechallenged after relapsing from an earlier response to bortezomib. Another proteasome inhibitor, NPI-0052, has already been described that can overcome previous bortezomib resistance,13 and its irreversible mechanism of inhibition suggested that other inhibitors that block proteasome function irreversibly merited study as well. This led us to evaluate the properties of the epoxomicin-related tetrapeptide carfilzomib and to determine its potential as an antimyeloma agent.
Carfilzomib shares structural similarity with epoxomicin. Cocrystal studies revealed that epoxomicin formed a unique 6-atom ring structure with the β5 subunit, arising from a 2-step process that ultimately led to intramolecular cyclization and morpholino adduction.36 It is a likely assumption that carfilzomib's interaction with the proteasome is similar in nature, given the identical keto-epoxide pharmacophore. Carfilzomib was found to specifically target the ChT-L proteasome activity (Figure 1B) by binding potently and specifically to the β5 constitutive proteasome and β5i immunoproteasome subunits (Figure 1C). This resulted in accumulation of ubiquitin-protein conjugates (Figure 1D) and proteasome substrates (Figure 1E), as well as inhibition of myeloma cell proliferation (Figure 2A-C) through induction of programmed cell death (Figure 2B-D). The molecular sequelae of carfilzomib are similar to those of bortezomib, in that activation of both the intrinsic and extrinsic apoptotic pathways was seen, and these converged on the common effector caspase-3 (Figure 3A). Caspase inhibitors dramatically blocked carfilzomib-stimulated apoptosis, similar to bortezomib-induced apoptosis (Figure 3B).23 JNK activation is well characterized in bortezomib-induced apoptosis,37 and we found a similarly marked increase in JNK activity with carfilzomib exposure (Figure 3E). Abrogation of JNK apoptotic signaling cascades with a DN-c-Jun construct correlated with a decrease in carfilzomib-induced apoptosis (Figure 3G). Lauricella et al had identical findings with bortezomib treatment in hepatoma cells.27
Interestingly, NPI-0052 activated apoptosis predominantly through a caspase-8-mediated pathway, with little dependence on caspase-9.13 Bortezomib preferentially inhibits ChT-L activity, as does carfilzomib, whereas NPI-0052 inhibits the ChT-L and the PGPH and T-L proteases. These findings suggest that proteasome inhibitors targeting predominantly the β5 and/or β5i subunits may trigger dual extrinsic and intrinsic apoptosis pathway activation irrespective of their chemical composition or their ability to form either reversible or irreversible bonds with the active-site threonine. The predominant reliance of NPI-0052 on caspase-8 would therefore seem to be partly related to the interaction of the proteasome active sites with the unique substituents on its bicyclic ring system, which do not induce Bax activation, oligomerization, and mitochondrial insertion. In further dissecting the activity and mechanism of action of proteasome inhibitors, it would therefore be of interest in the future to test agents that specifically target only the PGPH or the T-L proteases. In addition, the role of the immunoproteasome in contributing to the activation of distinct apoptotic pathways has not been defined as of yet, but all 3 classes of proteasome inhibitors, including bortezomib, carfilzomib, and NPI-0052, have been shown to bind and inhibit immunoproteasome-specific subunits. The use of specific immunoproteasome inhibitors that have been recently identified38 may be of help in determining the extent of immunoproteasome dependency to cellular survival.
Proteasome inhibition with carfilzomib resulted in potent antiproliferative and proapoptotic effects in myeloma cell lines (Figure 2) and in patient-derived models of myeloma (Figure 5). The presence of the deletion of chromosome 13 identifies a poor-prognosis subgroup of patients with myeloma who have good benefits with bortezomib-based therapy.39 Several of the cell lines and patient samples tested herein also harbored this cytogenetic abnormality and responded well to carfilzomib preclinically. This suggests the possibility that proteasome inhibitors as a class may be especially effective in this population, though larger prospective studies are needed to verify these findings for bortezomib, and carfilzomib is just entering clinical trials. Carfilzomib also showed evidence of activity against malignant cells from patients with NHL, and both AML and CLL, suggesting other possible areas for future clinical development. Stapnes et al also found that carfilzomib potently inhibited proliferation and induced apoptosis in CD34+ cells from patients with AML.40
One notable finding is that carfilzomib was more potent than bortezomib in proteasome inhibitor-naive models (Figure 3). Also of importance, carfilzomib overcame both primary and secondary resistance to bortezomib in both cell line models and clinical samples (Figure 6). The mechanism by which this occurs has not yet been elucidated, but previous studies with NPI-0052 suggested that it was this agent's ability to block several proteasome proteases that allowed it to overcome bortezomib resistance. Because carfilzomib has a similar property in overcoming resistance, but predominantly binds only the ChT-L subunit, our studies suggest that it may be the irreversibility of agents such as NPI-0052 and carfilzomib that confers this attractive property. If true, this suggests a possible mechanism, because one characteristic of cells that express primary resistance to bortezomib is the overexpression of proteasome subunits and other ubiquitin-proteasome pathway proteins.41-43 It may be that irreversible drugs would require the cell to synthesize and reassemble new proteasomes to recover their proteolytic capacity. Because this would take longer than recovery from reversible inhibitors, that in part would occur by dissociation of bortezomib without the need for new protein synthesis, irreversible inhibitors would provide a longer-lasting inhibition. Such prolonged inhibition could result in enhanced efficacy or the ability to overcome the cell's attempt at resistance by overproduction of proteasomes. In addition, the ability of carfilzomib to overcome bortezomib resistance does not seem to be linked to cellular transport mechanisms as the rate of onset of proteasome inhibition does not differ greatly in a variety of cell types exposed to carfilzomib and bortezomib (data not shown). However, it is important to note that these studies were conducted in proteasome inhibitor-naive cells. Further studies will be needed to elucidate the contribution of these possibilities or to determine whether some different pathway is responsible for the ability to partially overcome bortezomib resistance. These should be helped by the development of bortezomib-resistant myeloma cell models such as those described herein, which have not previously been reported.
Taken together, the above data provide a strong rationale for clinical evaluation of the proteasome inhibitor carfilzomib in patients with relapsed and refractory myeloma. One concern would be that the irreversible mechanism of action of carfilzomib could result in enhanced toxicities of the kind described for bortezomib, such as cytopenias, gastrointestinal events, constitutional symptoms, and peripheral neuropathy. It certainly is also possible that this agent's mechanism of action, and probably different clinical pharmacokinetics and pharmacodynamics, could result in new, heretofore uncharacterized dose-limiting toxicities. Phase I studies of carfilzomib are currently underway using 2 different schedules, however, and to date no untoward toxicities have been reported, and antitumor activity is beginning to be seen.44 This agent's ability to overcome bortezomib resistance preclinically suggests that it may provide another therapeutic option for patients whose disease has progressed through bortezomib or for those who have had responses of very brief duration. In addition, carfilzomib in combination with Dex leads to a synergistic antiproliferative effect, indicating that rationally designed combination regimens may further improve the therapeutic efficacy of carfilzomib in the relapsed/refractory and possibly front-line myeloma settings.
The online version of the article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
R.Z.O., a Leukemia and Lymphoma Society Mansbach Foundation Scholar in Clinical Research and a Jefferson Pilot fellow in Academic Medicine, was supported by the Leukemia and Lymphoma Society (grant 6096–07), the Multiple Myeloma Research Foundation, and the National Cancer Institute (grant RO1 CA102278). D.J.K. was supported by National Institutes of Health (grant T32 CA 09156).
National Institutes of Health
Authorship
Contribution: D.J.K. designed and performed the majority of the research and wrote the manuscript; K.D.S., C.M.S., S.D.D., and M.K.B provided reagents, helped with data analysis, and performed the ELISA experimentation; J.S.S. and P.M.V. received consent from patients and obtained their samples; Q.C. and P.M.V. were essential for helping with patient sample purifications; F.W.B.v.L. provided vital reagents and contributed to data analysis; A.A.C.-K. provided Opm-2 cell lines and manuscript critique; R.Z.O. supervised all of the research and offered valuable suggestions and manuscript editing.
Conflict-of-interest disclosure: K.D.S., C.M.C., S.D.D., and M.K.B. are employees of Proteolix and receive stock options as part of their employment. All other authors declare no competing financial interests.
Correspondence: Dr. Robert Z. Orlowski, University of Texas M. D. Anderson Cancer Center, Department of Lymphoma/Myeloma, 1515 Holcombe Boulevard, Unit 429, Houston, TX 77030; e-mail: rorlowsk@mdanderson.org.