Abstract
The use of arsenic trioxide (ATO) to treat multiple myeloma (MM) is supported by preclinical studies as well as several phase 2 studies, but the precise mechanism(s) of action of ATO has not been completely elucidated. We used gene expression profiling to determine the regulation of apoptosis-related genes by ATO in 4 MM cell lines and then focused on Bcl-2 family genes. ATO induced up-regulation of 3 proapoptotic BH3-only proteins (Noxa, Bmf, and Puma) and down-regulation of 2 antiapoptotic proteins Mcl-1 and Bcl-XL. Coimmunoprecipitation demonstrated that Noxa and Puma bind Mcl-1 to release Bak and Bim within 6 hours of ATO addition. Bak and Bim are also released from Bcl-XL. Silencing of Bmf, Noxa, and Bim significantly protected cells from ATO-induced apoptosis, while Puma silencing had no effect. Consistent with a role for Noxa inhibition of Mcl-1, the Bad-mimetic ABT-737 synergized with ATO in the killing of 2 MM lines. Finally, Noxa expression was enhanced by GSH depletion and inhibited by increasing GSH levels in the cells. Understanding the pattern of BH3-only protein response should aid in the rational design of arsenic-containing regimens.
Introduction
Multiple myeloma (MM) is an incurable malignancy of terminally differentiated B cells accounting for approximately 1% to 2% of all human cancers and 10% of all hematologic malignancies, with median survival times of 4 to 7 years.1,2 Traditional therapies include chemotherapy, steroids, and stem cell transplantation, which typically result in significant responses.2 However, all these therapies have a limited duration with eventual relapse of the disease and the development of resistance to treatment
Arsenic trioxide (ATO, As2O3, Trisenox) is a promising additional agent for progressive and refractory MM that induces growth inhibition and apoptosis in MM cell lines and freshly isolated human MM cells.3-8 The in vitro sensitivity of cultured MM cell lines to clinically achievable concentrations, as well as the modest activity observed as a single agent or in combination with ascorbic acid in phases 1 and 2 trials,9-11 led to the investigation of the mechanism independent of the PML-RARα.12,13 These studies demonstrated caspase activation by ATO in myeloma cells, although not all death was caspase dependent.14-17 Regardless of whether caspases are necessary, their activation is typically regulated by members of the Bcl-2 family.
The Bcl-2 family is divided in 2 main groups of proteins, antiapoptotic or proapoptotic.18,19 The antiapoptotic members display sequence conservation through all 4 Bcl-2 homology (BH) domains and include Bcl-2, Bcl-XL, Mcl-1, Bfl-1(A1), and Bcl-w. Proapoptotic members are further subdivided in 2 groups: in the first group are the more conserved multidomain members (Bax, Bak, and Bok) with homology in BH1-320-23 and the second group, the BH3-only proteins19 (eg, Bim, Bmf, Noxa), which contain only the BH3 domain, as indicated by the name. Several studies explaining how BH3-only proteins promote cell death have resulted in 2 models of BH3-only protein function.24 The direct activation model suggests that certain BH3-only proteins, named activators and including Bim and tBid, can bind to Bax and Bak directly and promote their activation.25-29 In this model, the remaining BH3-only proteins, termed sensitizers, bind to the antiapoptotic members, displacing bound Bim or tBid so they can activate Bax and Bak. The indirect activation model suggests that all the BH3-only proteins bind only their antiapoptotic relatives and prevent them from sequestering Bax or Bak.30,31 In this model, Bim and tBid are better inducers of apoptosis compared with the remaining BH3-only proteins, because they can bind all the antiapoptotic Bcl-2 members.30
In this study, we used gene expression profiling to determine the apoptotic response of myeloma cells to ATO. By correlating gene expression to response, we determined that 3 BH3-only proteins, Noxa, Bmf, and Puma, are up-regulated while Mcl-1 and Bcl-XL are down-regulated following ATO treatment. This prompted us to further investigate the role of the Bcl-2 subfamilies in this response.
Methods
Cell lines
Four multiple myeloma cell lines were used. U266 and 8226/S were obtained from ATCC (Manassas, VA). The MM.1s cell line was obtained from Dr Steven Rosen (Northwestern University, Chicago, IL) and the KMS-11 cell line was provided by Dr P. Leif Bergsagel (Mayo Clinic, Scottsdale, AZ). Cells were cultured at 37°C in a humidified atmosphere with 5% CO2, in RPMI-1640 medium, supplemented with 10% heat-inactivated fetal bovine serum, 100 U/mL penicillin, 100 μg/mL streptomycin, and 2 mM l-glutamine (all from Cellgro, Mediatech, Herndon, VA).
Reagents
Buthionine sulfoximine (BSO) and propidium iodide (PI) were purchased from Sigma-Aldrich (St Louis, MO); N-acetylcysteine (NAC) was purchased from Bedford Laboratories (Bedford, OH). Annexin-V–fluorescein isothiocyanate (FITC) was purchased from Biovision (Palo Alto, CA). ATO was kindly provided by Cell Therapeutics (Seattle, WA). ABT-737 and the less active enantiomer of ABT-737 ((-)ABT) were obtained from Abbott Laboratories (Abbott Park, IL). Bortezomib was kindly provided by Millennium Pharmaceuticals (Cambridge, MA). Melphalan was purchased from Sigma-Aldrich. Staurosporine was obtained from EMD Biosciences (La Jolla, CA).
Antibodies
The following primary antibodies were used for Western blot: rat anti-Bmf monoclonal antibody (mAb; Alexis Biochemicals, San Diego, CA); rabbit anti–COX IV polyclonal antibody (Abcam, Cambridge, MA); mouse anti-Noxa mAb (Abcam); rabbit anti-Puma polyclonal antibody (Cell Signaling, Danvers, MA); rabbit anti-Bim polyclonal antibody (CHEMICON International, Temecula, CA); rabbit anti–Mcl-1 polyclonal antibody (Stressgen Biotechnologies, Victoria, BC); rabbit anti–Bcl-XL polyclonal antibody (13.632 ); rabbit antiactin polyclonal antibody (Sigma-Aldrich); rabbit anti-Bak polyclonal antibody (Millepore, Billerica, MA); rabbit anti-Bax polyclonal antibody (Santa Cruz Biotechnology, Santa Cruz, CA); rabbit anti-Bid polyclonal antibody (Cell Signaling); and mouse anti–Bcl-2 mAb (Santa Cruz Biotechnology). For coimmunoprecipitation, the following antibodies were used: mouse anti–Mcl-1 mAb (BD Biosciences, San Jose, CA), mouse anti–Bcl-XL mAb (7B2.532 ), and mouse anti–Bcl-2 mAb. The following secondary antibodies were used: antimouse IgG1-HRP conjugate (Roche Applied Science, Indianapolis, IN) and the enhanced chemiluminescence (ECL) rabbit IgG, HRP-linked whole Ab (from donkey; GE HealthCare, Piscataway, NJ).
Cell viability by annexin-V–FITC and PI staining
Cell viability was measured by annexin-V–FITC and PI staining, following the manufacturer's instructions as previously described.5 Samples were acquired on a FACScan flow cytometer (Becton Dickinson, San Jose, CA) and analyzed with CellQuest software (Becton Dickinson).
Cellular assays
All cellular assays were set up as follows: cells were plated at 2.5 × 105 cells/mL in 1 mL (24-well plates), 5 mL (6-well plates), 10 mL (T-25 flasks), or 30 mL (T-75 flasks) in supplemented RPMI-1640 media and incubated with the indicated concentrations of ATO and/or BSO, NAC, or ABT-737. For ATO plus BSO and/or NAC experiments, cells were incubated with indicated concentrations of ATO, 100 μM BSO, and/or 10 mM NAC as indicated. For ATO plus ABT-737 cotreatment, cells were incubated with indicated concentrations of ABT-737, with and without 2 μM ATO for 24 hours. The less active enantiomer (-)ABT was used as a negative control at indicated concentrations.
Gene expression profiling
The 4 cell lines were treated for 0, 6, 24, and 48 hours with 2 μM ATO. Total RNA was isolated using the RNeasy Mini Kit (Qiagen, Valencia, CA), and the hybridization and initial data analysis were performed by Expression Analysis (Durham, NC). Briefly, total RNA quality was confirmed using an Agilent 2100 Bioanalyzer (Palo Alto, CA), and cRNA was generated for probing Affymetrix Hu133 2.0 Plus Chips (Santa Clara, CA) containing more than 50 000 (54 675 including controls) probe sets. Affymetrix GCOS software was used with statistical algorithms to determine a quantitative value (signal intensity) and a qualitative value (present [P] or absent [A] calls) for each transcript on the array. Signal intensities for each cell line were considered only if at least one present call, as qualitative value, was reported for any time point and at least one signal was higher than 100. Ratios versus baseline expression were calculated as positive or negative ratios for up- and down-regulated genes, respectively. Data were analyzed using Microsoft Excel (Redmond, WA) and the bioinformatic programs Cluster and TreeView (Eisen Laboratory, University of California, CA).
Subcellular fractionation
Mitochondrial-rich fractions (heavy membrane fractions) were obtained as follows: cells were harvested and washed using phosphate-buffered saline (PBS) and centrifuged at 500g for 5 minutes at room temperature. Cell pellets were then resuspended in isotonic buffer (5 mM HEPES, pH 7.2, 300 mM sucrose, 1 mM EDTA) supplemented with protease inhibitors (170 μg/mL PMSF, 2 μg/mL aprotinin, and 1 μg/mL leupeptin; Sigma-Aldrich) and homogenized. Homogenates were centrifuged at 500g for 5 minutes at 4°C, and the supernatant was centrifuged again at 500g for 5 minutes to remove all the remaining unbroken cells and nuclei. The resulting supernatant was centrifuged at 10 000g for 20 minutes at 4°C to obtain the heavy membrane (HM) or mitochondria-rich fractions and the supernatant was considered as the cytosolic fraction. Heavy membrane fractions were lysed using radioimmunoprecipitation assay (RIPA) buffer plus protease inhibitor mixture. Samples were kept at − 80°C for Western blot analysis.
Western blot analysis
Western blotting was performed using standard techniques as previously described.16 Briefly, cells were washed twice with PBS buffer and lysed in RIPA buffer supplemented with protease inhibitors. Protein concentration was determined using a BCA Protein Assay kit (Pierce Biotechnology, Rockford, IL). Total proteins (10-30 μg) were subjected to sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and transferred to nitrocellulose membranes (Protan Nitrocellulose Transfer Membrane; Whatman, Dassel, Germany).
Coimmunoprecipitation studies
Immunoprecipitation experiments were done using the ExactaCruz C Kit (Santa Cruz Biotechnology). Briefly, MM.1s and KMS11 cell lines were treated with 2 μM ATO at 2.5 × 105 cells/mL, 30 mL (T-75 flasks) in supplemented RPMI-1640 media for 6 and 24 hours. Control and treated cells were washed with PBS and protein lysates were obtained using CHAPS lysis buffer (10 mM HEPES, pH 7.2, 150 mM NaCl, 2% CHAPS). Total proteins (100 μg) were precleared first using 50 μL Protein-G Agarose Fast Flow (Millipore) for 1 hour rotating at 4°C and then using 50 μL Pre-clearing Matrix C (Santa Cruz Biotechnology) for 30 minutes rotating at 4°C. Immunoprecipitation (IP) antibody–IP matrix complexes were formed using 3 μg mouse anti–Mcl-1 mAb, 5 μL mouse anti–Bcl-XL mAb, or 3 μg mouse anti–Bcl-2 mAb and 50 μL IP-Matrix (Santa Cruz Biotechnology). Precleared lysates and IP antibody–IP matrix complex were incubated overnight rotating at 4°C. Pelleted matrix was washed twice with 500 μL ice-cold PBS, and proteins were eluted using reducing 2× electrophoresis sample buffer. Samples were resolved by SDS-PAGE and analyzed by Western blots.
Silencing studies using small interfering RNAs
Small interfering RNAs (siRNAs) were obtained from DHARMACON RNA Technologies (Chicago, IL) selecting the ON-TARGETplus SMARTpool duplexes as the RNAi-specific technology platform. siRNA against Bmf (BMF), Noxa (PMAIP1), Bim (BCL2L11), Puma (BBC3), Bid (BID), and the siCONTROL nontargeting siRNA (si(-)) were used. siRNAs were transfected into the cells by electroporation (Amaxa, Gaithersburg, MD) following the manufacturer's instructions. Briefly, 5 × 106 cells were electroporated in 100 μL cell line nucleofector solution (Amaxa Reagent V) with 15 μL (300 pmol) siRNA, using preselected Amaxa Program S-020 for MM.1s and G-015 for KMS11. Electroporated cells were plated in 6-well plates with 3 mL supplemented RPMI-1640 medium for 16 hours at 37°C. Then, cells were treated with 2 μM ATO and samples were harvested at 24 and 48 hours for apoptosis determination by annexin-V–FITC/PI staining and at 6 and 24 hours for protein expression analysis by Western blot.
Real-time PCR
MM.1s and KMS11 cells were transfected with si(-) control and siBmf. After 16 hours at 37°C in supplemented RPMI-1640, cells were treated with 2 μM ATO for 6 and 24 hours. Total RNA was isolated using the RNeasy Mini Kit (Qiagen). cDNA was synthesized using the GeneAmp RNA polymerase chain reaction (PCR) Kit (Applied Biosystems, Foster City, CA). Total RNA (1 μg) was reverse transcribed by the murine leukemia virus (MuLV) reverse transcriptase in a 20-μL reaction using random hexamer primers. cDNA was amplified using TaqMan Gene Expression Assay (Applied Biosystems) on the 7700 Sequence Detection System following the manufacturer's protocol and the 20× human Bmf Mix. TaqMan human GAPDH was used as internal control. Bmf mRNA expression was calculated as relative Bmf expression, normalized using GAPDH mRNA expression.
Results
Multiple myeloma cell lines displayed different sensitivity to ATO
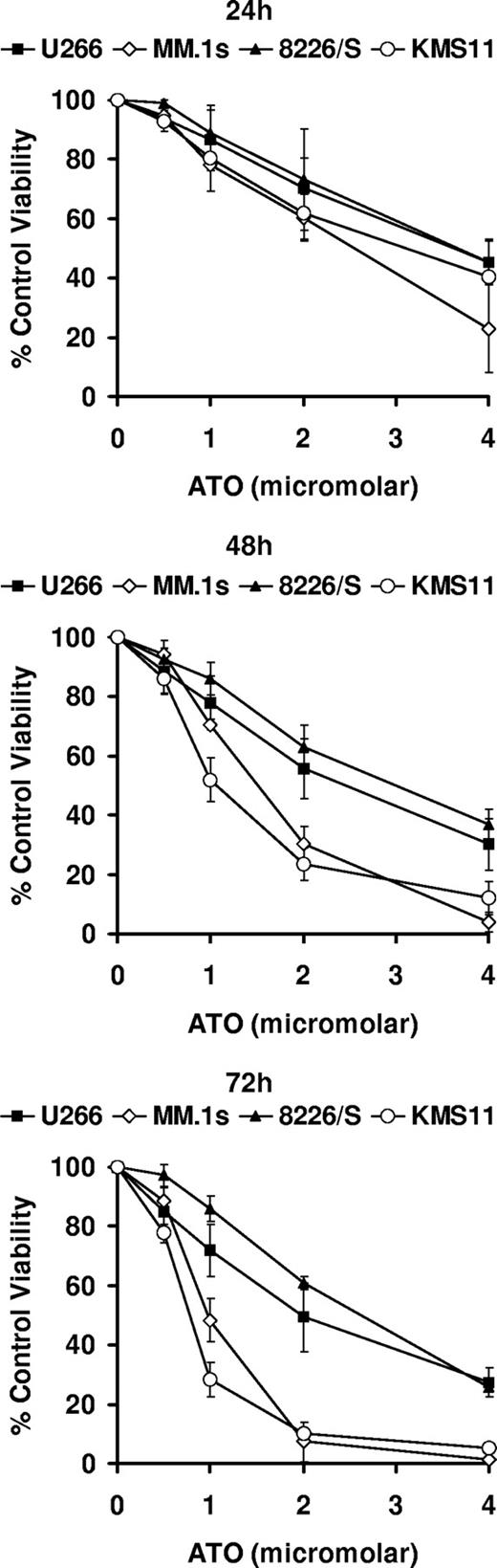
While numerous studies have examined the mechanisms by which ATO induces apoptosis in myeloma cells, it still remains unclear what the upstream signals are that result in caspase activation.3-8,33 To gain further insight into this process, we performed gene expression profiling of 4 multiple myeloma cell lines treated for 0, 6, 24, and 48 hours with 2 μM ATO. We then analyzed which apoptosis-related genes were either up- or down-regulated (increase [I] or decrease [D] calls) in all 4 cell lines regardless of the magnitude of the change. As can be seen in Table 1, this approach did not prove to be very informative. While the up-regulation of CD95, FADD, and caspase-8 is suggestive of a requirement for CD95 signaling, we have previously demonstrated that inhibition of this pathway through the transfection of the caspase-8 inhibitor CrmA had no effect on ATO toxicity.17 Up-regulation of Bcl-2 was surprising but unlikely to explain how ATO induces apoptosis. Moreover Bcl-2 up-regulation was modest (1.5- to 1.7-fold at 48 hours) and has been previously observed in myeloma cell lines treated with doxorubicin, etoposide, or hydrogen peroxide.34 In contrast, from all down-regulated genes in all 4 cell lines, only Bcl-X and survivin appeared to be likely candidates. However, we had previously demonstrated that overexpression Bcl-XL can only delay ATO-induced cell death,5,17 while survivin down-regulation is likely associated with ATO-induced cell-cycle arrest.35-37 Since changes in Bcl-2 and Bcl-X were observed in all 4 cell lines, we decided to determine the expression pattern in response to ATO of all Bcl-2 family members included in the array. For this analysis, we took into consideration the sensitivity of the 4 cell lines. As seen in Figure 1, all cell lines are killed by ATO in a time- and dose-dependent manner. However, the cell lines are not equally responsive. Two cell lines, U266 and 8226/S, were less sensitive to ATO than MM.1s and KMS11. This pattern of cell death was evident at 72 hours where percentage of control viabilities for cells treated with 2 μM ATO were 49.4 (± 11.5; mean [± SD]), 7.4 (± 6.7), 60.9 (± 2.3), and 10.1 (± 1.2) for U266, MM.1s, 8226/S, and KMS11, respectively (Figure 1).
Apoptosis-related genes either up- or down-regulated (I or D calls) in all 4 cell lines obtained using Affymetrix microarray following ATO treatment
| Gene descriptor . | Symbol . | U266 . | MM.1s . | 8226/S . | KMS11 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 h . | 24 h . | 48 h . | 6 h . | 24 h . | 48 h . | 6 h . | 24 h . | 48 h . | 6 h . | 24 h . | 48 h . | ||
| B-cell CLL/lymphoma 2 | BCL2 | 1.3 | 1.4 | 1.7 | 1.0 | 1.2 | 1.5 | 1.3 | 1.2 | 1.6 | 1.2 | 1.2 | 1.5 |
| Caspase 8, apoptosis-related cysteine protease | CASP8 | 1.0 | 1.3 | 1.5 | 1.6 | 1.4 | 1.4 | 1.5 | 1.1 | 1.3 | 2.1 | 1.8 | 1.6 |
| Fas (TNFRSF6)-associated via death domain | FADD | 1.5 | 1.3 | 1.6 | 1.6 | 1.0 | 1.4 | 1.3 | 1.1 | 1.1 | 1.4 | −1.2 | −1.1 |
| TNF receptor superfamily, member 6 (Fas/CD95) | TNFRSF6 | 1.1 | 1.5 | 1.4 | 4.6 | 3.7 | 2.6 | 1.1 | 1.1 | 1.5 | 1.0 | 1.7 | 1.4 |
| Baculoviral IAP repeat-containing 5 (survivin) | BIRC5 | −1.4 | −1.4 | −2.2 | −1.4 | −1.3 | −3.8 | −1.0 | −1.1 | −1.3 | −1.5 | 1.1 | −1.2 |
| BCL2-like 1 | BCL2L1 | −2.2 | −2.5 | −2.1 | −1.6 | −2.0 | −3.2 | −1.4 | −1.5 | −1.5 | −2.7 | −2.7 | −2.9 |
| Gene descriptor . | Symbol . | U266 . | MM.1s . | 8226/S . | KMS11 . | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 6 h . | 24 h . | 48 h . | 6 h . | 24 h . | 48 h . | 6 h . | 24 h . | 48 h . | 6 h . | 24 h . | 48 h . | ||
| B-cell CLL/lymphoma 2 | BCL2 | 1.3 | 1.4 | 1.7 | 1.0 | 1.2 | 1.5 | 1.3 | 1.2 | 1.6 | 1.2 | 1.2 | 1.5 |
| Caspase 8, apoptosis-related cysteine protease | CASP8 | 1.0 | 1.3 | 1.5 | 1.6 | 1.4 | 1.4 | 1.5 | 1.1 | 1.3 | 2.1 | 1.8 | 1.6 |
| Fas (TNFRSF6)-associated via death domain | FADD | 1.5 | 1.3 | 1.6 | 1.6 | 1.0 | 1.4 | 1.3 | 1.1 | 1.1 | 1.4 | −1.2 | −1.1 |
| TNF receptor superfamily, member 6 (Fas/CD95) | TNFRSF6 | 1.1 | 1.5 | 1.4 | 4.6 | 3.7 | 2.6 | 1.1 | 1.1 | 1.5 | 1.0 | 1.7 | 1.4 |
| Baculoviral IAP repeat-containing 5 (survivin) | BIRC5 | −1.4 | −1.4 | −2.2 | −1.4 | −1.3 | −3.8 | −1.0 | −1.1 | −1.3 | −1.5 | 1.1 | −1.2 |
| BCL2-like 1 | BCL2L1 | −2.2 | −2.5 | −2.1 | −1.6 | −2.0 | −3.2 | −1.4 | −1.5 | −1.5 | −2.7 | −2.7 | −2.9 |
Ratios versus baseline expression are shown. Positive values indicate up-regulation, while negative values indicate down-regulation. Two-fold changes are in bold.
ATO-induced apoptosis in 4 human myeloma cell lines. U266, MM.1s, 8226/S, and KMS11 were treated for 24, 48, and 72 hours with 0, 0.5, 1, 2, and 4 μM ATO. Cell viability was determined by flow cytometry following annexin-V–FITC/PI staining. Percentage (%) of control viability was plotted versus ATO concentration. Graphs are presented for each time point. The data are presented as the means (± SD) of 3 independent experiments.
ATO-induced apoptosis in 4 human myeloma cell lines. U266, MM.1s, 8226/S, and KMS11 were treated for 24, 48, and 72 hours with 0, 0.5, 1, 2, and 4 μM ATO. Cell viability was determined by flow cytometry following annexin-V–FITC/PI staining. Percentage (%) of control viability was plotted versus ATO concentration. Graphs are presented for each time point. The data are presented as the means (± SD) of 3 independent experiments.
Expression changes of Bcl-2 family genes correlated with induction of cell death
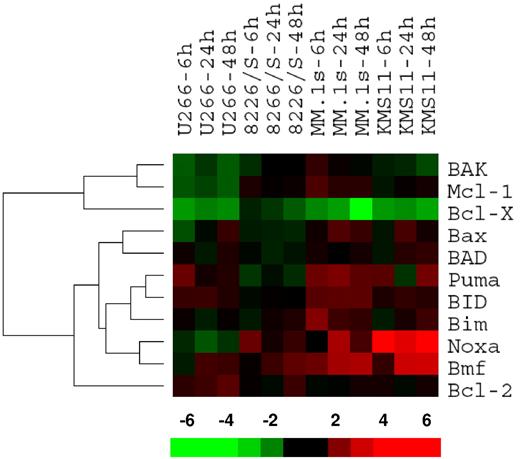
Bcl-2 family members were clustered in a supervised fashion to identify changes associated with ATO sensitivity. Interestingly, this approach resulted in 2 major gene clusters (Figure 2), the first containing the 2 antiapoptotic genes Mcl-1 and Bcl-X, while the second had 8 proapoptotic genes containing all 6 BH3-only proteins included in the analysis (all genes not expressed were excluded) and Bcl-2. Within this major cluster, it was revealed that the expression pattern of Noxa and Bmf correlates with ATO sensitivity. Puma expression, to a lesser extent, also correlates with sensitivity. Therefore, we initially focused on these 5 genes, by confirming changes at the protein level as Bcl-2 protein and mRNA expression do not always correlate.30
Gene expression profile for ATO-induced changes in the Bcl-2 family. U266, MM.1s, 8226/S, and KMS11 cell lines were treated with 2 μM ATO for 6, 24, and 48 hours. Total RNA was obtained and cRNA was probed on Affymetrix Hu133 2.0 Plus Chips. Signal intensity was used as gene expression. Ratios at 6, 24, and 48 hours versus baseline gene expression were calculated for each probe set and average ratios were obtained for genes. Bcl-2 family member genes were clustered using the bioinformatic programs Cluster and TreeView. The bar at the bottom represents the scale for fold changes.
Gene expression profile for ATO-induced changes in the Bcl-2 family. U266, MM.1s, 8226/S, and KMS11 cell lines were treated with 2 μM ATO for 6, 24, and 48 hours. Total RNA was obtained and cRNA was probed on Affymetrix Hu133 2.0 Plus Chips. Signal intensity was used as gene expression. Ratios at 6, 24, and 48 hours versus baseline gene expression were calculated for each probe set and average ratios were obtained for genes. Bcl-2 family member genes were clustered using the bioinformatic programs Cluster and TreeView. The bar at the bottom represents the scale for fold changes.
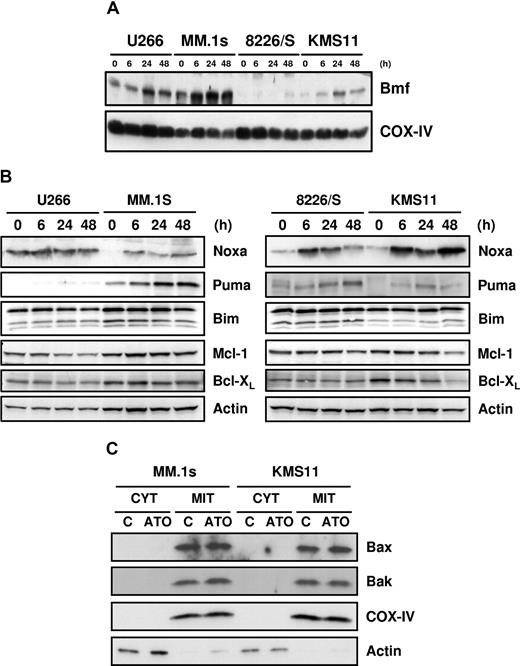
Consistent with the mRNA expression (Figure 2), Bmf protein was also up-regulated in all cell lines (Figure 3A), and Noxa protein was up-regulated in 3 of 4 cell lines (Figure 3B). Interestingly, in the fourth line, U266, Noxa mRNA is actually down-regulated by ATO. However, Noxa mRNA (not shown) and protein (Figure 3B) are expressed in a constitutive fashion in these cells, and protein expression was not altered by addition of ATO. Moreover, Mcl-1, one of the 2 antiapoptotic protein that Noxa binds,29,30,38 was down-regulated in this cell line, resulting in a net increase in Noxa relative to its target. Puma was also up-regulated in all the cell lines, but consistent with the array data, primarily in MM.1s, while Bim expression was relatively unchanged during ATO-induced apoptosis (Figure 3B). Similar to the mRNA expression pattern, Bcl-XL was down-regulated in all the cell lines, and Mcl-1 was down-regulated in U266 at 24 and 48 hours and in KMS11 at 48 hours (Figure 3B). Bcl-2 up-regulation at the messenger level did not correlate with protein expression, and no change in expression was obtained during ATO-induced apoptosis (not shown). Taken together, these data demonstrate that the combination of Noxa and Bmf up-regulation paired with Mcl-1 and Bcl-XL down-regulation could be sufficient to drive ATO-induced apoptosis. In addition, we tested the effects of different drugs on myeloma cells, including melphalan, bortezomib, and staurosporine. Of these agents, only bortezomib induced Noxa up-regulation at the protein level in U266, MM.1s, and 8226/S (data not shown). This suggests that Noxa up-regulation by ATO is not the result of a general mechanism for drug-induced apoptosis in myeloma. In addition, Bax and Bak expression was not regulated during ATO treatment, and they both localized in the heavy membrane fractions (mitochondria-rich fractions) as showed in Figure 3C. For the remaining studies, we focused on MM.1s and KMS11, since these cells were more responsive to ATO.
Up-regulation of Bmf, Noxa, and Puma and down-regulation of Mcl-1 and Bcl-XL by ATO. U266, MM.1s, 8226/S, and KMS11 were treated for 6, 24, and 48 hours with 2 μM ATO. (A) Mitochondrial-rich fractions were obtained and Bmf expression was determined by Western blot. Membranes were reprobed with rabbit anti–COX IV polyclonal antibody to determine loading. (B) Western blot analysis of total protein lysates from ATO-treated cells. Membranes were probed with specific antibodies for Noxa, Puma, Bim, Mcl-1, Bcl-XL, and actin. (C) Cytosolic and heavy membrane fractions were obtained for untreated and 24-hour ATO-treated MM.1s and KMS11 cells as indicated in “Subcellular fractionation.” Actin and COX IV were used as indicators of fraction purity.
Up-regulation of Bmf, Noxa, and Puma and down-regulation of Mcl-1 and Bcl-XL by ATO. U266, MM.1s, 8226/S, and KMS11 were treated for 6, 24, and 48 hours with 2 μM ATO. (A) Mitochondrial-rich fractions were obtained and Bmf expression was determined by Western blot. Membranes were reprobed with rabbit anti–COX IV polyclonal antibody to determine loading. (B) Western blot analysis of total protein lysates from ATO-treated cells. Membranes were probed with specific antibodies for Noxa, Puma, Bim, Mcl-1, Bcl-XL, and actin. (C) Cytosolic and heavy membrane fractions were obtained for untreated and 24-hour ATO-treated MM.1s and KMS11 cells as indicated in “Subcellular fractionation.” Actin and COX IV were used as indicators of fraction purity.
Noxa binding to Mcl-1 results in the release of Bak and Bim following addition of ATO
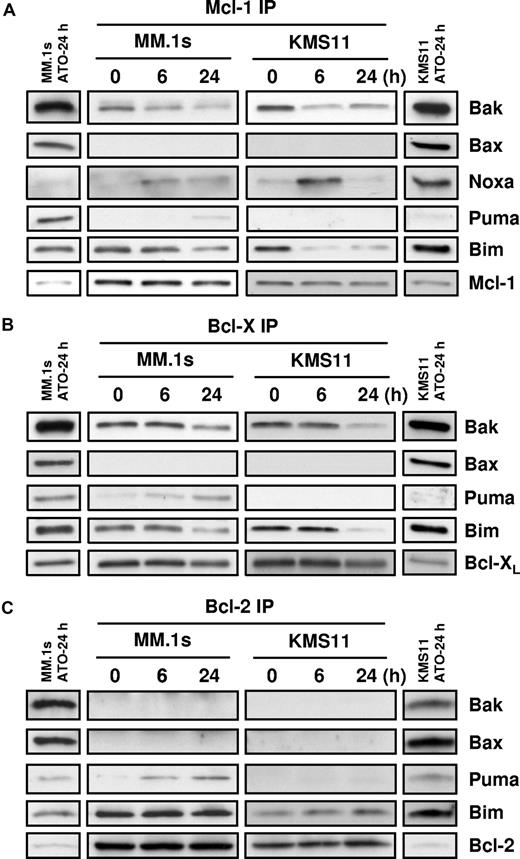
To elucidate interactions between Bcl-2 family members during ATO-induced apoptosis, coimmunoprecipitation experiments were performed in MM.1s and KMS11. Prior to ATO addition, coimmunoprecipitation revealed that both Bim and Bak were associated with Mcl-1 and Bcl-XL; however, only Bim was associated with Bcl-2 in the MM cell lines (Figure 4). Interestingly, Bax, while present in these cells and localized to the mitochondria, is not associated to Bcl-2, Bcl-XL, or Mcl-1. In addition, we did not see any evidence of conversion of Bid to tBid (not shown) and therefore we did not examine tBid binding. Within 6 hours of ATO treatment, Noxa was bound selectively to Mcl-1. Consistent with models of BH3-only protein function, Noxa binding is associated with release of both Bim and Bak from Mcl-1 in both cell lines (Figure 4A). Puma binding to Mcl-1 was also observed, albeit weakly and only in MM.1s. Unfortunately, because of limitations with available reagents, we could not determine whether Bmf was binding Bcl-XL, Mcl-1, or Bcl-2, therefore we focused on Bak, Bim, and Puma binding. As seen in Figure 4B, both Bak and Bim are released from Bcl-XL; however, the kinetics appear to be slower than from Mcl-1. Puma binding is observed, but again only in the MM.1s cells. Similar results were obtained for Bcl-2 Co-IP experiments; Puma binding is observed primarily in the MM.1s cells, consistent with the differences in Puma expression in these lines (Figure 3B). No change was observed for Bim binding in both cell lines, and surprisingly Bak is not bound to Bcl-2. These results suggest that Bcl-2 does not play a significant role in ATO-induced apoptosis. Together these data suggest that while expression of Bmf, Noxa, and Puma increases following ATO treatment, Noxa, Bim, and likely Bmf are more important than Puma. To further test the role of these BH3-only proteins, we determined the effect of silencing on ATO-induced apoptosis.
Coimmunoprecipitation of proapoptotic proteins with Mcl-1, Bcl-XL, and Bcl-2. MM.1s and KMS11 cell lines were treated with 2 μM ATO for 0, 6, and 24 hours. Protein lysates were prepared using 2% CHAPS buffer. (A) Mcl-1, (B) Bcl-XL, and (C) Bcl-2 proteins were immunoprecipitated with mAbs. Coimmunoprecipitated proteins were detected by Western blot using specific antibodies for Bak, Bax, Noxa, Bim, Puma, Mcl-1, Bcl-XL, and Bcl-2. The separated bands on each end of the panels represent the input for the IP. This lane contains 10% of the input, however only 60% of the immunoprecipitated protein was loaded in the gel. All bands represented are from the same experiment and same exposure of film.
Coimmunoprecipitation of proapoptotic proteins with Mcl-1, Bcl-XL, and Bcl-2. MM.1s and KMS11 cell lines were treated with 2 μM ATO for 0, 6, and 24 hours. Protein lysates were prepared using 2% CHAPS buffer. (A) Mcl-1, (B) Bcl-XL, and (C) Bcl-2 proteins were immunoprecipitated with mAbs. Coimmunoprecipitated proteins were detected by Western blot using specific antibodies for Bak, Bax, Noxa, Bim, Puma, Mcl-1, Bcl-XL, and Bcl-2. The separated bands on each end of the panels represent the input for the IP. This lane contains 10% of the input, however only 60% of the immunoprecipitated protein was loaded in the gel. All bands represented are from the same experiment and same exposure of film.
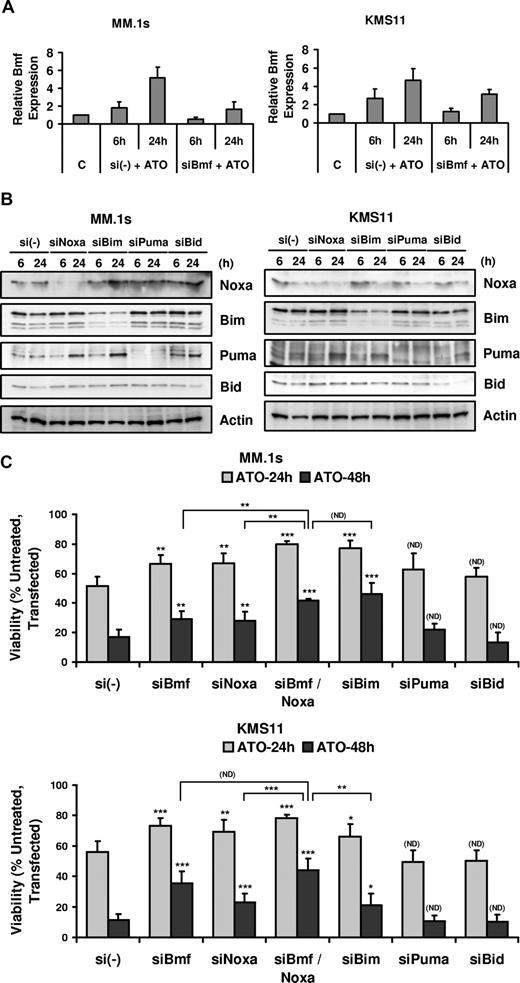
Silencing of Noxa and Bmf protected cells from ATO-induced apoptosis
MM.1s and KMS11 were transfected with siRNA for 5 different BH3-only proteins: Bmf, Noxa, Bim, Puma, and Bid, and the effect on ATO-induced apoptosis was determined. Bmf silencing was confirmed by real-time PCR comparing samples transfected with a nonsilencing control pool (si(-)) and siBmf (Figure 5A), while Noxa, Bim, Puma, and Bid silencing was verified by Western blot (Figure 5B). Silencing of Bmf and Noxa partially protected cells from ATO-induced apoptosis in MM.1s and KMS11 cells (Figure 5C). Consistent with these BH3-only proteins having distinct effects in cells, the combination of Bmf and Noxa silencing significantly increased the protection from ATO-induced apoptosis in MM.1s. An increase in viability was consistently observed in KMS11, however it is not statistically significant compared with Bmf silencing. This suggests that Bmf expression may be sufficient to inhibit both Mcl-1 and Bcl-XL in these cells and may explain the transient nature of Noxa binding to Mcl-1 observed in the coimunoprecipitations (Figure 4). Bim silencing is also protective in both cell lines. Interestingly, the loss of Bim in MM.1s cells has the most dramatic effect alone, yet is equal to the combination of Noxa and Bmf silencing. The effect of Bim silencing in KMS11 was less dramatic, which may be a result of incomplete silencing (Figure 5B). Consistent with the Co-IP results, Puma silencing did not protect MM.1s and KMS11 cell lines from apoptosis. Bid silencing also had no influence on ATO-induced apoptosis (Figure 5C), however silencing was poor in MM.1s. Regardless, the fact that this protein does not change in expression and that it is not cleaved is consistent with Bid not being important in ATO-induced apoptosis. This would also be consistent with the lack of a role for caspase-8 or CD95 in this death.
BH3-only proteins Bmf, Noxa, and Bim silencing protected cells from ATO-induced apoptosis. MM.1s and KMS11 cell lines were electroporated with siBmf, siNoxa, siBim, siPuma, and siBid (SmartPool; DHARMACON RNA Technologies). Nontargeting siRNA (si(-)) pool was used as a negative control. After 16 hours, cells were treated with 2 μM ATO for 6 and 24 hours for real-time PCR or Western blot analysis and for 24 and 48 hours for ATO-induced apoptosis analysis. (A) Total RNA was obtained for si(-) control and siBmf samples treated with ATO. Real-time PCR was used to determine Bmf transcript expression. Bmf relative expression refers to Bmf transcript expression relative to GAPDH transcript expression. (B) Protein lysates were obtained for si(-) control–, siNoxa-, siBim-, siPuma-, and siBid-transfected cells and silencing was determined by Western blot at 6 and 24 hours after ATO treatment. (C) siRNA electroporated cells were treated with 2 μM ATO for 24 and 48 hours. Viability was evaluated by annexin-V–FITC/PI staining. Percentage (%) of control (untreated, transfected) viability was plotted versus time (hours). The data are presented as the means (± SD) of 4 and 5 independent experiments for MM.1s and KMS11, respectively. t test was used to compare differences among samples, si(-), and experimental combinations unless otherwise indicated, with confidence intervals of 95%. ND indicates no difference; *P < .05; **P < .01; ***P < .001.
BH3-only proteins Bmf, Noxa, and Bim silencing protected cells from ATO-induced apoptosis. MM.1s and KMS11 cell lines were electroporated with siBmf, siNoxa, siBim, siPuma, and siBid (SmartPool; DHARMACON RNA Technologies). Nontargeting siRNA (si(-)) pool was used as a negative control. After 16 hours, cells were treated with 2 μM ATO for 6 and 24 hours for real-time PCR or Western blot analysis and for 24 and 48 hours for ATO-induced apoptosis analysis. (A) Total RNA was obtained for si(-) control and siBmf samples treated with ATO. Real-time PCR was used to determine Bmf transcript expression. Bmf relative expression refers to Bmf transcript expression relative to GAPDH transcript expression. (B) Protein lysates were obtained for si(-) control–, siNoxa-, siBim-, siPuma-, and siBid-transfected cells and silencing was determined by Western blot at 6 and 24 hours after ATO treatment. (C) siRNA electroporated cells were treated with 2 μM ATO for 24 and 48 hours. Viability was evaluated by annexin-V–FITC/PI staining. Percentage (%) of control (untreated, transfected) viability was plotted versus time (hours). The data are presented as the means (± SD) of 4 and 5 independent experiments for MM.1s and KMS11, respectively. t test was used to compare differences among samples, si(-), and experimental combinations unless otherwise indicated, with confidence intervals of 95%. ND indicates no difference; *P < .05; **P < .01; ***P < .001.
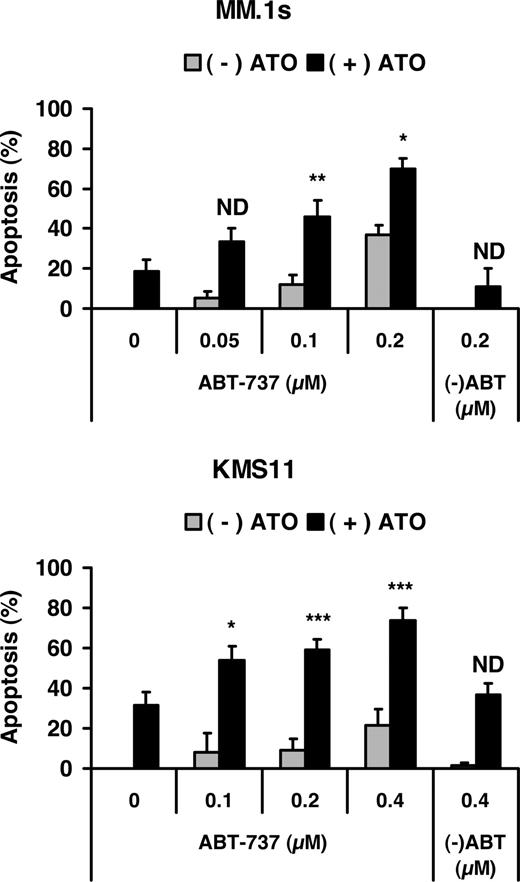
The Noxa/Mcl-1 interaction plays an important role during ATO-induced apoptosis
To further define the role of both BH3-only proteins (Noxa and Bmf) and antiapoptotic proteins (Mcl-1 and Bcl-X) during ATO-induced apoptosis, we examined the effect of combining ATO with the Bcl-2/Bcl-XL inhibitor ABT-737. Dose-response experiments with ABT-737 alone demonstrated that MM.1s was more sensitive to ABT-737 alone than KMS11, suggesting that these cells are likely to be “addicted” to Bcl-2 or Bcl-XL. However, addition of ABT-737 to ATO resulted in greater than additive activity in both cell lines (Figure 6). Similar results were observed in U266 cells, while ATO and ABT-737 were additive only in 8226/S (not shown). Together these data suggest that ATO has killing activity that is independent of Bcl-2 and Bcl-XL and confirm an important role for Noxa in this process.
Effect of Bcl-2/Bcl-XL inhibitor on ATO-induced apoptosis. MM.1s and KMS11 cell lines were treated with 2 μM ATO and indicated concentrations of ABT-737 or the less active enantiomer (-)ABT for 24 hours. Viability was measured by annexin-V–FITC/PI staining. Percentage (%) of apoptosis was plotted versus the concentration of ABT-737. The data are presented as the means (± SD) of 5 independent experiments. t test was used to compare differences between expected additive effect and actual cotreatment results, with confidence intervals of 95%. ND indicates no difference; *P < .05; **P < .01; ***P < .001.
Effect of Bcl-2/Bcl-XL inhibitor on ATO-induced apoptosis. MM.1s and KMS11 cell lines were treated with 2 μM ATO and indicated concentrations of ABT-737 or the less active enantiomer (-)ABT for 24 hours. Viability was measured by annexin-V–FITC/PI staining. Percentage (%) of apoptosis was plotted versus the concentration of ABT-737. The data are presented as the means (± SD) of 5 independent experiments. t test was used to compare differences between expected additive effect and actual cotreatment results, with confidence intervals of 95%. ND indicates no difference; *P < .05; **P < .01; ***P < .001.
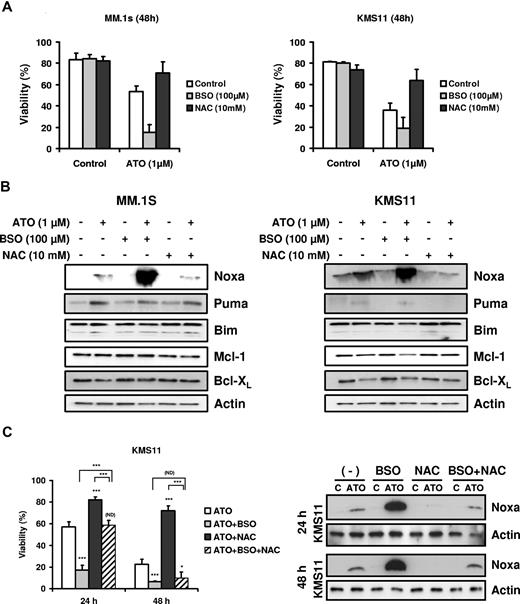
BSO potentiated sensitivity to ATO in MM and enhances up-regulation of Noxa
Finally, we tested the effect of glutathione availability on Bcl-2 family expression. As previously reported, glutathione depletion by BSO sensitized cell to ATO-induced apoptosis, while NAC, a precursor of glutathione synthesis, protected cells from ATO-induced apoptosis (Figure 7A).5,6,17,39-41 BSO and NAC alone did not induce any change in the Bcl-2 family members studied (Figure 7B). However, Noxa up-regulation by ATO was strongly enhanced when cells were cotreated with ATO and BSO and blocked by addition of NAC (Figure 7B). In contrast Bim, Puma, Mcl-1, and Bcl-XL expression was not affected by addition of BSO or NAC. In addition, the combination of BSO and NAC was not able to protect cells from ATO toxicity at 48 hours in the KMS11 cell line (Figure 7C). While NAC partially reversed the effect of BSO on viability at 24 hours, the protection was transient. Similar results were obtained for U266 and 8226/S (not shown). These results demonstrate that NAC protection is due primarily to an increase in intracellular glutathione and not simply an antioxidant effect. Surprisingly, while NAC could not provide protection against ATO-induced death in the presence of BSO, it was able to significantly effect the induction of Noxa in these cells.
BSO sensitized cells to ATO and enhanced up-regulation of the BH3-only protein Noxa. (A) MM.1s and KMS11 cells were treated with 1 μM ATO as a single agent or in combination with BSO (100 μM) or NAC (10 mM) for 48 hours. Viability was evaluated by annexin-V–FITC/PI staining. Data are presented as means (± SD) of 3 independent experiments. (B) MM.1s and KMS11 cells were treated for 24 hours. Protein expression was determined by Western blot. Membranes were reprobed with specific antibodies for Noxa, Puma, Bim, Mcl-1, Bcl-XL, and actin. (C) KMS11 cells were treated with 2 μM ATO as a single agent or in combination with BSO and/or NAC for 24 and 48 hours. Viability was evaluated by annexin-V–FITC/PI staining. Data are presented as means (± SD) of 4 independent experiments. Noxa expression was determined by Western blot. t test was used to compare differences among samples, ATO, and experimental combinations unless otherwise indicated, with confidence intervals of 95%. ND indicates no difference; *P < .05; **P < .01; ***P < .001.
BSO sensitized cells to ATO and enhanced up-regulation of the BH3-only protein Noxa. (A) MM.1s and KMS11 cells were treated with 1 μM ATO as a single agent or in combination with BSO (100 μM) or NAC (10 mM) for 48 hours. Viability was evaluated by annexin-V–FITC/PI staining. Data are presented as means (± SD) of 3 independent experiments. (B) MM.1s and KMS11 cells were treated for 24 hours. Protein expression was determined by Western blot. Membranes were reprobed with specific antibodies for Noxa, Puma, Bim, Mcl-1, Bcl-XL, and actin. (C) KMS11 cells were treated with 2 μM ATO as a single agent or in combination with BSO and/or NAC for 24 and 48 hours. Viability was evaluated by annexin-V–FITC/PI staining. Data are presented as means (± SD) of 4 independent experiments. Noxa expression was determined by Western blot. t test was used to compare differences among samples, ATO, and experimental combinations unless otherwise indicated, with confidence intervals of 95%. ND indicates no difference; *P < .05; **P < .01; ***P < .001.
Discussion
During the last decade, ATO attracted interest for its ability to induce complete remission in patients with APL, mainly through induction of apoptosis and differentiation.42-45 Activity in other hematologic diseases, including multiple myeloma, has been tested in clinical trials. Early clinical studies of ATO for patients with advanced refractory MM resulted in significant responses in approximately one-third of patients with daily dosing schedules9,10 or twice-weekly dosing schedule.46 Several preclinical studies demonstrated that ATO induces growth inhibition and apoptosis in a variety of lymphoid and myeloid malignant cells.41 Its antitumor activity has been associated with the generation of reactive oxygen species (ROSs),47,48 but the exact mechanism(s) for ATO-induced apoptosis is not well defined.
In this report, we studied 4 MM cell lines sensitive to ATO in a time- and a dose-dependent manner. MM.1s and KMS11 were more sensitive to ATO than U266 and 8226/S. Since the unsupervised clustering of genes and arrays did not provide sufficient information to determine how ATO kills the cells, we elected to take a supervised approach to look for gene expression patterns that correlated with this response. Supervised clustering has been very informative in the molecular classification of multiple myeloma as well as searching for genes that are involved in bone disease.49,50 We initially focused on the Bcl-2 family, since these proteins appear to be the initial sensors (BH3-only proteins), regulators (antiapoptotic), and effectors (Bax/Bak) of the intrinsic apoptotic pathway. Interestingly, the gene expression pattern of 3 BH3-only proteins, Bmf, Noxa, and Puma, correlated with ATO sensitivity in the 4 cell lines. Together with the up-regulation of these proapoptotic genes, 2 antiapoptotic Bcl-2 members, Mcl-1 and Bcl-X, were down-regulated by ATO. These changes would be consistent with changes in the balance of Bcl-2 proteins favoring Bax/Bak activation, resulting in permeability of the outer mitochondria membrane. It has been proposed that BH3-only proteins promote Bak activation by displacing it from Mcl-1 and Bcl-XL or by directly activating Bak following their release from antiapoptotic proteins.51-53 Our results seem to be consistent with aspects of both models and suggest that they do not need to be mutually exclusive. In untreated cells, Bak is associated with Mcl-1 and Bcl-XL, consistent with the former model. During ATO-induced apoptosis, BH3-only proteins Noxa, Puma, and possibly Bmf displace Bak from these 2 prosurvival antiapoptotic proteins. Bak is then free for oligomerization and induction of apoptosis. In addition, Bim also appears to play an important role during ATO-induced apoptosis. Bim is not induced by ATO, rather it is also released from Mcl-1 and Bcl-XL. The release of a BH3-only protein from an antiapoptotic protein is not consistent with a role as an inhibitor of these proteins. It is consistent with the latest model where activator BH3-only proteins are sequestered by antiapoptotic proteins to prevent them from directly activating Bak or Bax. This allows one to speculate that both models are correct and that cellular context may determine whether one or both are being used during the induction of cell death. For example, there are numerous reports of Bim induction by death signals, which is consistent with its role as a Bcl-2 inhibitor54-56 ; while in myeloma, disruption of Mcl-1/Bim complexes is consistent with death.57,58 Surprisingly, we do not see Bax bound to Bcl-XL, Mcl-1, or Bcl-2, yet it is found in the heavy membrane fraction and probably associated with mitochondria. While it is possible that Bax is maintained in an inactive state through association with other proteins, it is also possible that it does not need to be sequestered in cells where Bim is bound to antiapoptotic proteins and Bid is not activated. The regulation of Bax in these cells is focus of current studies.
Transient silencing of Noxa and Bmf partially protected cells from ATO-induced apoptosis, indicating that up-regulation of these 2 BH3-only proteins is part of the mechanism(s) for ATO toxicity. Surprisingly, Puma silencing did not protect cells from ATO-induced apoptosis. This result was not expected considering that Puma is up-regulated in all 4 cell lines following ATO treatment. In MM.1s, Puma was strongly up-regulated by ATO and this correlated with Puma binding to Mcl-1, Bcl-XL, and Bcl-2. However, the up-regulation and binding to these antiapoptotic proteins occurs with slower kinetics than Noxa. Noxa is up-regulated within 6 hours and is readily detected bound to Mcl-1 at this time. Moreover, Noxa binding to Mcl-1 correlated with Bak and Bim release.
A new class of drugs that directly interfere with Bcl-2 protein functions is currently being evaluated in preclinical and clinical studies. One drug, ABT-737, a Bad mimetic, blocks antiapoptotic proteins Bcl-2 and Bcl-XL and induces apoptosis in cells that are dependent on either of these proteins for survival.59 Previous experiments have demonstrated that ABT-737 should be effective on its own in tumors with low Mcl-1 expression, and may prove more widely efficacious when combined with agents that prevent Mcl-1 synthesis, promote its degradation, or induce BH3-only proteins that can inactivate it.60-62 ATO induces up-regulation of 3 BH3-only proteins Bmf, Noxa, and Puma that bind Mcl-1, therefore testing it in combination with a Bcl-XL inhibitor is warranted. Consistent with recently published results,63-65 ABT-737 was active in all myeloma cell lines tested in this study (data for U266 and 8226/S are not shown). Importantly, targeting Mcl-1 with Noxa up-regulation by ATO resulted in a synergistic effect when ATO was combined with ABT-737.
While our data clearly demonstrate that Noxa and Bmf are induced by ATO, they do not explain how they are induced. However, one experiment begins to shed light on this. We and others have previously shown that GSH levels are a key determinant of ATO action.5,6,17,39-41 Consistent with these findings, depletion of GSH results in increased Noxa expression, while increasing GSH blocks its induction. Since GSH can have multiple effects on arsenic, via direct conjugation or functioning as an antioxidant,66-69 it remains to be determined whether Noxa induction is due to ROS production or other effects of ATO. Our results suggest that the conjugation of ATO may be more important than the antioxidant effect, as NAC has only a transient effect on ATO-induced cell death in the absence of GSH synthesis. However the enhanced induction of Noxa observed in the absence of GSH synthesis is not observed when NAC is added. This implies that ROS may be important in the induction of the high level of Noxa. Noxa induction is not likely due to p53 as 3 of the 4 cell lines do not display p53 responses due to lack of expression or function. Finally, Puma expression does not appear to be altered by GSH manipulation, suggesting that its regulation is distinct from Noxa.
In summary our results present 3 BH3-only proteins Noxa, Bmf, and Bim as part of the mechanism(s) for ATO-induced apoptosis in multiple myeloma. Understanding the events important for the induction of apoptosis can allow for rational design of combinations of agents with modest activities as single agent in a disease. This is recently exemplified by the use of the AKT inhibitor perifosine in MM. While even less active than ATO in initial trials,70 its combination with bortezomib based on studies of the cellular response to bortezomib should be promising.71 As we understand more about ATO action, we will be in a better position to determine how to use it in effective combinations.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This study was supported in part by National Institutes of Health (NIH) grant R01 CA97243 and by a Senior Research Award from Multiple Myeloma Foundation.
The ABT-737 and enantiomer were a gift from Abbott Laboratories (Abbott Park, IL).
National Institutes of Health
Authorship
Contribution: A.A.M. contributed to the design, execution, data interpretation, and writing of the paper; D.G. contributed to the execution of this work and writing of the paper; K.P.L. was involved in data analyses; L.H.B. conceptualized the idea, designed the research, and contributed to result analyses, discussions, and writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Lawrence H. Boise, 1600 NW 10th Ave, RMSB, Room 3153, Miami, FL 33136; e-mail: lboise@med.miami.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal