Human herpesvirus-8 (HHV-8), also known as Kaposi sarcoma–associated herpesvirus (KSHV), is etiologically linked to primary effusion lymphoma (PEL). At least 10 KSHV-encoded proteins with potential roles in KSHV-associated neoplasia have been identified. However, with few exceptions, these putative oncogenes were analyzed in heterologous systems only using overexpression of single genes. Thus, the pathogenetic relevance of most of these putative oncogenes remains essentially unclear. We used RNA interference (RNAi) to knock down the expression of several KSHV genes in cultured PEL cells carrying the KSHV genome. The viral interferon-regulatory factor-3 (vIRF-3) was found to be required for proliferation and survival of cultured PEL cells. Knock-down of vIRF-3 expression by various RNAi approaches unequivocally resulted in reduced proliferation and increased activity of caspase-3 and/or caspase-7. Thus, vIRF-3 can be seen as a bona fide oncogene of KSHV-associated lymphoma. Surprisingly, although the related Epstein-Barr virus (EBV) is usually sufficient to immortalize human B lymphocytes, silencing of vIRF-3 reduced the viability of both EBV− and EBV+ PEL cells. This suggests that KSHV is the driving force in the pathogenesis of PEL.
Introduction
DNA sequences of human herpesvirus-8 (HHV-8), also known as Kaposi sarcoma–associated herpesvirus (KSHV), have first been identified in biopsy specimens from AIDS-associated Kaposi sarcoma (KS).1 It is now clear that KSHV DNA is regularly found in all epidemiologic forms of KS,2,,,–6 in primary effusion lymphomas (PELs),7,8 and certain forms of multifocal Castleman disease.9 In these diseases, only few cells spontaneously reactivate the virus, as shown by expression of lytic cycle genes.10,11 Thus, virtually all PEL cells and KS spindle cells of late KS lesions carry the KSHV genome in a latent state.12 Although not formally proven, a remarkably tight epidemiologic relationship clearly suggests a pathogenetic role of KSHV in these malignant disorders. However, the viral genes and pathogenetic mechanisms involved are only partially elucidated. The complete or nearly complete nucleotide sequences of this first human rhadinovirus have been determined from both a PEL cell line13 and from 2 KS biopsy specimens.14,15 These showed that the known oncogenes of related viruses like Epstein-Barr virus (EBV) or herpesvirus saimiri are not conserved in KSHV. However, several KSHV genes with transforming potential have since been identified in cell culture and animal models. These include the transmembrane protein K1,16 Kaposin A encoded by reading frame K12,17 the KSHV-encoded viral interleukin-8 receptor homolog (vIL8R), also known as viral G-protein–coupled receptor homolog (vGPCR),18,19 the viral interferon (IFN)–regulatory factor-1 (vIRF-1) encoded by K9,20,21 and vIRF-3 encoded by K10.5,22 as well as 3 proteins encoded by the latently expressed so-called “oncogenic cluster,” namely the latency-associated nuclear antigen-1 (LANA-1), the viral cyclin homolog, and the viral Flice-inhibitory protein.23,–25
The KSHV-encoded vGPCR, a constitutively active transmembrane protein mimicking the cellular IL-8 receptor,19,26,27 is considered to play a major role in the pathogenesis of KS. As vGPCR is primarily expressed by the few lytically infected cells in a KS lesion, it is thought to exert an essential role by paraendocrine mechanisms (eg, by inducing expression of vascular endothelial growth factor).18,26 Even before the discovery of KSHV, endocrine mechanisms mediated by inflammatory cytokines and angiogenetic factors had been thought to play a major role in the pathogenesis of this heterogeneous sarcoma-like disorder.28 Viral factors involved in lymphomagenesis by KSHV remain more enigmatic. In contrast to KS, all the cells of KSHV-related PELs harbor the KSHV genome. Even after prolonged propagation in culture, KSHV is usually maintained in the cells and loss of the KSHV genome results in cell death,29 demonstrating that KSHV genes play a role in PEL cell survival. With respect to KSHV-associated lymphoproliferative disorders, the list of putatively oncogenic viral factors currently under debate includes K1,16 vIL-6,30 vIRF-1,31 K12/Kaposin,32 vFLIP,24 vCyclin r25, LANA-1,23 and K15 (reviewed in Schulz33 ). There are also data pointing at a possible oncogenic role of vIRF-3, also known as LANA-2.22 Work by Rivas and colleagues showed that vIRF-3 is expressed in latently infected PEL cells and inhibits p53-induced transcription in cotransfection experiments. Furthermore, it has been described that vIRF-3 is able to inhibit both p53-22 and protein kinase R (PKR)34 –induced apoptosis. Data pointing at a role of vIRF-3 in IFN signal transduction are more controversial. It may be either IFN-inhibitory in lytically infected cells35 or IFN-stimulatory in latency.36 In contrast to the related EBV and herpesvirus saimiri, de novo transformation of cultured primary lymphocytes by KSHV infection has not been possible so far. Thus, most studies aimed at elucidating mechanisms of KSHV lymphoma genesis rely on overexpression of isolated KSHV genes in cells not infected with KSHV. Only 2 groups made use of RNA interference (RNAi) to knock down KSHV genes in cultured primary effusion lymphoma cells, thus examining the role of KSHV genes in the context of KSHV-transformed lymphocytes.24,29 Transcripts originating from the “oncogenic cluster” encoding LANA-1, vCyclin, and vFlip were targeted by both groups. Essentially, they were consistently able to show that vFLIP and/or vCyclin are required for PEL cell survival. Similar studies aimed at other candidate oncogenes have not been published so far.
In the current study, we used RNAi to identify additional viral genes essential for the proliferation of cultured PEL cells. By using both stably expressed small hairpin RNAs (shRNAs) and transiently transfected synthetic small interfering RNAs (siRNAs), we show for the first time that knock-down of the vIRF homolog vIRF-3/K10.5/LANA-2 results in decreased proliferation and increased apoptosis. Expression of vIRF-3 was required for the survival of both EBV+ and EBV− PEL cells. This is the first demonstration that vIRF-3 functions as a bona fide oncogene in PEL cells. In addition, we confirm that KSHV is the essential oncogenic virus in dually infected PEL cells.
Methods
Cell lines and culture conditions
Suspension cultures of KSHV carrying BCBL-1,7,37 JSC-1,38 and BC-339 cells, as well as the KSHV− Burkitt lymphoma cell line BJAB and the multiple myeloma cell line INA-6,40 were maintained at 37°C with 7.5% CO2 in RPMI 1640 supplemented with 100 mg/mL gentamycin, 350 mg/mL l-glutamine, 1 mM sodium pyruvate (Sigma Chemical, St Louis, MO), 0.05 mM beta-mercaptoethanol (cell culture grade; Gibco BRL, Carlsbad, CA), and 10% (BJAB, BCBL-1) or 20% (BC-3, JSC-1) heat-inactivated fetal calf serum (FCS; PAA Laboratories, Pasching, Austria). INA-6 cells were cultured in the presence of 500 U/mL human IL-6 (Strathmann Biotech, Hannover, Germany). Expression of viral genes in BCBL-1, JSC-1, and BC-3 cells was stimulated with either 12-O-tetradecanoylphorbol 13-acetate (TPA; Sigma Chemical) at 25 ng/mL or 0.3 mM sodium butyrate (Sigma Chemical) for 3 to 4 days. Human embryonic kidney cell lines HEK293 and HEK293T were obtained from ATCC (Manassas, VA) and grown in Dulbecco modified Eagle medium (DMEM) supplemented with 10% fetal calf serum. The GP2–293 packaging cell line was obtained from Clontech (Palo Alto, CA) and maintained in DMEM supplemented with 10% FCS and penicillin/streptomycin.
Expression of recombinant vIRF-3 and generation of mAbs
A N-terminal fragment of the cDNA coding for vIRF-3 was amplified from the human PEL cell line BCBL-1 using oligonucleotides K10.1-Bam-2 (5′GATCGGATCCATGGCGGGACG-CAGGCTTACCTG-3′) and K10.1-Hind900R (5′GATCAAGCTTTTACCACGCCGCT-TGCATCC-3′). The polymerase chain reaction (PCR) fragment was cloned into the procaryotic expression vector pQE9 (Qiagen, Valencia, CA) via BamHI and HindIII restriction sites (italicized). The resulting plasmid pQK10-5N was verified by DNA sequencing. It encodes the N-terminal 298 amino acids of vIRF-3/K10.5 fused to an N-terminal histidine tag. Recombinant protein was expressed in Escherichia coli strain JM109 and purified under denaturing conditions as described previously.41
Following purification, approximately 50 μg of HIS-tagged K10.5-N protein was injected both intraperitoneally and subcutaneously into Lou/C rats. After an 8-week interval, a final boost was given intraperitoneally and subcutaneously 3 days before fusion. Fusion of the myeloma cell line P3 × 63-Ag8.653 with rat spleen cells was performed essentially as described.42 Hybridoma supernatants were tested in a solid-phase immunoassay using K10.5-N protein (3 μg/mL) adsorbed to polystyrene microtiter plates. Following incubation with culture supernatants for 1 hour, bound mAbs were detected using peroxidase-labeled goat anti-rat IgG antibodies and O-phenylenediamine as chromogen. Solid-phase enzyme-linked immunosorbent assay (ELISA) on microtiter plates coated with mouse anti-rat Ig antibodies was used to determine the immunoglobulin type with rat Ig class–specific (anti-IgM) and IgG subclass–specific mouse mAbs (Zymed, San Francisco, CA). 3G7 (rat IgG1) was selected for its reaction pattern in Western blot, immunoprecipitation (IP), and immunofluorescence.
Immunoblotting
Cells were harvested by centrifugation (10 minutes at 400g), washed twice in phosphate-buffered saline (PBS), and lysed in 1% Triton, 500 mM NaCl, 2 mM EDTA, and 2 mM EGTA in PBS. SDS sample buffer (5 ×; 4% SDS, 10% beta-mercaptoethanol, 20% glycerol, 2 mM EDTA, 120 mM Tris-HCl pH 6.8, and 0.1 mg bromphenol blue/mL) was added to the samples and an equivalent of 105 cells was loaded per lane. Proteins were separated on 8% discontinuous SDS–polyacrylamide gels. Western blot analyses were carried out as described previously.41 Briefly, proteins were transferred from 8% discontinuous SDS–polyacrylamide gels onto nitrocellulose membranes using the Hoefer SemiPhor TE70 blotting apparatus as described by the manufacturer (Pharmacia Biotech, Uppsala, Sweden). Membranes were probed with antibodies against vIRF-3/K10.5 (3G7 cell culture supernatant; dilution 1:100), β-actin (catalog no. A5441, dilution 1:5000; Sigma Chemical), cellular IRF-3 (cIRF-3; dilution 1:100; BD Biosciences PharMingen, San Jose, CA) or cIRF-4 (1:1000; Cell Signaling Technologies, Beverly, MA), respectively. Secondary antibodies were conjugated with horseradish peroxidase. Peroxidase activity was detected by electrochemoluminescence using ImageReader LAS-1000 (Fujifilm, Tokyo, Japan) CCD camera, and software. AIDA image analysis software (version 3.1; Raytest Germany, Straubenhardt, Germany) was used for relative quantification of bands.
Immunofluorescence
BCBL-1 and BJAB cells were washed twice in PBS. A cell suspension in PBS was applied to glass slides and allowed to dry at room temperature. Cells were then fixed on immunofluorescence assay slides using 3% paraformaldehyde in PBS for 10 minutes at room temperature. A total of 0.2% Triton X-100 in PBS was used to permeabilize the cells. Prior to incubation with antibodies, fixed cells were incubated with PBS containing 1% bovine serum albumin and 5% fetal calf serum for 30 minutes at room temperature. Incubation with the first antibody was performed for 30 minutes at room temperature followed by washing 3 times for 5 minutes in PBS. As a first antibody, rat monoclonal antibody 3G7 directed against KSHV protein vIRF-3 was used. For the detection of rat monoclonal antibodies, cells were then incubated with a sheep anti–rat CY3 conjugate (C2181; Sigma Chemical) diluted 1:300 in PBS followed by 3 washing steps in PBS as described. Cells were then visualized using a Zeiss Axioplan microscope with fluorescent lamp (Carl Zeiss Microimaging, Göttingen, Germany). The objective lens magnification was ×40 (Zeiss Plan Neofluar, NA(air): 0.75) with total final magnification of ×400. Images were taken using a Spot Diagnostic Imaging camera (Spot 1) and Metaview version 6.0 software (Diagnostic Instruments, Burroughs, MI).
Construction of shRNA expression vectors
For stable knock-down of viral genes by RNAi, the retroviral shRNA expression vector pSIREN-IRES-EGFP-RetroQ was constructed. It allows continuous expression of shRNA in transduced cells. In addition, cells successfully transduced by pSIREN-IRES-EGFP-RetroQ–derived retroviral particles can be selected by puromycin resistance and detected via expression of enhanced green fluorescent protein (EGFP). pSIREN-RetroQ-IRES-EGFP was derived from pSIREN-RetroQ (Clontech) by insertion of an internal ribosome entry site (IRES)–EGFP expression cassette downstream of the puromycin gene. For this purpose, a 1.3-kb BamHI/NotI restriction fragment containing the IRES-EGFP cassette was isolated from plasmid pIRES2-EGFP (Clontech). Ends were blunted with T4 DNA polymerase and the DNA fragment was ligated into pSIREN-RetroQ digested with EcoRV. Oligonucleotides for shRNAs were designed with the siRNA Hairpin Oligonucleotide Sequence Designer Tool (Clontech). They contained (5′ to 3′) a BamHI site, the respective siRNA sequence (italicized), a loop region, the complementary siRNA sequence (italicized), an RNA polymerase III termination sequence, and an EcoRI cloning site (sh976 sense: 5′GATCCCGTGTAGCAGGGGAATATGTGTTCAAGAGACACATATTCCCCTGCTACATTTTTTG-3′; sh976 antisense: 5′AATTCAAAAAATGTAGCAGGGGAATATGTGTCTCTTGAAC-ACATATTCCCCTGCTACACGG-3′). Oligonucleotides were annealed in 10 mM Tris and 20 mM NaCl (pH 7.6) by heating to 95°C for 2 minutes followed by slowly cooling to room temperature. Double-stranded oligonucleotides were then inserted into the retroviral vector pSIREN-RetroQ-IRES-EGFP via BamHI and EcoRI restriction sites. The resulting shRNA expression plasmids were called pSiren-IRES-EGFP-sh976 (target at position 976 of the K10.5 coding sequence) and pSiren-IRES-EGFP-shN (random sequence). The correct sequence of all constructs was confirmed by automated DNA sequencing on an ABI3700 genetic analyzer (Applied Biosystems, Foster City, CA) according to the manufacturer's instructions.
Transient transfection
BC-3 and JSC cells were maintained at a density of 350 000 cells/mL for 3 days prior to transfection. A total of 106 PEL cells were transfected with 2.6 μg siRNA using HiPerFect transfection reagent (Qiagen) according to the manufacturer's instructions.
Production of pseudotyped retrovirus and shRNA expression by retroviral transduction
GP2–293 cells were seeded in 10-cm cell-culture dishes 1 day prior to transfection. At 70% density, cells were transfected with 10 μg pSiren-IRES-EGFP vector and 5 μg pVSV-G (Clontech) for pseudotyping using the calcium phosphate transfection method. In short, media was changed to chloroquine-containing media (25 μM) 1 hour before transfection. DNA was mixed with 180 μL CaCl2 (2 M) and distilled water was added up to 1.5 ml. A total of 1.5 mL HBS buffer (0.28 M NaCl, 0.05 M HEPES, and 1.5 mM Na2HPO4 pH 7) was added and the solution was mixed vigorously. After 10 minutes of incubation at 20°C, the transfection mixture was added drop-wise to the culture dish. On days 2 and 3 after transfection, media containing retroviral particles were collected. Particles were concentrated 40-fold by ultracentrifugation. A total of 2 mL of this concentrated retroviral particle solution (supplemented with 2 μg/mL polybrene) was used to infect 106 PEL cells. Both transfection efficiency of packaging cells and transduction efficiencies of PEL cells were monitored by EGFP expression. Transduced cells were selected with puromycin (2 μg/mL) from day 2 after transduction.
Synthetic siRNAs and RNAi assay
Synthetic siRNAs targeted at vIRF-3 were selected using the Qiagen siRNA design tool. The following target sequences were used: AAUGUAGCAGGGGAAUAUGUG, called si976; and AAGGCCAUUUGUGGGUGAAAA, called si463. As control, nonsense siRNA (called siN), which shows no homology to any known human or KSHV gene, was used (sequence: AAGACUACCGUUGUAUAGUAG). In order to determine the percentage of successfully transfected cells, siN RNA oligonucleotides were partially labeled with FAM at the 3′ end. Transfection efficiency was determined by fluorescence-activated cell sorting (FACS) analysis.
Cell proliferation assay
3H-thymidine incorporation assays were performed for measuring proliferation of PEL cells. Cells were incubated for 24 hours with media containing 0.37 MBq (10 μCi) 3H-thymidine/mL. Cells were then transferred onto a nitrocellulose sheet, and radioactivity was counted with a beta counter.
Apoptosis assay
For simultaneous determination of caspase-3 and caspase-7 activities in the cells, Caspase-Glo 3/7 Assay (Promega, Madison, WI) was used according to the manufacturer's instructions. Briefly, cell suspensions were incubated for 1 hour with an equal volume of Caspase-Glo 3/7 reagent. Luciferase activity of the sample, which corresponds to activities of effector caspase-3/7, was then measured with a microplate luminometer (Berthold Detection Systems, Pforzheim, Germany).
Results
Knock-down of vIRF-3 expression by shRNA represses proliferation of PEL cells
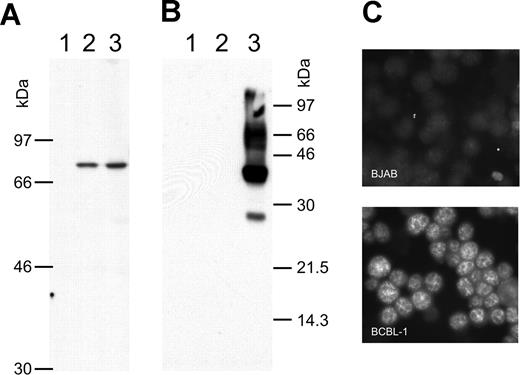
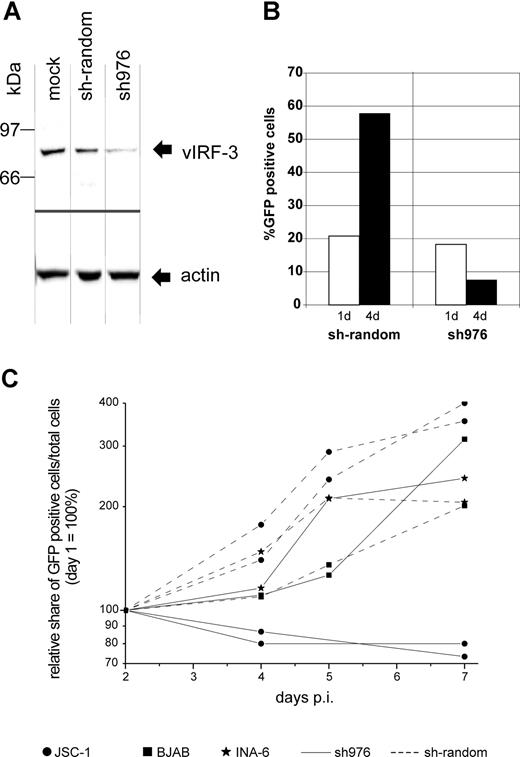
There is still a controversial debate on whether vIRF-3 is expressed in KSHV latency22,36 or not.35,43,44 We therefore examined the pattern of vIRF-3 expression in cultured PEL cells. By immunoblotting, anti–vIRF-3 monoclonal antibody 3G7 detected a single protein band in uninduced BCBL-1 cells (Figure 1A lane 2) that were negative for the lytic KSHV virion glycoprotein K8.141 (Figure 1B lane 2). Whereas stimulation of the KSHV lytic cycle by the phorbolester TPA strongly induced expression of glycoprotein K8.1 (Figure 1B lane 3), expression of vIRF-3 was not found to be increased (Figure 1A lane 3). Expression of vIRF-3 in latently infected PEL cells could also be confirmed by immunofluorescence (Figure 1C). Similar results as shown in Figure 1C using BCBL-1 cells were obtained for 2 other PEL cell lines, namely BC-1 and BC-3 (data not shown). This confirms that, at least in cultured PEL cells, vIRF-3 is the second latent nuclear antigen of KSHV.22 We first sought to determine which of the putative KSHV oncogenes may be required for the typical phenotype and continuous proliferation of PEL cells by stable knock-down via retroviral shRNA expression. We were able to generate several cell lines with stable knock-down of a viral gene by using retroviral shRNA expression constructs based on vector pSIREN-RetroQ-IRES-EGFP. These included the viral genes K1, K2 (vIL-6), K12, and vGPCR (Nadine Rohland, A.H., Chawaree Chaipan, Angela Holzer, E.W., and F.N., manuscript in preparation). Similar experiments were undertaken using retroviral vectors expressing sh976 targeted at the vIRF-3 encoded by K10.5. Roughly 20% of JSC-1 cells could be infected with pseudotyped retroviral particles, as was concluded from EGFP expression analysis by FACS 24 hours after infection. As shown in Figure 2A, this resulted in about 80% reduction of vIRF-3 expression compared with mock-treated cells or cells transduced by a retroviral vector expressing randomized shRNA (Figure 2A left lane). Expression of actin was not altered (Figure 2A bottom). Cells were then cultured in the presence of puromycin to select for cells carrying the retroviral vector. The proportion of EGFP-expressing cells increased from 20% to almost 60% 3 days later in sh-random transduced cells, but declined from 20% to less than 10% in sh976-transduced cells in the same time (Figure 2B). In a next series of experiments the KSHV− and EBV− B-cell lines BJAB and INA-6 were included as controls to exclude nonspecific effects of shRNAs or retroviral vectors. BJAB, INA-6, and JSC-1 cells were transduced with either sh976 or random shRNA retroviral vectors. Cells were then kept under puromycin selection and monitored for EGFP expression on days 1, 4, and 7 after transduction by FACS analysis. Both BJAB and INA-6 cells transduced with either sh-random or anti–vIRF-3 shRNA vectors all continued to expand at similar rates (Figure 2C squares and asterisks, respectively). KSHV+ JSC-1 cells transduced with a retroviral vector expressing nonsense shRNA also expanded about 3- to 4-fold within 1 week. However, KSHV+ JSC-1 cells expressing an shRNA against vIRF-3 failed to proliferate (Figure 2C continuous lines labeled with filled circles).
vIRF-3 is constitutively expressed in primary effusion lymphoma cells. Whole-cell lysates from the human KSHV− B-cell line BJAB (lane 1), KSHV+ BCBL-1 cells (lane 2), and BCBL-1 cells activated by TPA for 4 days (lane 3) were separated on 8% (A) or 12% (B) discontinuous SDS–polyacrylamide gels. Separated proteins were transferred onto nitrocellulose membranes and incubated with either monoclonal antibody 3G7 generated against a N-terminal fragment of vIRF-3 (A), or antibody BS555 directed against the virion glycoprotein K8.1.45 Whereas the various forms of gpK8.1 were abundantly expressed in BCBL-1 cells induced with TPA (panel B lane 3) and not detectable without induction (panel B lane 2), the relatively moderate expression level of vIRF-3 remains essentially unaltered by TPA treatment (panel A lanes 2-3). The immunofluorescence in panel C shows that vIRF-3 is present in the nuclei of latently infected BCBL-1 cells. BCBL-1 cells (lower photomicrograph) or KSHV− BJAB cells were fixed onto glass slides and incubated with monoclonal antibody 3G7 directed against recombinant vIRF-3, followed by detection with fluorescein-labeled antibodies against rat IgG. A fine-grained nuclear fluorescence was clearly detectable in virtually all BCBL-1 cells, but not in the KSHV− human B-cell line BJAB.
vIRF-3 is constitutively expressed in primary effusion lymphoma cells. Whole-cell lysates from the human KSHV− B-cell line BJAB (lane 1), KSHV+ BCBL-1 cells (lane 2), and BCBL-1 cells activated by TPA for 4 days (lane 3) were separated on 8% (A) or 12% (B) discontinuous SDS–polyacrylamide gels. Separated proteins were transferred onto nitrocellulose membranes and incubated with either monoclonal antibody 3G7 generated against a N-terminal fragment of vIRF-3 (A), or antibody BS555 directed against the virion glycoprotein K8.1.45 Whereas the various forms of gpK8.1 were abundantly expressed in BCBL-1 cells induced with TPA (panel B lane 3) and not detectable without induction (panel B lane 2), the relatively moderate expression level of vIRF-3 remains essentially unaltered by TPA treatment (panel A lanes 2-3). The immunofluorescence in panel C shows that vIRF-3 is present in the nuclei of latently infected BCBL-1 cells. BCBL-1 cells (lower photomicrograph) or KSHV− BJAB cells were fixed onto glass slides and incubated with monoclonal antibody 3G7 directed against recombinant vIRF-3, followed by detection with fluorescein-labeled antibodies against rat IgG. A fine-grained nuclear fluorescence was clearly detectable in virtually all BCBL-1 cells, but not in the KSHV− human B-cell line BJAB.
Knock-down of vIRF-3 expression in JSC-1 cells by retroviral shRNA is associated with reduced proliferation. KSHV+ JSC-1 cells as well as KSHV− BJAB and INA-6 cells were transduced with VSV-G pseudotyped retrovirus harvested from the supernatant of transfected GP2–293 cells and kept under puromycin selection for up to 7 days. Data from 3 independent experiments are shown. (A) Western blot performed using monoclonal antibody against vIRF-3 (3G7). JSC-1 cells were harvested 3 days after transduction with retroviral shRNA expression constructs or mock treatment. Equal amounts of total cell protein (5 × 105 cells) were loaded per lane and separated on 8% discontinuous SDS-PAGE. Expression of sh976 targeted at vIRF-3 resulted in an about 80% reduction of vIRF-3 expression. Expression of actin, however, remained unchanged. (B) Expansion of vector-carrying cells as quantified via EGFP expression analysis by FACS; the percentage of EGFP-expressing cells was determined 24 hours and 4 days after retrovirus transduction. Cells were maintained in media containing puromycin. Cells transduced with retroviral vector and thus EGFP+ and puromycin resistant continued to proliferate in the case of sh-random (left). However, when vIRF-3 expression was knocked down by sh976, the percentage of EGFP+ transduced cells in the culture declined despite puromycin selection (right). (C) PEL cells (2 independent experiments represented by 2 pairs of dotted lines with ● and 2 solid lines with ●), BJAB cells (■), and cIRF-4+ INA-6 cells (*) were transduced with retroviral particles expressing sh976 targeted at vIRF-3 (solid lines) or nonsense shRNA (interrupted lines). Cells were kept under puromycin selection and the relative growth of EGFP-expressing cells was monitored for 7 days by FACS analysis. Whereas JSC-1 cells transduced with nonsense shRNA–expressing retrovirus continued to grow under puromycin selection (● connected by dotted lines), JSC-1 cells expressing the shRNA targeted at vIRF-3 failed to expand (solid lines with ●). The KSHV− cell lines BJAB and INA-6 were used as a control (■ and *, respectively). BJAB and INA-6 cells continued to proliferate with both nonsense (■ or * and continuous lines) and shRNA (■ and * and dotted lines). p.i. indicates postinfection.
Knock-down of vIRF-3 expression in JSC-1 cells by retroviral shRNA is associated with reduced proliferation. KSHV+ JSC-1 cells as well as KSHV− BJAB and INA-6 cells were transduced with VSV-G pseudotyped retrovirus harvested from the supernatant of transfected GP2–293 cells and kept under puromycin selection for up to 7 days. Data from 3 independent experiments are shown. (A) Western blot performed using monoclonal antibody against vIRF-3 (3G7). JSC-1 cells were harvested 3 days after transduction with retroviral shRNA expression constructs or mock treatment. Equal amounts of total cell protein (5 × 105 cells) were loaded per lane and separated on 8% discontinuous SDS-PAGE. Expression of sh976 targeted at vIRF-3 resulted in an about 80% reduction of vIRF-3 expression. Expression of actin, however, remained unchanged. (B) Expansion of vector-carrying cells as quantified via EGFP expression analysis by FACS; the percentage of EGFP-expressing cells was determined 24 hours and 4 days after retrovirus transduction. Cells were maintained in media containing puromycin. Cells transduced with retroviral vector and thus EGFP+ and puromycin resistant continued to proliferate in the case of sh-random (left). However, when vIRF-3 expression was knocked down by sh976, the percentage of EGFP+ transduced cells in the culture declined despite puromycin selection (right). (C) PEL cells (2 independent experiments represented by 2 pairs of dotted lines with ● and 2 solid lines with ●), BJAB cells (■), and cIRF-4+ INA-6 cells (*) were transduced with retroviral particles expressing sh976 targeted at vIRF-3 (solid lines) or nonsense shRNA (interrupted lines). Cells were kept under puromycin selection and the relative growth of EGFP-expressing cells was monitored for 7 days by FACS analysis. Whereas JSC-1 cells transduced with nonsense shRNA–expressing retrovirus continued to grow under puromycin selection (● connected by dotted lines), JSC-1 cells expressing the shRNA targeted at vIRF-3 failed to expand (solid lines with ●). The KSHV− cell lines BJAB and INA-6 were used as a control (■ and *, respectively). BJAB and INA-6 cells continued to proliferate with both nonsense (■ or * and continuous lines) and shRNA (■ and * and dotted lines). p.i. indicates postinfection.
Knock-down of vIRF-3 expression by synthetic siRNAs represses proliferation of PEL cells
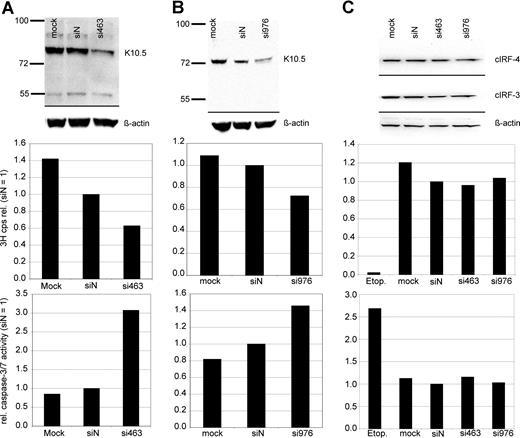
Experiments on vIRF-3 silencing with retrovirally expressed shRNAs were controlled with synthetic siRNAs. In order to exclude potential sequence-related off-target effects, 2 different siRNA sequences were used. The target sequences started at positions 463 and 976 of the vIRF-3 coding area, respectively. More than 90% of PEL cells could be transfected with these small molecules, as determined by FACS analysis after transfection with fluorescently labeled RNA oligonucleotides (data not shown). JSC-1 cells were transfected with either nonsense siRNA, mock treated, or 1 of 2 siRNAs targeted at vIRF-3, si463 (Figure 3A) or si976 (Figure 3B). At 2 days after transfection of PEL cells, 3H-thymidine was added to the cell culture media and proliferation was measured by 3H incorporation into DNA 24 hours later. Knock-down of vIRF-3 expression was checked by immunoblot and quantified by densitometry. Figure 3 shows representative examples. The amount of vIRF-3 protein was reduced by 47% 2 days after transfection with si463 (Figure 3A top). Actin levels remained essentially unchanged, and there was no marked effect of the nonsense siRNA on vIRF-3 expression (Figure 3A top). Knock-down of vIRF-3 by si463 was associated with about 40% reduction of cell proliferation as measured via incorporation of 3H-thymidine (Figure 3A middle). Knock-down of vIRF-3 expression by si976 in JSC-1 cells was slightly less effective (Figure 3B top), and resulted in roughly 30% reduction of 3H-thymidine incorporation (Figure 3B middle). Both cellular IRF-3 (cIRF-3) and IRF-4 (cIRF-4) are expressed in cells of the human myeloma cell line INA-6 (Figure 3C top). Although the target sequences of neither si463 nor si976 are conserved in cellular IRF-3 (cIRF-3) or IRF-4 (cIRF-4), INA-6 cells were included as a control to exclude any effect of transfected siRNAs on expression of cIRF-3 or cIRF-4, the closest cellular homolog of vIRF-3. As shown in Figure 3C, transfection of si463 or si976 did not influence cIRF-3 or cIRF-4 expression (Figure 3C top). Accordingly, neither proliferation rate nor caspase-3/7 activity was altered in si463- or si973-transfected cells compared with siN transfected INA-6 cells (Figure 3C middle and bottom). In a series of 10 experiments, dually EBV+ and KSHV+ JSC-1 cells were used (data not shown). To assess the influence of vIRF-3 silencing on proliferation, only experiments where at least 40% silencing of vIRF-3 was achieved were included for further analysis (6 of 10 independent experiments; mean vIRF-3 knock-down of 60%). si463–mediated silencing of vIRF-3 expression was associated with a mean reduction of cell proliferation by 44% (compared with siN; P = .002). Although the mean rate of proliferation of cells transfected with nonsense siRNA was also reduced in comparison to mock-transfected cells, this reduction was not statistically significant (P = .064; 2-sided t test).
Reduction of PEL cell proliferation and induction of caspase activity by transient knock-down of vIRF-3 expression. JSC-1 cells were transfected with synthetic RNA oligonucleotides directed against vIRF-3 (panel A, si463; panel B, si976), nonsense siRNA (siN), or mock-transfected (mock) and cultured for 2 days. The KSVH− cIRF-4 expression–positive myeloma cell line INA-6 was included as a control (C). At 2 days after transfection, two-thirds of the cells were harvested for both analysis of vIRF-3 expression by Western blot and enzymatic assay of caspase-3/7 activity. 3H-thymidine was added to the remaining cells, and proliferation was measured via 3H incorporation into DNA 24 hours later. Transfection of synthetic siRNAs resulted in 47% (panel A top) or 40% (panel B top) knock-down of vIRF-3 protein by si463 or si976, respectively. This was associated with a reduction of 3H-thymidine incorporation into DNA by 37% (panel A middle) or 28% (panel B middle), respectively. Reduced 3H-thymidine incorporation was accompanied by a pronounced increase of caspase-3/7 activity (panels A,B bottom). Expression of both cIRF-3 and cIRF-4 was not altered by any of the siRNAs in INA-6 cells (C). Cell proliferation and caspase-3/7 activity remained unchanged in these KSHV− cells, whereas apoptosis could be effectively induced by etoposide treatment (etop.).
Reduction of PEL cell proliferation and induction of caspase activity by transient knock-down of vIRF-3 expression. JSC-1 cells were transfected with synthetic RNA oligonucleotides directed against vIRF-3 (panel A, si463; panel B, si976), nonsense siRNA (siN), or mock-transfected (mock) and cultured for 2 days. The KSVH− cIRF-4 expression–positive myeloma cell line INA-6 was included as a control (C). At 2 days after transfection, two-thirds of the cells were harvested for both analysis of vIRF-3 expression by Western blot and enzymatic assay of caspase-3/7 activity. 3H-thymidine was added to the remaining cells, and proliferation was measured via 3H incorporation into DNA 24 hours later. Transfection of synthetic siRNAs resulted in 47% (panel A top) or 40% (panel B top) knock-down of vIRF-3 protein by si463 or si976, respectively. This was associated with a reduction of 3H-thymidine incorporation into DNA by 37% (panel A middle) or 28% (panel B middle), respectively. Reduced 3H-thymidine incorporation was accompanied by a pronounced increase of caspase-3/7 activity (panels A,B bottom). Expression of both cIRF-3 and cIRF-4 was not altered by any of the siRNAs in INA-6 cells (C). Cell proliferation and caspase-3/7 activity remained unchanged in these KSHV− cells, whereas apoptosis could be effectively induced by etoposide treatment (etop.).
Reduced vIRF-3 expression is associated with activation of caspase-3 and/or caspase-7 in PEL cells
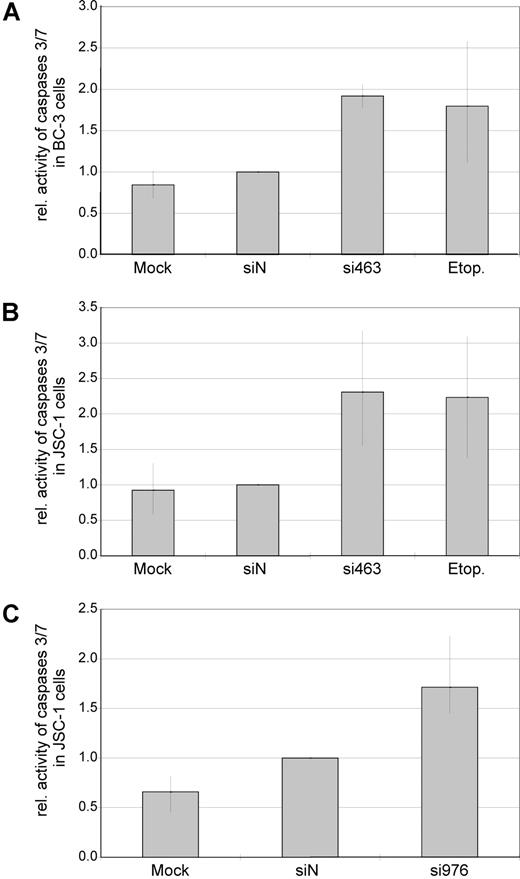
Reduced proliferation in cell culture can be caused by either cell-cycle arrest, increased apoptosis, or both. To assess whether silencing of vIRF-3 affected apoptosis, activity of caspase-3 and/or caspase-7 was measured in transiently siRNA-transfected PEL cells. Data from 2 representative experiments are shown in Figure 3A (JSC-1 cells transfected with si463) and Figure 3B (JSC-1 cells transfected with si976). Silencing of vIRF-3 by approximately 40% was accompanied by a 3-fold (si463) or 1.5-fold (si976) increase of caspase-3/7 activity (Figure 3 bottom). In a larger series of experiments, both the dually EBV+ and KSHV+ PEL cell line JSC-1 and the KSHV+ but EBV− PEL cell line BC-3 were used. At least 40% reduction of vIRF-3 expression was seen in 13 of 19 independent experiments. Data from these 13 experiments were included in the analysis. Arithmetic means for caspase activity from all 13 experiments are shown in Figure 4. Mean induction of caspase-3/7 activity for both JSC-1 and BC-3 cells with si463 was 2-fold (Figure 4A,B, respectively). Compared with si463, the vIRF-3 knock-down by si976 was slightly less efficient (Figure 3B), which correlated with a lower increase of caspase activity (Figure 4C). The 2-fold induction of caspase 3/7 activity by si463 was comparable to the effect of the strong apoptosis inducer etoposide (Figure 4A,B). Thus, induction of apoptosis is likely to be the main reason for the observed reduction in 3H-thymidine incorporation following vIRF-3 silencing.
Knock-down of expression is associated with increased caspase activity in both EBV− and EBV+ PEL cells. EBV−, KSHV+ PEL cells (BC-3; A) and dually EBV/KSHV-positive PEL cells (JSC-1; B,C) were transfected with synthetic siRNAs targeted at vIRF-3 (si463, A,B; si976, C). Mock-transfected cells (HiPerFect only) or scrambled siRNAs (siN) were used as negative controls. Cells treated with the apoptosis-inducing agent etoposide (etop.) served as a positive control for caspase induction. Cells were harvested 2 days after transfection, and activity of caspase-3/7 was measured in duplicates. Arithmetic means and total range of values are given from 4 (A), 6 (B), or 3 (C) independent transfection experiments (data were normalized to siN in individual experiments). Although a slight increase of caspase activity could be observed by transfection of scrambled siRNAs as compared with mock-transfected cells, this difference was statistically not significant (P = .713). In contrast, transfection of si463 as well as si976 in either BC-3 or JSC-1 cells resulted in an increase of caspase activity (siN compared with si463: range, 1.5- to 3.1-fold; P < .001 2-sided t test for paired samples. Absolute values were used for statistical calculations).
Knock-down of expression is associated with increased caspase activity in both EBV− and EBV+ PEL cells. EBV−, KSHV+ PEL cells (BC-3; A) and dually EBV/KSHV-positive PEL cells (JSC-1; B,C) were transfected with synthetic siRNAs targeted at vIRF-3 (si463, A,B; si976, C). Mock-transfected cells (HiPerFect only) or scrambled siRNAs (siN) were used as negative controls. Cells treated with the apoptosis-inducing agent etoposide (etop.) served as a positive control for caspase induction. Cells were harvested 2 days after transfection, and activity of caspase-3/7 was measured in duplicates. Arithmetic means and total range of values are given from 4 (A), 6 (B), or 3 (C) independent transfection experiments (data were normalized to siN in individual experiments). Although a slight increase of caspase activity could be observed by transfection of scrambled siRNAs as compared with mock-transfected cells, this difference was statistically not significant (P = .713). In contrast, transfection of si463 as well as si976 in either BC-3 or JSC-1 cells resulted in an increase of caspase activity (siN compared with si463: range, 1.5- to 3.1-fold; P < .001 2-sided t test for paired samples. Absolute values were used for statistical calculations).
Discussion
We show here for the first time that vIRF-3, also known as LANA-2 or K10.5, is essential for the survival of cultured PEL cells and can thus be considered a bona fide oncogene of KSHV. Silencing of vIRF-3 expression by RNAi using 2 different target sequences (463 and 976) deployed into 2 different PEL cell lines (BC-3, JSC-1) by 2 different methods (lipofection and retroviral transduction) was unequivocally associated with an impaired growth of cultured PEL cells and increased apoptosis. Notably, this included both EBV− and EBV+ PEL cells, confirming the essential oncogenic role of KSHV in this highly malignant lymphoma. RNAi is certainly the most efficient method of gene silencing available. However, it is still debated when and where nonspecific or off-target effects must be expected.46 These include induction of an IFN response through dsRNA,47 nonspecific effects through saturation of the cellular RNAi machinery, or activation of toll-like receptors (TLRs), especially TLR7.48 In addition, sequences selected for silencing of 1 particular gene might also match to other genes sufficient for gene silencing inducing so-called off-target effects.
To address these concerns, all sequences used for silencing were thoroughly checked for similarity to KSHV, EBV, and the human genome. Accordingly, expression of cIRF-3 or cIRF-4 was not altered when siRNAs targeted at vIRF-3 were transfected in KSHV− INA-6 cells (Figure 3C) or KSHV+ JSC-1 cells (data not shown). Sequence motifs known to be required for induction of an IFN response via TLRs49 were also not present in any of the siRNA sequences used here. In addition, 2 different target sequences were selected for vIRF-3 (si463 and si976). Silencing with both siRNAs yielded comparable results. To exclude nonspecific effects related to the route of RNAi application, we used transient gene silencing by liposomal delivery of synthetic siRNAs as well as retroviral gene transfer for continuous expression of shRNAs. Again, results obtained by either method were consistent. In addition, we were not able to detect an IFN response in transfected or transduced cells by IFN-α ELISA (data not shown). Although a modest growth reduction was often seen with scrambled siRNAs as compared with mock-treated cells, this growth reduction was statistically not significant and much less pronounced than the growth reduction seen in cells with knock-down of vIRF-3 expression. In addition, cell viability of KSHV− B cells was not reduced by siRNAs against vIRF-3 (Figure 2C). In summary, silencing of vIRF-3 expression with 2 different target sequences always resulted in reduced cell viability and increased apoptosis compared with cells treated with randomized siRNAs or shRNAs, respectively. This proves that reduced cell viability and increased caspase activity were due to vIRF-3 silencing and not to other effects possibly associated with RNAi.
We confirmed by immunoblot and immunofluorescence that vIRF-3 is latently expressed in virtually all PEL cells, as first shown by Rivas and coworkers22 in contrast to several other groups.35,43,44 IRFs are a growing family of transcription factors that regulate expression of IFNs and IFN-regulated genes at a transcriptional level (for review, see Mamane et al50 and Nguyen et al51 ). Cellular IRFs bind to conserved cis-acting sequence elements frequently present in promoters of IFN-regulated genes. Induction of an antiviral state is only 1 characteristic feature of IFNs. The IFN system is also critical for cell proliferation and differentiation,52 especially of lymphoid cells. For example, whereas IRF-1 inhibits cell proliferation53 and acts in an antioncogenic manner,54 IRF-2 acts antagonistic to IRF-1 and has oncogenic potential.54 The lymphocyte-specific IRF-4, also known as multiple myeloma oncogene 1 (mum-1) due to its possible involvement in multiple myeloma pathogenesis,55 is another example for the role of IRFs in proliferation and cell differentiation. Mice deficient for IRF-4 do not develop a humoral immune response, lack mature B cells, and show reduced proliferation of T cells.56 Among the mammalian IRFs identified so far, vIRF-3 shares the most pronounced (but still weak) homology with the lymphocyte-specific IRF-4. IRF-4 has also been shown to have oncogenic activity in vitro.55 Cellular IRFs are thus exemplary for the tight connection between pathways of innate immunity and tumor suppression. Thus, by inhibiting tumor suppressor pathways also involved in innate immunity, viruses may eventually promote oncogenesis.57 Initially, vIRF-3 has been shown to inhibit IFN-α signaling via inactivation of cIRF-3 and cIRF-7.35 More recent work, however, showed the contrary.36 Work by 2 groups associates vIRF-3 with regulation of apoptosis. Using heterologous expression systems, Rivas et al showed that vIRF-3 is able to inhibit p53-mediated apoptosis,22 and Esteban and colleagues reported inhibition of PKR-induced activation of caspase-3.34 The latter is in good agreement with our findings. However, it is considered unlikely and remains to be shown whether PKR is activated in latently infected PEL cells without interferon stimulation. It is thus unclear which pathway is responsible for the activiation of caspase-3/7 by vIRF-3 knock-down in PEL cells. In addition, in contrast to these 2 reports,22,34 a third group claimed induction of apoptosis by vIRF-3 via inhibition of NFκB.58 It is likely that this discrepancy of findings published so far on vIRF-3 is at least in part due to the fact that different cell systems and high levels of vIRF-3 by transfection of expression plasmids were used. In contrast to previous work, we did not rely on heterologous expression of vIRF-3 in cells not infected by KSHV, but used RNAi-mediated silencing of vIRF-3 in naturally infected PEL cells. This showed that vIRF-3 is required for continuous proliferation of PEL cells and knock-down of vIRF-3 induces apoptosis. Until today, the function of only few other KSHV genes has been analyzed in PEL cells by RNAi: LANA-1, vCyclin, and vFLIP, which are translated from alternatively spliced or bicistronic mRNAs. Although LANA-1 has been shown to inhibit apoptosis in transfected cells, silencing of LANA-1 by RNAi did not induce apoptosis but reduced the number of copies of the viral episome.29 In contrast, targeting the bicistronic transcript encoding both vFlip and vCyclin resulted in growth arrest.24,29 In summary, we show that vIRF homolog K10.5/vIRF-3/LANA-2 is the second identified latently expressed bona fide oncogene in KSHV-associated lymphoproliferation which—like the vFLIP/vCyclinlocus—might serve as a target for future therapeutic strategies.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by Deutsche Forschungsgemeinschaft grant SFB 643, the IZKF Erlangen, the European Community research project TargetHerpes, the Frieda-Marohn Foundation, and the Mainzer Akademie der Wissenschaften und der Literatur.
Authorship
Contribution: E.W. designed the research plan and performed key laboratory experiments; Y.M. performed key laboratory experiments, including expression analysis and protein purification; A.H. performed laboratory experiments on cell growth and apoptosis; E.K. generated monoclonal antibodies targeted at vIRF-3; M.S. and B.F. provided logistical support and approved the final version of the manuscript; and F.N. designed the study, approved the data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Frank Neipel, Institut für Klinische und Molekulare Virologie, Universität Erlangen-Nürnberg, Schloßgarten 4, D-91054 Erlangen, Germany; e-mail: frank.neipel@viro.med.uni-erlangen.de.