Abstract
Peripheral T-cell lymphomas (PTCLs) are aggressive tumors in which the current therapy based on multiagent chemotherapy is not successful. Since cytochrome P450 3A subfamily (CYP3A) enzymes are involved in the inactivation of chemotherapy drugs, we hypothesized that CYP3A and P-glycoprotein (MDR1) expression in these lymphomas could result in a poor clinical response. We measured tumoral CYP3A and MDR1 mRNA content in 44 T-cell lymphomas, finding a large variation in CYP3A expression. Multiplex polymerase chain reaction (PCR) analysis and fluorescence in situ hybridization (FISH) analysis showed genomic gains affecting CYP3A and MDR1 genes in T-cell lines and primary tumors, suggesting that this could be the mechanism underlying the tumoral expression variation. To test whether the tumoral expression of CYP3A and/or MDR1 could influence PTCL treatment outcome, their expression levels were compared with the clinical response and survival of the patients, finding that a high tumoral expression of CYP3A4 was significantly associated with a lower complete remission rate. This was further investigated with cell lines stably expressing CYP3A4 that exhibited an increased resistance to doxorubicin and etoposide. In conclusion, a high CYP3A4 tumoral expression could be useful to predict poor response to the standard PTCL chemotherapy; in these cases alternative chemotherapy combinations or doses should be explored.
Introduction
Peripheral T-cell lymphomas (PTCLs) constitute a complex group of tumors that are still a challenge for medicine. More than half of PTCLs remain classified as unspecified (PTCLu), a group that includes histologically and clinically heterogeneous tumors.1,2 Other frequent PTCL subtypes in the current classification are the angioimmunoblastic T-cell lymphomas (AITLs) and anaplastic large-cell lymphomas (ALCLs).3,4 Regardless of the histologic subtype, multiagent chemotherapy is the treatment of choice for most PTCL patients. However, PTCLs are clinically aggressive tumors with poorer response to treatment and shorter survival times than diffuse large B-cell lymphomas, typically showing less than 30% 5-year overall survival.5-7 To date, the most effective therapy is a combination chemotherapy regimen, in many cases CHOP (cyclophosphamide, doxorubicin, vincristine, and prednisone), although a variety of other regimens (CHOEP [CHOP with etoposide], VACPE [vincristine, adriamycin, cyclophosphamide, prednisone, and etoposide]) that add other drugs with important antilymphoma activity, such as etoposide, have demonstrated similar activity. However, the poor clinical outcome of most patients is disappointing and clearly reveals the need to improve the therapy by identifying factors affecting the response
Cytochromes P450 (CYPs) of families 1 to 3 are drug-metabolizing enzymes involved in the activation and detoxification of a large number of chemotherapeutic drugs.8 Among them, the CYP3A subfamily, constituted in humans by 4 enzymes of similar substrate specificity (CYP3A4, CYP3A5, CYP3A7, and CYP3A43), is involved in the metabolism of more than 50% of all therapeutic drugs in clinical use.9-11 Both genetic polymorphisms and environmental factors have been shown to alter CYP activities, resulting in interindividual differences in drug effects.12,13 In addition to the variations in the constitutive expression, the CYP3A enzymes, which are mainly expressed in the liver, have been found in different tumors.14-18 Because CYP3As inactivate many anticancer drugs, an overexpression of CYP3As in tumors could result in an increased intratumoral drug inactivation and consequently decreased drug efficacy.17,19 Similarly, the ABC efflux drug transporter p-glycoprotein (Pgp) can confer tumoral drug resistance and because of the overlapping substrate specificity of CYP3A and Pgp, functional interactions have been suggested. Interestingly, microarray expression-profiling analysis of PTCLs in our group20,21 showed large differences in the tumoral CYP3A expression. Furthermore, CYP3A and MDR1 (encoding Pgp) genes are closely located in 7q21, a region recurrently gained in T-cell lymphomas.22 Thus, in this study, we measured the expression of CYP3As and MDR1 in a panel of 44 PTCL tumors and compared it with clinical parameters such as therapy response and survival.
Patients, materials, and methods
Tumor samples
Frozen and paraffin-embedded tumor biopsy samples from a group of 44 peripheral T-cell and natural killer (NK) lymphomas including 23 PTCLu's, 14 AILTs, and 7 NK lymphomas (NKs) were collected through the Centro Nacional de Investigaciones Oncológicas (CNIO) tumor bank network from pathology departments of different hospitals in Spain. These lymphomas have been previously analyzed for their expression profiles using cDNA microarrays.20,21 Most of the cases were treated with similar therapy protocols based on combination chemotherapy, typically CHOP or CHOP-like protocols. Five reactive lymphadenopathy samples were used as controls. Total RNA and DNA were extracted from frozen samples with standard methods. This study has the approval of the Bioethics Committee of the Instituto de Salud Carlos III and fulfills all ethical requirements.
Eleven T-cell lymphoma cell lines were used to analyze genomic gains in chromosome 7q: Karpas-45, HSB-2, Jurkat, KE-37, Molt-4, Molt-13, Molt-16, HPB-ALL, Peer, Hut-78, and Karpas 299.
Quantitative reverse transcriptase–polymerase chain reaction analysis
One μg of total RNA was reverse transcribed using MMLV Reverse Transcriptase (Invitrogen, Carlsbad, CA) and a random primer. The cDNAs were subjected to quantitative real-time polymerase chain reaction (PCR) assay with the use of gene-specific double-fluorescent–labeled probes and the TaqMan Universal PCR Mix in an ABI prism 7900 system (Applied Biosystems, Foster City, CA) under manufacturer's recommendations. The PCR amplification was carried out with 10 minutes at 95°C, followed by 50 cycles of 15 seconds at 95°C and 1 minute at 60°C, using the oligonucleotides shown in Table S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Assays-on-Demand Taqman MGB probes (Applied Biosystems) were used for β-actin quantification, which was then used as an internal standard and allowed normalization of the samples. To perform relative quantification of the expression of the genes, standard curves were constructed with serial 10-fold dilutions of a human liver cDNA. All samples were analyzed in triplicates.
Specific multiplex-PCR analysis
A multiplex-PCR analysis (MPA) was designed to determine the number of copies of CYP3A4 and MDR1 genes, both located at 7q21. Using a multiplex-PCR kit and following standard recommendations (QIAGEN, Hilden, Germany), 10 pairs of primers were designed and labeled (at the 5′ end with 6-FAM) to obtain uniquely sized amplification fragments: 4 for the CYP3A4 gene (501, 364, 385, and 419 bp) and 1 for MDR1 (220 bp; Table S1). Five additional fragments were used as controls and were located at 11q23, 5q33, 15q25, 17q21, and 7p14. Briefly, multiplex amplifications were performed using the Qiagen Multiplex PCR kit (QIAGEN) with 25 μL of a mixture containing 1 × multiplex-PCR master mix, 0.2 μM of each primer, and 50 to 100 ng of genomic DNA, which was amplified by 22 cycles of 30 seconds at 94°C, 90 seconds at 60°C, and 90 seconds at 72°C. The PCR amplification products obtained were analyzed on an ABI PRISM 310 capillary sequencer (Applied Biosystems) and using GeneScan v3.1 software (Applied Biosystems). For each sample, the peak area of all fragments was determined and normalized with the values of the control peaks from a control sample (ie, genomic DNA from a healthy volunteer).23
FISH
To assess the presence of CYP3A4 gene gains we performed fluorescence in situ hybridization (FISH) analysis on several tissue samples. Ensembl Cytoview24 was used to select the 4 bacterial artificial chromosome (BAC) clones covering the CYP3A locus (RP11–757A13, RP11–150A16, RP11–977H6, and RP11–268P20) and 3 BAC clones covering the MDR1 locus (RP11–212B1, RP11–1149O20, and RP11–647N21). The BACs were obtained from BACPAC Resource Center (BPRC) at the Children's Hospital Oakland Research Institute (Oakland, CA). Additionally, a commercial Texas Red–labeled probe for the centromeric region of chromosome 7 (Vysis, Downers Grove, IL) was used for ploidy control. FISH assays were carried out as described elsewhere and according to the manufacturer's instructions.25 In brief, all BACs were labeled directly by nick translation with SpectrumGreen (Vysis) according to the manufacturer's specifications. The probes were blocked with Cot-1 DNA (Vysis) to suppress repetitive sequences. Paraffinated tissue slides were deparaffinated and boiled in a pressure cooker with 1 mM EDTA (pH 8.0) for 5 to 10 minutes and incubated with pepsin at 37°C for 30 minutes and dehydrated. The probe was denatured at 75°C for 2 minutes and hybridized overnight at 37°C in a humid chamber. After posthybridization washes, the tissue samples were counterstained with 4′,6-diamidino-2-phenylindole (DAPI II; Vysis) for chromatin counterstaining before microscopy. Cell images were captured using an Olympus BX61 microscope with a 100×/1.30 NA oil objective and a charge-coupled device (CCD) camera (Photometrics SenSys; Spectra Service, Ontario, NY) connected to a computer running the Chromofluor image analysis system (Cytovision; Applied Imaging, San Jose, CA). FISH scoring of CYP3A fluorescence signals was carried out in each sample by counting the number of single copy gene and control probe signals in an average of 100 nuclei. Gain or polyploidy status was considered as positive for a sample when the green-red signal ratio was over 1.5 in greater than 30% of tumor cells.
Statistical analysis of the clinical data
Since different subgroups of T-cell lymphomas could have different clinical behavior, clinical analysis was carried out only in the group of PTCLu's. After the initial therapy, patients were classified as nonresponders (NRs) or as having partial remission (PR) or complete remission (CR). Pearson chi-square and Fisher exact tests were performed to carry out comparisons between the rates of clinical responses in cases with high or low expression of CYP3A4. Kaplan-Meier survival curves were used to compare overall survival (OS) between lymphomas with higher or lower expression of CYP3As and MDR1 genes. Overall survival time was measured from the date of diagnosis to the date of death or last follow-up. The log-rank test was used to compare survival curves. The SPSS version 12.0 software (Chicago, IL) was used for these analyses.
Construction and cytotoxicity analysis of HEK293 cells stably overexpressing CYP3A4
Based upon sequences from their corresponding cDNAs, we designed primers CYP3A4FW 5′-GAAAGCTAGCATGGCTCTCATCCCAGACTTGGCCA-3′ and CYP3A4RV 5′-CTGCGGCCGCTTCAGGCTCCACTTACGGTGC-3′ to amplify the CYP3A4 coding sequence from human liver cDNA and introduce NheI and NotI cleavage sites. The product was cloned into pIRESpuro2 vector (Clontech, Palo Alto, CA) and sequenced. To generate stable cell lines, HEK293 cells were electroporated and 24 hours later grown with puromycin (Sigma, St Louis, MO) for selection. CYP3A4-resistant clones were analyzed for CYP3A4 expression by Western blot using a primary antibody recognizing CYP3A4 (299223; Daiichi Pure Chemicals, Tokyo, Japan). Control cells (HEK293-pIRES) were also selected with puromycin and all resistant clones were pooled together. Stable transfectants were maintained in DMEM supplemented with 10% FBS, penicillin, and streptomycin with puromycin at a concentration of 0.5μg/ mL.
Cytotoxicity tests for doxorubicin, etoposide, and vincristine (Sigma) were performed in HEK293-CYP3A4 and control cells. Briefly, 24 hours after trypsinization the cells were incubated with the drugs for 96 hours and each concentration was assayed in triplicate and then incubated with MTT substrate. The resulting absorbance was measured by means of a microplate reader (Bio-Rad, Hercules, CA), and the cytotoxic effect of each treatment was assessed by IC50 value (concentration of the drug leading to 50% cell survival).
Results
PTCLs show a variable expression of CYP3As and MDR1
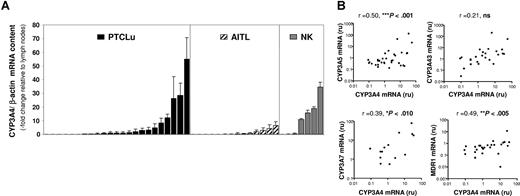
In a previous study by our group,20 expression-profiling analysis using microarrays allowed us to detect overexpression of different CYP3A genes in a subset of PTCLs (data not shown). To confirm that CYP3As and MDR1 are expressed in PTCL tumors, quantitative reverse transcriptase–polymerase chain reaction (RT-PCR) was used in 44 PTCL tumors and compared with 5 reactive lymph nodes. A large intersample variability in the expression of the CYP3As was found, with about one third of the cases showing CYP3A4, CYP3A5, CYP3A7, and CYP3A43 mRNA levels at least 2-fold higher than the reactive lymph nodes. On the contrary, MDR1 mRNA levels showed less variation and most tumors (85%) had lower mRNA content than the reactive lymph nodes. Because T-cell lymphomas include a range of different histologic subtypes, we investigated whether the heterogeneity found in the CYP3A expression could be associated to a specific tumor subtype. In general, the proportion of tumors overexpressing CYP3A genes was similar for PTCLu and AITL tumors, on average 35% and 20%, respectively, whereas NK tumors had a higher CYP3A expression, with 57% of the tumors showing increased levels of CYP3A4 and CYP3A43 but only 29% and 14% showing increased CYP3A7 and CYP3A5 expression, respectively. Figure 1. A shows the expression levels of CYP3A4 in the different tumor subtypes.
CYP3As and MDR1 mRNA quantification in PTCL tumors. (A) CYP3A mRNA content in PTCL tumors. CYP3A4 mRNA expression was quantified by real-time RT-PCR in 45 PTCL tumors: 23 PTCLu's, 15 AITLs, and 7 NKs. Results are expressed relative to CYP3A4 expression in control tissue (reactive lymph nodes). Error bars represent SD of 3 independent experiments. (B) Correlation between CYP3As and MDR1 expression in PTCL tumors. The mRNA expression of CYP3A4, CYP3A5, CYP3A7, CYP3A43, and MDR1 in 45 PTCLs was compared by correlation analysis. The results obtained for CYP3A4 are shown with the correlation coefficients and P values.
CYP3As and MDR1 mRNA quantification in PTCL tumors. (A) CYP3A mRNA content in PTCL tumors. CYP3A4 mRNA expression was quantified by real-time RT-PCR in 45 PTCL tumors: 23 PTCLu's, 15 AITLs, and 7 NKs. Results are expressed relative to CYP3A4 expression in control tissue (reactive lymph nodes). Error bars represent SD of 3 independent experiments. (B) Correlation between CYP3As and MDR1 expression in PTCL tumors. The mRNA expression of CYP3A4, CYP3A5, CYP3A7, CYP3A43, and MDR1 in 45 PTCLs was compared by correlation analysis. The results obtained for CYP3A4 are shown with the correlation coefficients and P values.
Since the different CYP3A genes were simultaneously overexpressed in specific tumors, we performed correlation analysis and found that CYP3A4 expression correlated with CYP3A5 and CYP3A7 (Figure 1B). When MDR1 expression was analyzed, a significant correlation was found with CYP3A7, CYP3A43, and CYP3A4. Therefore, the 4 CYP3A enzymes and MDR1, although showing differences in their mRNA levels, have a coordinated expression in PTCL tumors, which suggests a common regulatory mechanism.
T-cell lines and PTCLs show recurrent gains of the 7q21 region containing CYP3A and MDR1 genes
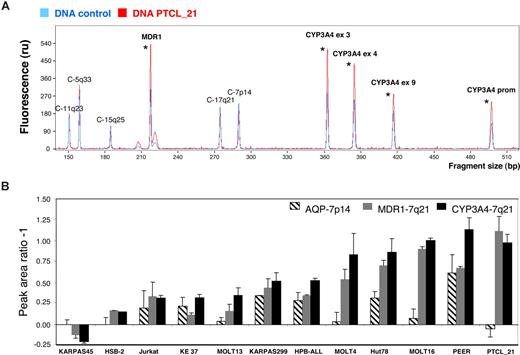
To identify the molecular mechanisms involved in the altered CYP3A4 tumoral expression, we set up an MPA technique able to detect changes in CYP3A4 and MDR1 copy numbers using genomic DNA (Figure 2A). Initially, we used T-cell lymphoma cell lines because they are homogeneous tumoral cell populations with no contaminating nontumoral genomic DNA, which facilitates the technique. We included 11 cell lines in the study and found that 9 showed simultaneous increases in CYP3A4 and MDR1 values, indicating that a region containing both genes (located at 7q21 and separated by 12 million base pairs (Mbp) was gained. Most cell lines also showed 7p gains, although to a lower degree. In general, the cell lines followed 2 different gain patterns: (i) gain of both CYP3A4 and MDR1 genes in 7q21 but minimal changes in the 7p AQP1 gene, such as MOLT4 and MOLT16; and (ii) gain of all 7q21 and 7p genes, such as Karpas 299, HPB-ALL, Hut-78, and Peer (Figure 2B). When we used this technique to study PTCL tumors with a high content of tumoral cells, we could also detect in some cases clear gains affecting CYP3A4 and MDR1 genes by MPA. For example, PTCL-21 had an increased number of the 7q21 genes, whereas no changes in 7p could be detected (Figure 2A,B, last column).
CYP3A4 and MDR1 copy number analysis by multiplex-PCR analysis (MPA). Ten DNA fragments with sizes ranging from 150 to 501 bp were amplified by PCR using genomic DNA and specific primers: 4 fragments corresponded to CYP3A4 (7q21), 1 to MDR1 (7q21), and 5 to additional fragments used as controls (C-) and located in different chromosomes. (A) The chromatogram of a DNA sample corresponding to PTCL-21 (red) was normalized and compared with a control DNA sample (blue). Fragments showing gains are marked with an asterisk. (B) MPA data were obtained for 11 T-cell lines, the chromatograms for each cell line were normalized with a control DNA sample, and the mean peak fluorescence area was calculated. The values represent the ratio between the T-cell line peak area divided by the control peak area after subtracting 1 plus or minus SD (ie, no gains result in a ratio of 1 and subtraction of 1 will result in a value of 0). Error bars represent SD of 3 independent experiments.
CYP3A4 and MDR1 copy number analysis by multiplex-PCR analysis (MPA). Ten DNA fragments with sizes ranging from 150 to 501 bp were amplified by PCR using genomic DNA and specific primers: 4 fragments corresponded to CYP3A4 (7q21), 1 to MDR1 (7q21), and 5 to additional fragments used as controls (C-) and located in different chromosomes. (A) The chromatogram of a DNA sample corresponding to PTCL-21 (red) was normalized and compared with a control DNA sample (blue). Fragments showing gains are marked with an asterisk. (B) MPA data were obtained for 11 T-cell lines, the chromatograms for each cell line were normalized with a control DNA sample, and the mean peak fluorescence area was calculated. The values represent the ratio between the T-cell line peak area divided by the control peak area after subtracting 1 plus or minus SD (ie, no gains result in a ratio of 1 and subtraction of 1 will result in a value of 0). Error bars represent SD of 3 independent experiments.
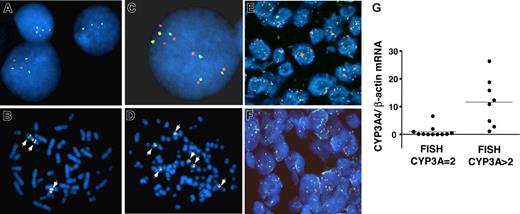
We selected 4 T-lymphoblastic cell lines (Jurkat, KE-37, MOLT 16, and Peer) and 1 peripheral T-cell line (Hut-78) to confirm the gains in CYP3A4 copies using FISH analysis. Between 3 and 6 CYP3A copies were found by FISH in these cell lines; in contrast, Karpas 45 cells, which by MPA showed a normal number of CYP3A copies, were also normal by FISH (data not shown). As shown in Figure 3A–D, MOLT 16 FISH analysis showed 3 CYP3A copies due to an isochromosome 7, whereas Peer cells had 6 CYP3A copies, likely by an increased number of chromosome 7, since 6 centromeres were also detected. We could also confirm by FISH the CYP3A4 gains in primary T-cell lymphomas with a high percentage of tumor cells, demonstrating that the 7q21 gains are not specific for the cell lines but rather a common event in T-cell tumorigenesis. As shown in Figure 3E, about 8 CYP3A copies could be observed in the tumor cells of PTCL-21, whereas the number of chromosome 7 centromeres was lower. The relatively small changes seen by MPA for PTCL-21 are probably caused by contamination of the total genomic DNA by nontumoral cells that decrease the effect observed by MPA. An additional PTCL case FISH is shown in Figure 3F. We obtained FISH data from 19 PTCL cases for which we had measured CYP3A content (9 PTCLu's, 6 AITLs, and 4 NK cell lymphomas). We observed that 7 (87%) of 8 of the cases with increased CYP3A copies showed high CYP3A4 mRNA expression, whereas only 1 (9%) of 11 of the cases with no alterations in CYP3A copy number had a high CYP3A4 mRNA expression (Figure 3G).
FISH analysis of CYP3A. Hybridization of the CYP3A probe (labeled in green) and a chromosome 7 centromeric probe (labeled in red) in tumoral T cells. (A,B) Representative FISH images of MOLT16 cells. (C,D) FISH images of PEER cells. (E,F) FISH images from 2 paraffin-embedded PTCLs, showing cells with multiple CYP3A and centromeric 7 signals. The case PTCL-21 corresponds to panel E. (G) CYP3A4 mRNA content of 19 PTCL cases classified according to the number of CYP3A copies per cell, as assessed by FISH (CYP3A = 2 corresponds to 2 copies per cell; CYP3A > 2 corresponds to cases with more than 2 copies per cell). The mean CYP3A4 mRNA content for PTCL cases with 2 CYP3A copies and for those with more than 2 CYP3A copies is shown with horizontal dashed lines (0.9- and 11.6-fold change relative to lymph nodes, respectively; P < .001).
FISH analysis of CYP3A. Hybridization of the CYP3A probe (labeled in green) and a chromosome 7 centromeric probe (labeled in red) in tumoral T cells. (A,B) Representative FISH images of MOLT16 cells. (C,D) FISH images of PEER cells. (E,F) FISH images from 2 paraffin-embedded PTCLs, showing cells with multiple CYP3A and centromeric 7 signals. The case PTCL-21 corresponds to panel E. (G) CYP3A4 mRNA content of 19 PTCL cases classified according to the number of CYP3A copies per cell, as assessed by FISH (CYP3A = 2 corresponds to 2 copies per cell; CYP3A > 2 corresponds to cases with more than 2 copies per cell). The mean CYP3A4 mRNA content for PTCL cases with 2 CYP3A copies and for those with more than 2 CYP3A copies is shown with horizontal dashed lines (0.9- and 11.6-fold change relative to lymph nodes, respectively; P < .001).
CYP3A4 expression influences chemotherapy response and survival
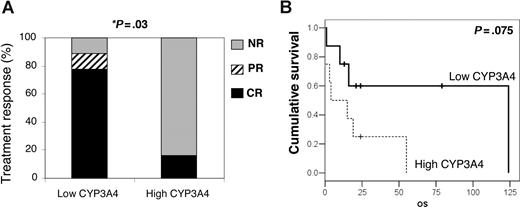
Because CYP3As and Pgp can influence therapy outcome, we studied whether the expression levels of CYP3A and MDR1 genes could predict the treatment response and/or survival of T-cell lymphoma patients. Since the different histologic T-cell lymphoma subtypes have differences in treatment response and survival rates, to avoid affecting the results by the intrinsic features of the tumors, we performed survival analysis only in the PTCLu subgroup. Clinical data and CYP3A4, CYP3A5, CYP3A7, CYP3A43, and MDR1 expression were compared, finding that CYP3A4 expression had a significant association with treatment response, whereas no significant differences were found for the other genes. In the group of patients with tumors overexpressing CYP3A4 (over 2-fold the expression in reactive tissue), only 1 (16%) of 6 cases had complete remission in contrast with 7 (77%) of 9 complete remissions and 1 (11%) of 9 partial remission in the group with low CYP3A4 expression (P = .011). As shown in Figure 4A, in the high-CYP3A4 group all patients except 1 did not respond to the treatment, whereas in the low-CYP3A4 group only 1 did not respond. No significant differences were found in the distribution of common clinical variables such as age, sex, stage of disease, or lactate dehydrogenase (LDH) levels in the group of PTCLu cases showing low or high expression of CYP3A4 (Table 1), indicating that difference in response was not likely effected by clinical parameters typically influencing PTCL survival. Most of the cases with high CYP3A4 had advanced disease stages and did not respond to therapy; however, in the low-CYP3A4 expression group, 3 of 5 cases in stages III and IV reached complete remission.
CYP3A4 tumoral expression predicts therapy response and survival of PTCLu patients. Twenty-three patients with PTCLu were divided into 2 groups: those with CYP3A4 mRNA tumoral expression less than 2-fold the control sample content (low CYP3A4), and those with CYP3A4 mRNA greater than 2-fold the control sample content (high CYP3A4). (A) After the initial therapy, patients were classified as nonresponders (NR) or as having partial remission (PR) or complete remission (CR). The therapy response of the patient groups was compared by Pearson chi-square (P = .02) and Fisher exact (P = .041) tests. (B) Kaplan-Meier curves were constructed to analyze the survival of the 2 groups (P = .075).
CYP3A4 tumoral expression predicts therapy response and survival of PTCLu patients. Twenty-three patients with PTCLu were divided into 2 groups: those with CYP3A4 mRNA tumoral expression less than 2-fold the control sample content (low CYP3A4), and those with CYP3A4 mRNA greater than 2-fold the control sample content (high CYP3A4). (A) After the initial therapy, patients were classified as nonresponders (NR) or as having partial remission (PR) or complete remission (CR). The therapy response of the patient groups was compared by Pearson chi-square (P = .02) and Fisher exact (P = .041) tests. (B) Kaplan-Meier curves were constructed to analyze the survival of the 2 groups (P = .075).
Clinical features of PTCLu cases with low or high expression of CYP3A4
| Clinical variables . | Low CYP3A4, n/total (%) . | High CYP3A4, n/total (%) . | P . |
|---|---|---|---|
| Age, y | |||
| Younger than 60 | 4/14 (29) | 2/7 (29) | — |
| Older than 60 | 10/14 (71) | 5/7 (71) | 1.0 |
| Sex | |||
| F | 5/14 (36) | 3/9 (33) | — |
| M | 9/14 (64) | 6/9 (67) | .907 |
| Stage | |||
| I-II | 5/10 (50) | 1/6 (17) | — |
| III-IV | 5/10 (50) | 5/6 (83) | .182 |
| LDH level | |||
| Low | 3/8 (38) | 2/4 (50) | — |
| High | 5/8 (63) | 2/4 (50) | .679 |
| Clinical variables . | Low CYP3A4, n/total (%) . | High CYP3A4, n/total (%) . | P . |
|---|---|---|---|
| Age, y | |||
| Younger than 60 | 4/14 (29) | 2/7 (29) | — |
| Older than 60 | 10/14 (71) | 5/7 (71) | 1.0 |
| Sex | |||
| F | 5/14 (36) | 3/9 (33) | — |
| M | 9/14 (64) | 6/9 (67) | .907 |
| Stage | |||
| I-II | 5/10 (50) | 1/6 (17) | — |
| III-IV | 5/10 (50) | 5/6 (83) | .182 |
| LDH level | |||
| Low | 3/8 (38) | 2/4 (50) | — |
| High | 5/8 (63) | 2/4 (50) | .679 |
— indicates not applicable.
Furthermore, Kaplan-Meier curves of cases in the high-CYP3A4 expression group showed a tendency to shorter survival times than patients with tumors with low CYP3A4 expression (P = .075; Figure 4B). Although these analyses were not carried out for other histologic subtypes, it is remarkable that the NK tumors, which have the highest CYP3A4 expression, usually do not respond to the standard treatments and have very short survival times.
CYP3A4 expression increases cell resistance to doxorubicin and etoposide
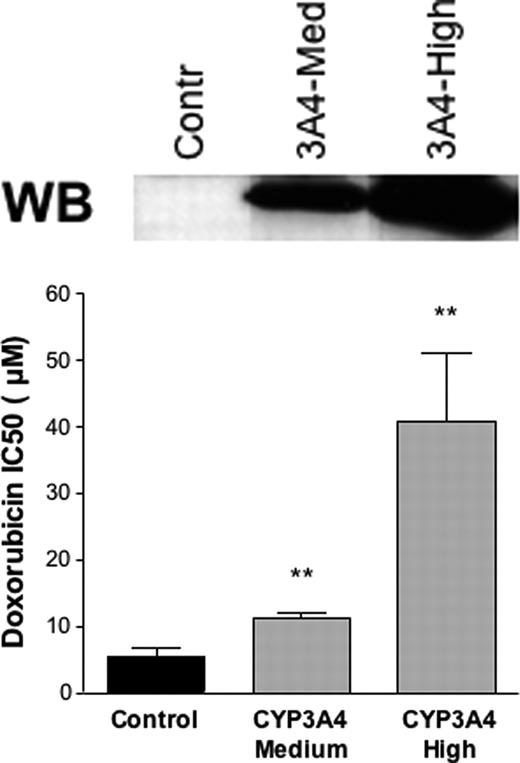
To test whether an increased CYP3A4 expression could result in altered chemotherapy sensitivity, we constructed HEK293 cells stably expressing CYP3A4 and incubated these and control cells with drugs commonly used in T-lymphoma treatment: vincristine, doxorubicin, and etoposide. The HEK293 cells showed a constitutive resistance to vincristine (data not shown) and, thus, the effect of CYP3A4 could not be assessed in this system. However, previous data had already shown that vincristine is a CYP3A4 substrate and that cells overexpressing CYP3A were more resistant to vincristine.26 As shown in Table 2, the different stable transfected clones used in the study had a large difference in CYP3A4 protein content and were classified as medium and high CYP3A4 expression. As shown in Figure 5, we found that the concentration of doxorubicin and etoposide that caused 50% cell death (IC50) was higher in the cells overexpressing CYP3A4 than in the control cells. In the case of doxorubicin, the expression levels of CYP3A4 modulated the response with 2- and 7.5-fold increases in cell-death resistance for medium and high CYP3A4-expressing cells, respectively. For etoposide, the full effect was already achieved at intermediate CYP3A4 expression levels, and in average CYP3A4 caused a 39-fold increase in drug resistance. These data, therefore, support the idea that an increased CYP3A4 expression can cause chemoresistance.
IC50 values
| Clones and drug used . | IC50, μM . |
|---|---|
| Doxorubicin | |
| Control | 5.4 ± 3.2 |
| CYP3A4-medium | 11.2 ± 1.8 |
| CYP3A4-high | 40.7 ± 25.3 |
| Etoposide | |
| Control | 0.03 ± 0.01 |
| CYP3A4-medium | 1.40 ± 0.42 |
| CYP3A4-high | 0.55 ± 0.31 |
| Clones and drug used . | IC50, μM . |
|---|---|
| Doxorubicin | |
| Control | 5.4 ± 3.2 |
| CYP3A4-medium | 11.2 ± 1.8 |
| CYP3A4-high | 40.7 ± 25.3 |
| Etoposide | |
| Control | 0.03 ± 0.01 |
| CYP3A4-medium | 1.40 ± 0.42 |
| CYP3A4-high | 0.55 ± 0.31 |
Data are means plus or minus SD of 3 replicates.
Effect of CYP3A4 expression on doxorubicin and etoposide toxicity. The survival of HEK293 cells stably expressing medium or high levels of CYP3A4 and control cells was measured for doxorubicin and etoposide at various concentrations by MTT assay. The concentration of the drug causing 50% of cell death (IC50) is plotted. Asterisks (**) indicate that the difference with the control is statistically significant (P < .05). A Western blot (WB) showing the different CYP3A4 expression levels in the cells used is shown.
Effect of CYP3A4 expression on doxorubicin and etoposide toxicity. The survival of HEK293 cells stably expressing medium or high levels of CYP3A4 and control cells was measured for doxorubicin and etoposide at various concentrations by MTT assay. The concentration of the drug causing 50% of cell death (IC50) is plotted. Asterisks (**) indicate that the difference with the control is statistically significant (P < .05). A Western blot (WB) showing the different CYP3A4 expression levels in the cells used is shown.
Discussion
To date, there are very limited data available regarding the biologic factors that affect prognosis and PTCL survival. Thus, it is clear that in addition to the classical factors included in the International Prognostic Index (IPI), more molecular markers are needed to accurately predict the course of PTCL disease and treatment response after chemotherapy. The low incidence of PTCL and the difficulty to collect simultaneously frozen and paraffined tissues and clinical data of the patient contributes to the scarcity of these types of studies. In this study, we gain insight into the mechanisms underlying the poor response rates to the common chemotherapy regimens used in these lymphomas.
Overexpression of Pgp and CYP3A enzymes in tumoral tissue has been associated with resistance to drug treatment: p-glycoprotein by pumping cytotoxic drugs out of the tumor cells, and CYP3A enzymes by inactivation of anticancer drugs.17,19,27 Interestingly, previous work from our group20 showed by microarray expression-profiling analysis of this PTCL series that CYP3A expression was variable among the samples. In this work we used quantitative RT-PCR for the 4 different CYP3A enzymes to validate the microarray data. We found that all CYP3A enzymes showed a large tumoral expression variation in all histologic PTCL subtypes and that some PTCL tumors had an overexpression of the CYP3A genes with respect to the nontumoral tissue (Figure 1). In addition, there was a correlation in the tumoral expression of CYP3As and MDR1, suggesting that there is a common mechanism responsible for the overexpression and that a synergistic drug-resistance mechanism could be expected in the tumors with high expression; in fact, both MDR1 and CYP3A are detoxifying proteins that have a large substrate specificity overlap. Since CYP3As and MDR1 genes are located in 7q21 and gains involving this region have been previously detected in 30% of T-cell tumors examined,22 we hypothesized that this could be the mechanism underlying the overexpression. Gains of 7q were found in an important proportion of T-cell lines. Although these cells mainly corresponded to lymphoblastic T-cell lines, which are not representative of PTCL, 2 peripheral T-cell lines (Hut-78 and Karpas 299) were also analyzed and the 7q21 gains occurred in both lymphoblastic and peripheral T cells. Despite the fact that MPA and FISH techniques are hampered by the contamination of the tumoral samples with normal cells, we were able to detect 7q21 gains not only in the T-cell lines but also in PTCL primary tumors. Furthermore, FISH analysis showed that 87% of the cases with CYP3A4 mRNA expression 2-fold higher than the normal tissue had more than 2 CYP3A signals per cell, whereas only 9% of the cases with low CYP3A4 mRNA expression showed an increased number of CYP3A signals by FISH (Figure 3G). These data support that somatic chromosomal aberrations in 7q21 are frequent and could be involved in the tumoral overexpression of CYP3A genes in PTCLs.
We hypothesized that tumoral CYP3A and P-glycoprotein28 expression could result in resistance to treatment. In fact, disease progression of PTCLs has been frequently observed during anthracycline-based chemotherapy, suggesting an inherent resistance of these tumors to conventional systemic therapy.28 When we compared the treatment response and survival of PTCLu patients with the tumoral MDR1 and CYP3A expression, we found that patients with tumors of high CYP3A4 expression had a significantly lower response to treatment and a tendency to shorter survival times when compared with tumors with low CYP3A4 expression. These data suggest that CYP3A4 tumoral expression could be a marker predicting treatment response and that it could likely influence PTCL survival. Interestingly, in the NK/T-cell lymphomas, which are one of the most aggressive lymphomas,29 the expression of CYP3A4 was the highest (Figure 1). To further investigate the effect of CYP3A4 on drug treatment, we constructed cell lines overexpressing CYP3A4 and showed that they exhibited an increased resistance to doxorubicin and etoposide, in agreement with previous studies performed with other cytotoxic drugs such as vincristine.26 Because these drugs are commonly used to treat lymphomas, we propose that tumoral CYP3A4 expression has a direct effect on PTCL's response to chemotherapy.
In conclusion, CYP3A4 is a key drug-metabolizing enzyme differently expressed in T-cell tumors. A poor response to standard chemotherapy was found in patients with tumors with high CYP3A4 expression, suggesting that tumoral CYP3A4 expression could be a marker for treatment response. Thus, patients with an increased CYP3A4 tumoral expression might benefit from a different chemotherapy regimen, such as increased doses of cytotoxic drugs or drugs that follow an inactivation pathway independent from CYP3A4/Pgp.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank all people from different hospitals in Spain who sent us tumors included in this study: C. Rivas, Fundacion Jiménez Diaz (Madrid); J. Alves Ferreira, Hospital La Paz (Madrid); J. Menarguez, Hospital Gregorio Maranon (Madrid); M. Mollejo, Hospital Virgen de la Salud (Toledo); C. Bellas, Hospital Puerta de Hierro (Madrid); E. Campo, Hospital Clinico (Barcelona); A. Saez, F. Jimenez, Hospital Clinico San Cecilio (Granada); S. Nieto, A. Acevedo, Hospital La Princesa (Madrid); J. L. Rodriguez Peralto, Hospital 12 Octubre (Madrid); M. Garcia Cosio, C. Montalbán, Hospital Ramón y Cajal (Madrid); J. Briones, I. Espinosa, Hospital San Pau (Barcelona); T. Alvaro, Hospital Verge de la Cinta (Tortosa); P. Dominguez, Hospital de Alcorcon (Madrid); J. Gonzalez Carralero, Hospital Xeral-Cies (Vigo); J. Forteza, Hospital Clinico Universitario (Santiago de Compostela); T. Contra, Hospital Nino Jesus (Madrid). This study was supported by grants from the Comunidad Autónoma de Madrid (CAM GR/SAL/0203/2004) and Fondo de Investigaciones Sanitarias (PI 061071). M.C. is a fellow of the Fondo de Investigaciones Sanitarias (FIS). C.R.-A. has a Fellowship from the “Ramon y Cajal” program from the Spanish Ministry of Education.
Authorship
Contribution: B.M.-D., C.R.-A., and J.B. designed the research; S.L., M.Z., C.R.-A., M.C., M.V.M., and C.M. performed the research; J.A. contributed to the collection and analysis of clinical data; J.C.C., A.C., and M.R. analyzed data; and B.M.-D. and C.R.-A. wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Beatriz Martinez-Delgado, Human Genetics Group, Human Cancer Genetics Program, Spanish National Cancer Centre, C/Melchor Fernández Almagro n °3, 28029, Madrid, Spain; e-mail: bmartinez@cnio.es