Abstract
Spontaneous somatic reversions of inherited mutations are poorly understood phenomena that are thought to occur uncommonly in a variety of genetic disorders. When molecularly characterized, revertant cells have rarely exhibited more than one revertant genotype per patient. We analyzed individual allospecific T-cell clones derived from a Wiskott-Aldrich syndrome (WAS) patient identified by flow cytometry to have 10% to 15% revertant, WAS protein–expressing lymphocytes in his blood. Genotypic analysis of the clones revealed a remarkable diversity of deletions and base substitutions resulting in at least 34 different revertant genotypes that restored expression of WASp. A large fraction of these revertant genotypes were also identified in primary T cells purified from peripheral blood. These data suggest that the use of sensitive methods may reveal the presence of wide arrays of individual genotypic revertants in WAS patients and offer opportunities for further understanding of their occurrence.
Introduction
Somatic reversion has recently been described in several disorders.1 Reversion has been particularly prominent in severe immunodeficiency diseases, including Wiskott-Aldrich syndrome (WAS).2-10 One common feature of most diseases for which reversion has been observed is a strong selective advantage for corrected cells in vivo. When characterized molecularly, revertant cells have exhibited only one or rarely a few revertant genotypes per patient.1
WAS is an X-linked primary immunodeficiency disease resulting from defects in a single gene (WAS). WAS is composed of 12 exons encoding a cytoplasmic protein (WASp) that is involved in signal transduction pathways in hematopoietic cells and that is a key regulator of the actin cytoskeleton.11 In addition to thrombocytopenia, WAS patients present with both defective T- and B-cell function.11
We have been following a WAS patient with a nonsense mutation in exon 10 (995C>T; Arg321Stop). The 995C>T mutation results in no detectable WASp expression and is generally associated with severe clinical presentation.12,13 Two affected brothers and 2 affected nephews of our patient exhibited death within the first decade of life or the requirement for stem cell transplantation, whereas the clinical history of the proband is notable for far milder symptoms. In this report, we describe that this patient harbors WASp-expressing peripheral blood lymphocytes with a remarkable diversity of WAS revertant T-cells.
Methods
Peripheral blood was obtained under informed consent from Wiskott-Aldrich patients seen at the NIH under an NIH-approved protocol and in accordance with the Declaration of Helsinki.
Patient WAS4 was diagnosed at birth because of thrombocytopenia occurring in the context of 2 older brothers with WAS. The first blood draw analyzed in this report was obtained in March 2006 with the patient exhibiting his usual mild symptomatology. Approximately 3 months later, the patient was diagnosed with diffuse large B-cell lymphoma of the small intestine. He was treated with 6 cycles of rituximab plus cyclophosphamide/doxorubicin/vincristine/prednisone (R-CHOP) and is in complete remission 22 months after presentation. The second blood draw was obtained October 2006 between the third and fourth cycles of chemotherapy.
Peripheral blood mononuclear cells (PBMC) were cultured as described.14 Immunomagnetic beads (Dynal Biotech, Lake Success, NY) were utilized for isolation of CD3+, CD8+, or CD3+/CD8− cells from PBMCs. PBMCs, CD8+, or CD3+/CD8− cells (first blood draw) or CD3+cells (second blood draw) were seeded at low density in 96-well plates with allogeneic cells also as described.14 Clones were stimulated in Y10 media with 1 μg/mL phytohemagglutinin A and 10 ng/mL each of IL-2, IL-4, and IL-7 (YP247). Cytoplasmic WASp expression was analyzed as previously described.2
A 2.8-kb polymerase chain reaction (PCR) product was obtained by amplification of DNA from T-cell clones (Figure 1C) and either sequenced or seeded in a nested PCR reaction to yield a 1.3-kb product (Figure 1C) for sequencing. Genomic DNA from primary CD3+or CD3+/CD8− cells was subjected to nested PCR; the resultant 1.3-kb products were cloned and sequenced.
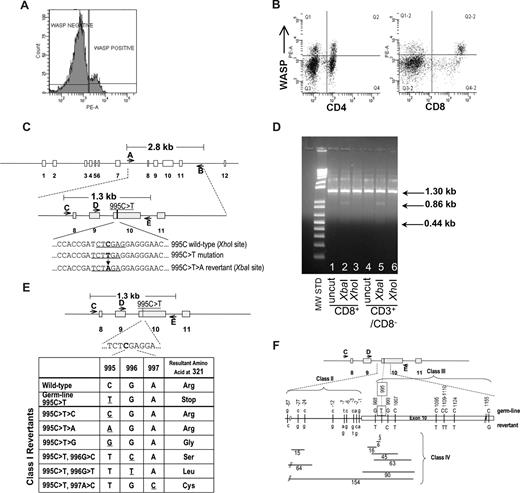
Identification of revertant cells in patient's T-lymphocytes. (A) FACS analysis showing presence of WASp in approximately 14% of patient's lymphocytes. (B) FACS analysis showing presence of WASp-expressing cells at 21% frequency in CD4+ and 44% frequency in CD8+ lymphocyte populations. The top right quadrants contain WASp+ cells coexpressing CD4 or CD8. (C) Schematic of WAS gene structure and relevant genotypes. The 995C>T>A reversion introduces a novel XbaI restriction enzyme site allowing detection of revertant genotype. (D) Examination of 995C>T>A revertant genotype in lymphocyte populations. XbaI digestion confirmed the presence of the 995C>T>A revertant in CD8+ and CD3+/CD8− T cells. The 1.3 kb PCR product for the 995C>T>A genotype yields 2 bands (0.86 kb, 0.44 kb) when digested with XbaI. (E) Class I revertants, identified in allospecific T-cell clones, involved single base pair substitutions (positions 995-997) resulting in replacement of the mutant 321stop codon with either the wild-type arg321 or the other amino acids indicated. (F) Schematic of class II (intron 9 base pair substitutions just upstream of the beginning of exon 10), class III (single or double base pair substitutions within exon 10), and class IV revertants (deletions of various size in intron 9 and/or exon 10).
Identification of revertant cells in patient's T-lymphocytes. (A) FACS analysis showing presence of WASp in approximately 14% of patient's lymphocytes. (B) FACS analysis showing presence of WASp-expressing cells at 21% frequency in CD4+ and 44% frequency in CD8+ lymphocyte populations. The top right quadrants contain WASp+ cells coexpressing CD4 or CD8. (C) Schematic of WAS gene structure and relevant genotypes. The 995C>T>A reversion introduces a novel XbaI restriction enzyme site allowing detection of revertant genotype. (D) Examination of 995C>T>A revertant genotype in lymphocyte populations. XbaI digestion confirmed the presence of the 995C>T>A revertant in CD8+ and CD3+/CD8− T cells. The 1.3 kb PCR product for the 995C>T>A genotype yields 2 bands (0.86 kb, 0.44 kb) when digested with XbaI. (E) Class I revertants, identified in allospecific T-cell clones, involved single base pair substitutions (positions 995-997) resulting in replacement of the mutant 321stop codon with either the wild-type arg321 or the other amino acids indicated. (F) Schematic of class II (intron 9 base pair substitutions just upstream of the beginning of exon 10), class III (single or double base pair substitutions within exon 10), and class IV revertants (deletions of various size in intron 9 and/or exon 10).
Results and discussion
WAS4 was first noted in 2004 to express normal levels of WASp in approximately 10% of his peripheral blood lymphocytes. A similar frequency of WASp+ lymphocytes was present in 2005 (14%; Figure 1A,B) and 2006 (data not shown). Direct sequencing of PCR-amplified exon 10 sequences from sorted WASp-expressing CD3+ T cells revealed the presence of a 995C>T>A genotypic reversion (Figure 1C,D), restoring Arg321 and the full-length wild-type WASp coding sequence.
In 2006, we prepared individual, primary allospecific T-cell clones from total PBMCs or T-cell subsets and WAS-specific DNA sequences (Figure 1C) were amplified by PCR and directly sequenced. T-cell clones, obtained from a single blood draw, contained the germline mutant 995C>T genotype, revertant 995C>T>A genotype, or 24 other putative revertant genotypes (Table 1 and Figure 1E,F). Sequencing of this same exon 10 region in 47 allospecific T-cell clones from a healthy control and 45 allospecific T-cell clones from patient WAS1 (carrying a 434insACGAGG mutation2,15 ) yielded no differences from wild-type sequence. Further, sequencing of genomic DNA encompassing exons 3 to 5 from 45 allospecific T-cell clones from this patient WAS4 revealed no differences from the wild-type sequence.
Genotype and phenotype of allospecific T-cell clones from WAS 995C>T patient
| Genotype (WT CGA-Arg) . | No. of clones carrying specified genotype . | WASp expression +/− . | Present in primary cells*CD3 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| First blood draw . | Second blood draw . | ||||||||
| CD4† . | CD8‡ . | ND . | Total . | % of total . | Total . | % of total . | |||
| 995C>T germline mutation (Stop) | 112§ | 8 | 15 | 135 | 50.6 | 69 | 50.0 | − | + |
| % CD4 or CD8 clones with germline mutation | 65 | 18 | |||||||
| Class I | |||||||||
| 995C>T>A (Arg) | 33 | 6 | 15 | 54 | 20.2 | 31 | 22.5 | + | + |
| 995C>T>C (Wt-Arg) | 0 | 0 | 2 | 2 | 0.7 | 0 | 0.0 | ND | + |
| 995C>T>G (Gly) | 3 | 1 | 3 | 7 | 2.6 | 3 | 2.2 | + | + |
| 995C>T, 996G>C (Ser) | 9 | 8 | 3 | 20 | 7.5 | 7 | 5.1 | + | + |
| 995C>T, 996G>T (Leu) | 0 | 0 | 0 | 1∥ | 0.4 | 0 | 0.0 | + | + |
| 995C>T, 997A>C (Cys) | 0 | 3 | 0 | 3 | 1.1 | 0 | 0.0 | + | + |
| CD4 or CD8 clones with class I genotype, % | 26 | 40 | |||||||
| Total class I | 86 | 32.2 | 41 | 29.7 | |||||
| Class II | |||||||||
| 995C>T, IVS9 −1G>A | 0 | 5 | 3 | 8 | 3.0 | 3 | 2.2 | + | + |
| 995C>T, IVS9 −2A>C | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, IVS9 −2A>G | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, IVS9 −3C>G | 1 | 11 | 2 | 14 | 5.2 | 4 | 2.9 | + | + |
| 995C>T, IVS9 −3C>T | 1 | 0 | 0 | 1 | 0.4 | 3 | 2.2 | + | + |
| 995C>T, IVS9 −6C>G | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, IVS9 −7T>G | 0 | 0 | 1 | 1 | 0.4 | 0 | 0.0 | ND | + |
| 995C>T, IVS9 −7T>A | 2 | 0 | 0 | 2 | 0.7 | 0 | 0.0 | + | − |
| 995C>T, IVS9 −12C>G | 2 | 0 | 0 | 2 | 0.7 | 2 | 1.4 | + | + |
| 995C>T, IVS9 −24C>G | 2 | 0 | 0 | 2 | 0.7 | 3 | 2.2 | + (weak) | + |
| 995C>T, IVS9 −27C>G | 0 | 0.0 | 1 | 0.7 | + (weak) | − | |||
| 995C>T, IVS9 −84G>C | 0 | 0.0 | 1 | 0.7 | + (weak) | + | |||
| CD4 or CD8 clones with class II genotype, % | 5 | 36 | |||||||
| Total class II | 30 | 11.2 | 20 | 14.5 | |||||
| Class III | |||||||||
| 995C>T, 988G>T | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, 999G>C | 0 | 0.0 | 1 | 0.7 | + (weak) | − | |||
| 995C>T, 1007C>T | 2 | 0 | 0 | 2 | 0.7 | 0 | 0.0 | + | − |
| 995C>T, 1095C>T | 0 | 0 | 1 | 1 | 0.4 | 0 | 0.0 | ND | + |
| 995C>T, 1109C>T, 1110C>T | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | ND | − |
| 995C>T, 1124C>T | 0 | 0 | 1 | 1 | 0.4 | 2 | 1.4 | + (weak) | + |
| 995C>T, 1155C>G | 0 | 0.0 | 1 | 0.7 | + (weak) | − | |||
| CD4 or CD8 clones with class III genotype, % | 2 | 0 | |||||||
| Total class III | 5 | 1.9 | 5 | 3.6 | |||||
| Class IV | |||||||||
| 995C>T, del IVS9 −84 to −21 (64 bp) | 0 | 1 | 0 | 1 | 0.4 | 0 | 0.0 | + | − |
| 995C>T, del IVS9 −32 to −18 (15 bp) | 0 | 0.0 | 2 | 1.4 | + | − | |||
| del IVS9 −74 to exon 10 1045 (154 bp) | 0 | 1 | 0 | 1 | 0.4 | 0 | 0.0 | + | + |
| del exon 10 968 to 1057 (90 bp, 30 aa) | 0 | 0 | 1 | 1 | 0.4 | 0 | 0.0 | ND | − |
| 995C>T, del exon 10 976 to 991 (16 bp) | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | + | − |
| del exon 10 984 to 1028 (45 bp, 15 aa) | 0 | 1 | 1 | 2 | 0.7 | 1 | 0.7 | + | − |
| del exon 10 988 to 1050 (63 bp, 21 aa) | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | ND | − |
| del exon 10 992 to 997 (6 bp, 2 aa) | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | + | − |
| 995C>T, del exon 10 996 to 998 (3 bp) | 2 | 0 | 0 | 2 | 0.7 | 0 | 0.0 | ND | − |
| CD4 or CD8 clones with class IV genotype, % | 3 | 7 | |||||||
| Total class IV | 10 | 3.7 | 3 | 2.2 | |||||
| Total no. of clones | 173 | 45 | 48 | 267 | 100.0 | 138 | 100.0 | ||
| CD4 or CD8 clones, % | 65 | 17 | |||||||
| Genotype (WT CGA-Arg) . | No. of clones carrying specified genotype . | WASp expression +/− . | Present in primary cells*CD3 . | ||||||
|---|---|---|---|---|---|---|---|---|---|
| First blood draw . | Second blood draw . | ||||||||
| CD4† . | CD8‡ . | ND . | Total . | % of total . | Total . | % of total . | |||
| 995C>T germline mutation (Stop) | 112§ | 8 | 15 | 135 | 50.6 | 69 | 50.0 | − | + |
| % CD4 or CD8 clones with germline mutation | 65 | 18 | |||||||
| Class I | |||||||||
| 995C>T>A (Arg) | 33 | 6 | 15 | 54 | 20.2 | 31 | 22.5 | + | + |
| 995C>T>C (Wt-Arg) | 0 | 0 | 2 | 2 | 0.7 | 0 | 0.0 | ND | + |
| 995C>T>G (Gly) | 3 | 1 | 3 | 7 | 2.6 | 3 | 2.2 | + | + |
| 995C>T, 996G>C (Ser) | 9 | 8 | 3 | 20 | 7.5 | 7 | 5.1 | + | + |
| 995C>T, 996G>T (Leu) | 0 | 0 | 0 | 1∥ | 0.4 | 0 | 0.0 | + | + |
| 995C>T, 997A>C (Cys) | 0 | 3 | 0 | 3 | 1.1 | 0 | 0.0 | + | + |
| CD4 or CD8 clones with class I genotype, % | 26 | 40 | |||||||
| Total class I | 86 | 32.2 | 41 | 29.7 | |||||
| Class II | |||||||||
| 995C>T, IVS9 −1G>A | 0 | 5 | 3 | 8 | 3.0 | 3 | 2.2 | + | + |
| 995C>T, IVS9 −2A>C | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, IVS9 −2A>G | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, IVS9 −3C>G | 1 | 11 | 2 | 14 | 5.2 | 4 | 2.9 | + | + |
| 995C>T, IVS9 −3C>T | 1 | 0 | 0 | 1 | 0.4 | 3 | 2.2 | + | + |
| 995C>T, IVS9 −6C>G | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, IVS9 −7T>G | 0 | 0 | 1 | 1 | 0.4 | 0 | 0.0 | ND | + |
| 995C>T, IVS9 −7T>A | 2 | 0 | 0 | 2 | 0.7 | 0 | 0.0 | + | − |
| 995C>T, IVS9 −12C>G | 2 | 0 | 0 | 2 | 0.7 | 2 | 1.4 | + | + |
| 995C>T, IVS9 −24C>G | 2 | 0 | 0 | 2 | 0.7 | 3 | 2.2 | + (weak) | + |
| 995C>T, IVS9 −27C>G | 0 | 0.0 | 1 | 0.7 | + (weak) | − | |||
| 995C>T, IVS9 −84G>C | 0 | 0.0 | 1 | 0.7 | + (weak) | + | |||
| CD4 or CD8 clones with class II genotype, % | 5 | 36 | |||||||
| Total class II | 30 | 11.2 | 20 | 14.5 | |||||
| Class III | |||||||||
| 995C>T, 988G>T | 0 | 0.0 | 1 | 0.7 | + | − | |||
| 995C>T, 999G>C | 0 | 0.0 | 1 | 0.7 | + (weak) | − | |||
| 995C>T, 1007C>T | 2 | 0 | 0 | 2 | 0.7 | 0 | 0.0 | + | − |
| 995C>T, 1095C>T | 0 | 0 | 1 | 1 | 0.4 | 0 | 0.0 | ND | + |
| 995C>T, 1109C>T, 1110C>T | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | ND | − |
| 995C>T, 1124C>T | 0 | 0 | 1 | 1 | 0.4 | 2 | 1.4 | + (weak) | + |
| 995C>T, 1155C>G | 0 | 0.0 | 1 | 0.7 | + (weak) | − | |||
| CD4 or CD8 clones with class III genotype, % | 2 | 0 | |||||||
| Total class III | 5 | 1.9 | 5 | 3.6 | |||||
| Class IV | |||||||||
| 995C>T, del IVS9 −84 to −21 (64 bp) | 0 | 1 | 0 | 1 | 0.4 | 0 | 0.0 | + | − |
| 995C>T, del IVS9 −32 to −18 (15 bp) | 0 | 0.0 | 2 | 1.4 | + | − | |||
| del IVS9 −74 to exon 10 1045 (154 bp) | 0 | 1 | 0 | 1 | 0.4 | 0 | 0.0 | + | + |
| del exon 10 968 to 1057 (90 bp, 30 aa) | 0 | 0 | 1 | 1 | 0.4 | 0 | 0.0 | ND | − |
| 995C>T, del exon 10 976 to 991 (16 bp) | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | + | − |
| del exon 10 984 to 1028 (45 bp, 15 aa) | 0 | 1 | 1 | 2 | 0.7 | 1 | 0.7 | + | − |
| del exon 10 988 to 1050 (63 bp, 21 aa) | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | ND | − |
| del exon 10 992 to 997 (6 bp, 2 aa) | 1 | 0 | 0 | 1 | 0.4 | 0 | 0.0 | + | − |
| 995C>T, del exon 10 996 to 998 (3 bp) | 2 | 0 | 0 | 2 | 0.7 | 0 | 0.0 | ND | − |
| CD4 or CD8 clones with class IV genotype, % | 3 | 7 | |||||||
| Total class IV | 10 | 3.7 | 3 | 2.2 | |||||
| Total no. of clones | 173 | 45 | 48 | 267 | 100.0 | 138 | 100.0 | ||
| CD4 or CD8 clones, % | 65 | 17 | |||||||
ND indicates not determined.
Indicates whether genotype, first identified in allospecific T-cell clone was also confirmed in uncultured, immunomagnetically isolated CD3+ (including CD3+/CD8−) cells.
Allospecific T-cell clones initiated either with CD3+/CD8− cells or with total PBMCs and subsequently identified by fluorescence-activated cell sorting (FACS) to be CD4+.
Allospecific T-cell clones initiated with CD8+ cells or with total PBMCs and subsequently identified by FACS to be CD8+.
Indicated is the number of primary T-cell clones with the specified genotypes.
This 996G>T clone was of CD4−/CD8− phenotype.
A second blood draw was obtained 7 months later. Ten of the original 25 revertant genotypes were again observed. The relative frequency and distribution of revertant genotypes was quite stable between these 2 time points (Table 1). In addition, 9 new revertant genotypes were recovered, raising to 34 the revertant genotypes identified.
All sequence differences between putative revertant clones and the patient WAS4 mutant WAS sequence were clustered in 275 base pairs (bp), approximately centered about the original 995C>T mutation. This nonrandom positioning suggests that the revertants encode for some functional restoration of WASp expression. Indeed, 995C>T>A revertants demonstrated normal levels of WASp expression while germline mutant 995C>T clones exhibited negligible WASp levels (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Of 26 additional putative revertants examined, all demonstrated some level of restoration of WASp expression (Table 1 and Figure S1).
The class I revertants likely restore full-length WASp expression by replacing the 321Stop codon with various amino acids. Class II revertants likely restore expression of either an internally deleted or frame-shifted WASp through altered splicing of the WAS mRNA and skipping of the 321Stop codon. Many of the class IV deletions represent in-frame deletion of 2 to 30 amino acids encompassing the 321Stop codon, whereas other deletions possibly affect the mRNA splicing pattern.
We also PCR-amplified, cloned, and sequenced WAS sequences directly from the patient's freshly purified CD3+ cells (first blood draw) and detected 16 of the allospecific T-cell clone revertant genotypes (Table 1). Thus the extensive diversity of WAS genotypes observed in allospecific T-cell clones reflects a WAS genotype diversity already present in CD3+ T-lymphocytes in vivo. Of the recombinant DNA clones sequenced, 13.4% (20 of 149) had the predominant 995C>T>A reversion while 50.3% (75 of 149) had the germline 995C>T—similar to the observed frequencies of these genotypes in allospecific T-cell clones (Table 1; first blood draw).
We can only speculate regarding possible mechanism(s) responsible for the diversity of genotypic revertants in this patient. It is formally possible that the high frequency of genetic changes observed reflects the average DNA mutation rate for lymphocytes. There is no known involvement of the normal WASp in chromosomal DNA replication, proofreading or DNA repair. However, we cannot rule out the possibility that this patient has an intrinsic defect in fidelity of DNA repair with an increased DNA mutation rate.
We have been unable to find other reports of such a diverse range of revertant genotypes—neither in WAS nor in other diseases. Reversions in WAS are typically first identified through the identification of WASp-expressing cells in the peripheral blood, followed by direct sequencing of PCR-amplified genomic WAS DNA sequences. Identification of individual revertant genotypes likely requires at least a 5% frequency to be detected; thus, minor revertant genotypes would be missed. In contrast, analysis of the genotypes of expanded individual blood lymphocytes easily revealed the remarkable diversity of genetic revertants present. It is also likely that the nature and location of the primary WAS mutation might also have a major effect on the frequency of revertants capable of providing some restored function and therefore allow revertant cells to accumulate over time.
These data suggest that development of individual genotypic revertants in WAS patients may occur relatively frequently, challenging the current notion that spontaneous reversions are rare events and that their occurrence is restricted to long-lived progenitors.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank patients WAS1 and WAS4 who generously participated in this study.
This work was supported by grants to B.D. from the United States Immunodeficiency Network (USIDNET) and National Institute for Allergy and Infectious Diseases (NIAID), to M.B. from National Foundation of Cancer Research (NFCR) and Malcolm H. Wiener Foundation, and to F.C. from the Intramural Research Program of NHGRI/NIH.
National Institutes of Health
Authorship
Contribution: D.M., L.M., F.C., and M.B. obtained peripheral blood samples from the WAS1 and WAS4 patients. D.M. performed the experiments described in Figure 1A,B. B.D., together with M.D., N.P., and J.R., performed the experiments described in Figure 1C-F, Table 1, and Figure S1. B.D., F.C., and M.B. designed the experiments and, together with significant assistance from N.P., wrote the paper. All authors contributed to the review and final editing of the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Brian R. Davis, Centre for Stem Cell Research, Brown Foundation Institute of Molecular Medicine, University of Texas Health Science Center, 1825 Pressler St, Houston, TX 77030; e-mail: Brian.R.Davis@uth.tmc.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal