Abstract
Low-density lipoprotein receptor–related protein (LRP-1) functions in endocytosis and in cell signaling directly (by binding signaling adaptor proteins) or indirectly (by regulating levels of other cell-surface receptors). Because recent studies in rodents suggest that LRP-1 inhibits inflammation, we conducted activity-based protein profiling experiments to discover novel proteases, involved in inflammation, that are regulated by LRP-1. We found that activated complement proteases accumulate at increased levels when LRP-1 is absent. Although LRP-1 functions as an endocytic receptor for C1r and C1s, complement protease mRNA expression was increased in LRP-1–deficient cells, as was expression of inducible nitric oxide synthase (iNOS) and interleukin-6. Regulation of expression of inflammatory mediators was explained by the ability of LRP-1 to suppress basal cell signaling through the IκB kinase–nuclear factor-κB (NF-κB) pathway. LRP-1–deficient macrophages, isolated from mice, demonstrated increased expression of iNOS, C1r, and monocyte chemoattractant protein-1 (MCP-1); MCP-1 expression was inhibited by NF-κB antagonism. The mechanism by which LRP-1 inhibits NF-κB activity involves down-regulating cell-surface tumor necrosis factor receptor-1 (TNFR1) and thus, inhibition of autocrine TNFR1-initiated cell signaling. TNF-α–neutralizing antibody inhibited NF-κB activity selectively in LRP-1–deficient cells. We propose that LRP-1 suppresses expression of inflammatory mediators indirectly, by regulating TNFR1-dependent cell signaling through the IκB kinase–NF-κB pathway.
Introduction
Low-density lipoprotein (LDL) receptor–related protein (LRP-1) is a member of the LDL receptor gene family, which includes type I transmembrane proteins that function in receptor-mediated endocytosis and cell signaling.1 LRP-1 was first recognized as a receptor for apolipoprotein E and for the protease inhibitor, α2-macroglobulin (α2M)2,3 ; however, it is now clear that LRP-1 binds more than 40 different ligands, well beyond the more limited scope of the LDL receptor.1 LRP-1 ligands include proteases, such as tissue-type plasminogen activator (tPA) and matrix metalloprotease-9, growth factors, extracellular matrix proteins, and foreign toxins. LRP-1 also mediates the endocytosis of plasma membrane proteins, including the urokinase receptor (uPAR), amyloid precursor protein, and tissue factor.4 By regulating cell-surface uPAR, LRP-1 controls the activity of cell signaling factors downstream of uPAR, including extracellular signal-regulated kinase/mitogen-activated protein (ERK/MAP) kinase and Rac1.5,6 In mice, systemic LRP-1 gene deletion is embryonic lethal.7
LRP-1 is synthesized as a 600-kDa single-chain precursor and processed by a furin-like protease into the mature 2-chain form.8 The 85-kDa β-chain includes a small ectodomain, a single transmembrane domain, and the cytoplasmic tail. The 515-kDa α-chain is entirely extracellular but coupled to the β-chain through strong noncovalent forces.8 The α-chain includes 4 clusters of cysteine-rich complement-type repeats (CRs).1 The second and fourth clusters of CRs mediate most ligand-binding interactions, with the exception of the protease inhibitor, α2M, which binds to the first and second CR clusters.9
In addition to its role as a regulator of cell signaling downstream of uPAR, LRP-1 directly activates cell signaling upon binding of ligands, including tPA, activated α2M, and apolipoprotein E.10-12 The exact mechanism by which ligand binding to LRP-1 initiates cell signaling is unclear; however, the LRP-1 β-chain contains 2 NPxY motifs, the more C-terminal of which binds signaling adaptor proteins, such as Shc, c-Jun amino-terminal kinase (JNK)-interacting protein-1/2 (JIP-1/JIP-2), Dab-1, and FE65.13-15 Adaptor protein binding to the LRP-1 β-chain is controlled by tyrosine phosphorylation, which occurs in response to ligand binding or the activity of nonreceptor tyrosine kinases.11,13,16 LRP-1-JIP complex sequesters JNK and thereby prevents its translocation to the nucleus.15 Binding of Shc to LRP-1 provides a mechanism for regulation of downstream factors, such as ERK/MAP kinase and phosphatidylinositol 3-kinase.13
Recent studies suggest that LRP-1 may regulate cell survival,17 angiogenesis,18,19 and inflammation.20 When the LRP-1 gene is selectively disrupted in vascular smooth muscle cells21 or in macrophages and neutrophils,20 atherosclerosis is promoted. Overton et al20 proposed that the activity of LRP-1 in atherosclerosis may be linked to its ability to suppress local inflammation. Similarly, we have collected evidence suggesting that LRP-1 may inhibit inflammation in peripheral nerve injury.22 Thus, understanding the mechanisms by which LRP-1 may regulate inflammation emerges as an important goal.
Because proteases are important mediators in inflammation, we used an approach called activity-based protein profiling (ABPP)23 to identify active proteases that are regulated by LRP-1. We determined that the complement proteases, C1r and C1s, which are members of the first component of complement,24 are more abundant in conditioned medium from LRP-1–deficient cells. In our effort to identify the responsible mechanism, we determined that LRP-1 inhibits complement protease expression by suppressing cell signaling through the IκB kinase (IKK)/nuclear factor-κB (NF-κB) pathway. NF-κB is a ubiquitous regulator of inflammation and cell survival in diverse cell types.25-27 In macrophages, NF-κB drives expression of potent and sometimes cytotoxic cytokines while allowing the cells to survive in this self-created milieu. Our data demonstrate that LRP-1 controls the IKK/NF-κB pathway indirectly, by down-regulating cell-surface expression of tumor necrosis factor receptor-1 (TNFR1). This ability of LRP-1 to regulate cell signaling downstream of TNFR1 represents a novel mechanism by which LRP-1 controls cell physiology.
Methods
Reagents and proteins
Sulfo-NHS-LC-biotin was purchased from Pierce Chemical (Rockford, IL). JNK inhibitor, PD98059, JSH-23, activated C1r, and C1-INH were purchased from EMD Biosciences (San Diego, CA). Rat tumor necrosis factor-α (TNF-α), TNF-α-neutralizing antibody, and C1r-specific antibody were purchased from R&D Systems (Minneapolis, MN). Specific antibodies that detect total IKK, phosphorylated IKK, NF-κB subunit p65/RelA, phosphorylated p65, total IκB-α, and phosphorylated IkB-α were purchased from Cell Signaling Technology (Danvers, MA). TNFR1-specific antibody was purchased from Abcam (Cambridge, MA). Glutathione-S-transferase receptor-associated protein (GST-RAP) was expressed in bacteria and purified as previously described.28 TaqMan quantitative polymerase chain reaction (qPCR) primers, probes, and reagents were purchased from Applied Biosystems (Foster City, CA). Smart-pool siRNA targeting mouse LRP-1 and nontargeting pooled control (NTC) siRNA were from Dharmacon RNA Technologies (Lafayette, CO).
Cell culture
RAW 264.7 macrophage-like cells, murine embryonic fibroblasts (MEFs) that are genetically deficient in LRP-1 (MEF-2 cells), and control LRP-1(+/−) MEFs (PEA-10 cells) were obtained from the ATCC (Manassas, VA). PEA-10 and MEF-2 cells were cloned from the same MEF culture, heterozygous for LRP-1 gene disruption, and selected with the LRP-1-selective toxin, Pseudomonas exotoxin A.7 B-41 cells are MEF-2 cells that were transfected for stable expression of full-length human LRP-1.29 RAW 264.7 were cultured in RPMI 1640 supplemented with 10% fetal bovine serum (FBS) and penicillin/streptomycin. MEFs were cultured in Dulbecco modified Eagle medium (DMEM) with 10% FBS. To silence LRP-1, RAW 264.7 cells were transfected with siRNA targeting LRP-1 (2 μg) using nucleofector technology. Control cells were transfected with NTC siRNA. Transfections were executed 36 hours before performing experiments. CL-16 cancer cells, which are derived from MDA-MB-435 cells and transfected to express LRP-1-specific shRNA (shLRP-1 cells) or with the empty vector, pSUPER (control CL-16 cells), have been previously described.30
ABPP and MudPIT analysis
MEFs were cultured until 80% confluent in DMEM with 10% FBS (complete medium), washed 3 times in 20 mM of sodium phosphate, 150 mM NaCl, pH 7.4 (phosphate-buffered saline [PBS]), and then cultured in serum-free DMEM for 48 hours. Conditioned medium (CM) was harvested and cleared by centrifugation at 2400g for 5 minutes. Proteins in CM samples were precipitated with ammonium sulfate (0.561 g/mL CM) overnight at 4°C. The precipitated proteins then were resuspended in 50 mM Tris-HCl, pH 7.5, and desalted on a PD-10 column (GE Healthcare, Little Chalfont, United Kingdom). To detect active serine hydrolases, samples were adjusted to a final protein concentration of 1 mg/mL in 50 mM Tris-HCl, pH 7.5, and treated with 1 μM rhodamine-coupled fluorophosphonate for 1 hour at 22°C.31 A portion of each sample was treated with PNGaseF (New England Biolabs, Ipswich, MA) to dissociate N-linked glycans. Reactions were quenched with one volume of standard 2× sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) loading buffer. Samples were then subjected to SDS-PAGE and visualized in-gel with a Hitachi FMBio IIe flatbed fluorescence scanner (Hitachi Software Engineering, Yokohama, Japan).
For multidimensional protein identification technology (MudPIT) analysis, desalted CM samples, containing 1.0 mg total protein in 1.0 mL, were treated with fluorophosphonate-polyethylene glycol-biotin (FP-PB; 5 μM) for 2 hours at 22°C. Preparations were then subjected to avidin-affinity chromatography, as previously described.31 While immobilized on avidin, FP-PB-labeled proteins were washed successively with 1% SDS, 6 M urea, and then 50 mM Tris-HCl, pH 8.0 (2 washes each). The beads and associated proteins were suspended in 200 μL of 8 M of urea, 50 mM Tris-HCl, pH 8.0, and treated with the reductant, tris(2-carboxylethyl) phosphine (10 mM), followed by the alkylating agent, iodoacetamide (12 mM). “On-bead protein digestion” was performed overnight by incubation with trypsin at 37°C in 2 M urea, 50 mM Tris-HCl, pH 8.0, and 2 mM CaCl2. Solutions of tryptic peptides were acidified to a final concentration of 5% formic acid, loaded onto a biphasic capillary column, and analyzed by 2D liquid chromatography in combination with tandem mass spectrometry, as previously described.32
C1r internalization
Activated C1r was radiolabeled with Na125I using iodo-Beads (Pierce Chemical). C1r-C1INH complex was formed by mixing equal concentrations of 125I-labeled C1r and C1-INH for 1 hour at 20°C. Complex formation was assessed by SDS-PAGE. Cells were seeded in 24-well plates and cultured until confluent. To begin an experiment, the cells were washed and then preincubated for 1 hour in binding buffer, which consists of DMEM, 20 mM N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 15 mg/mL BSA, and insulin-transferrin-selenium complement (Invitrogen, Carlsbad, CA). The medium was then removed and replaced by fresh binding buffer containing 5 nM 125I-labeled C1r or 125I-labeled C1r-C1INH complex. Incubations were allowed to proceed for 3 hours at 37°C. In some wells, GST-RAP (0.5 μM) was added. To determine cellular internalization, the cultures were washed twice with cold binding buffer and then treated for 15 minutes with 0.25% pronase in DMEM at 4°C. The detached cells were pelleted by centrifugation at 3000g for 3 minutes. Radioactivity associated with the cell pellet was measured in a gamma counter.
Electrophoretic mobility shift assay
Cells were grown in serum-free medium (SFM) for 18 hours and harvested in PBS containing 5 mM ethylenediaminetetraacetic acid. Nuclear extracts were prepared from 2 × 106 cells, using Panomics extraction reagents (Panomics, Fremont, CA). Nuclear NF-κB was determined by electrophoretic mobility shift assay (EMSA) using NF-κB–specific biotin-labeled and unlabeled probes, provided by Panomics.
Luciferase assay
PEA-10 and MEF-2 cells were cotransfected with the promoter-reporter construct, pNF-κB-Luc (Stratagene, La Jolla, CA) and with pRL-TK (Promega, Madison, WI) using FuGENE HD (Roche Diagnostics, Indianapolis, IN). pRL-TK drives expression of Renilla luciferase and serves as a transfection efficiency control. The ratio of pNF-κB-Luc/pRL-TK, in the transfection mixture, was 25:1. Transfected cells were cultured in SFM for 18 hours and then cultured for an additional 6 hours in SFM, or in SFM with TNF-α (50 ng/mL), lipopolysaccharide (LPS; 100 ng/mL), TNF-α-neutralizing antibody, or control IgG (2 μg/mL). Firefly and Renilla luciferase activity was determined using the Dual-Luciferase reporter assay system (Promega).
Quantitative PCR
Total RNA was extracted from confluent cultures that were maintained for 18 hours in SFM, using the RNAeasy kit (QIAGEN, Hilden, Germany). In some experiments, cells were grown in the presence of JNK inhibitor (10 μM), JSH-23 (10 μM), PD98059 (40 μM), or DMSO as a control. cDNA was synthesized using the iScript cDNA synthesis kit (Bio-Rad, Hercules, CA). qPCR was performed using a System 7300 instrument (Applied Biosystems) and a one-step program: 95°C, 10 minutes; 95°C, 30 seconds, 60°C, 1 minute for 40 cycles. Hypoxanthine phosphoribosyl transferase gene expression was measured as a normalizer for each sample. Results were analyzed by the relative quantity (ΔΔCt) method, as previously described.33 All experiments were performed in triplicate with internal triplicate determinations.
Surface-protein labeling and affinity precipitation
Cell-surface proteins were biotinylated using the membrane-impermeable reagents, sulfo-NHS-LC-biotin and sulfo-NHS-SS-biotin (1 mg/mL), as previously described.34 The biotin-labeled cells were washed with 100 mM glycine in PBS supplemented with 0.5 mM CaCl2 and 0.5 mM MgCl2. Cell extracts were prepared in 1% Triton X-100, 0.5% deoxycholate, 0.1% SDS in PBS with 2 mM phenylmethylsulfonyl fluoride, 2 mM ethylenediaminetetraacetic acid, and 2 mM sodium orthovanadate (radioimmunoprecipitation [RIPA] buffer). Cellular debris was removed by centrifugation at 13 000 g at 4°C for 15 minutes. Equal amounts of cellular protein were incubated with streptavidin-agarose, overnight at 4°C. The streptavidin beads were washed 2 times with RIPA buffer and again with 20 mM Tris-HCl and 150 mM NaCl, pH 7.4 (Tris-buffered saline). Proteins were eluted with SDS sample buffer for SDS-PAGE and immunoblot analysis.
SDS-PAGE and immunoblotting
Cells were extracted in RIPA buffer. Equal amounts of cellular protein were subjected to SDS-PAGE35 and electrotransferred to polyvinylidene fluoride membranes (Bio-Rad). Proteins were visualized using 0.2% Ponceau-S in 3% trichloroacetic acid before immunoblot analysis. Membranes then were blocked with 5% nonfat dry milk in Tris-buffered saline, 0.1% Tween 20. Purified primary antibodies and horseradish peroxidase–conjugated secondary antibodies (GE Healthcare) were diluted in the same buffer. Detection was performed using Western Lightning horseradish peroxidase chemiluminescence (PerkinElmer Life and Analytical Sciences, Waltham, MA) and Kodak Biomaxlight Films (Rochester, NY).
FACS analysis
Cell-surface TNFR1 was detected using the Fluorokine Kit (R&D Systems). Briefly, MEFs were detached by scraping and incubated with biotin-labeled human TNF-α for 1 hour at 4°C. Human TNF-α binds only to TNFR1 on murine cells.36 Cells were incubated for an additional 30 minutes at 4°C with fluorescein isothiocyanate-labeled streptavidin, washed, and subjected to fluorescence-activated cell sorting (FACS) analysis using FACSCanto (BD Biosciences, San Jose, CA). The data were analyzed using FlowJo software (TreeStar, Ashland, OR).
TNF-α binding
Rat recombinant TNF-α was radiolabeled with Na125I using iodo-Beads (Pierce Chemical). Cells were seeded in 24-well plates and cultured until confluent. To begin an experiment, cells were cultured for 1 hour in binding-buffer (Earle balanced salt solution, 20 mM of N-2-hydroxyethylpiperazine-N′-2-ethanesulfonic acid, pH 7.4, 0.1% BSA). 125I-labeled TNF-α (1 nM) in binding buffer was incubated with the cells for 3 hours at 4°C. Cells were then washed 3 times and extracted in 0.1 M of NaOH, 1% SDS. Radioactivity associated with the extract was measured in a gamma counter. Cellular protein recovery was determined by bicinchoninic acid assay.
Peritoneal macrophage collection
Peritoneal macrophages were obtained from mice in which LRP-1 is conditionally deleted in neutrophils and macrophages and from wild-type mice in the same genetic background (C57BL/6).20 In brief, 1 mL of thioglycolate broth was injected intraperitoneally in each mouse. Three days later, cells were recovered by flushing with 10 mL ice-cold PBS. Preparations of intraperitoneal cells, which consist primarily of macrophages, were washed twice with ice-old PBS and frozen for qPCR analysis. All experiments were conducted according to the guidelines and under approval of the Vanderbilt University Institutional Animal Care and Usage Committee.
Results
LRP-1 regulates multiple serine hydrolases in MEFs
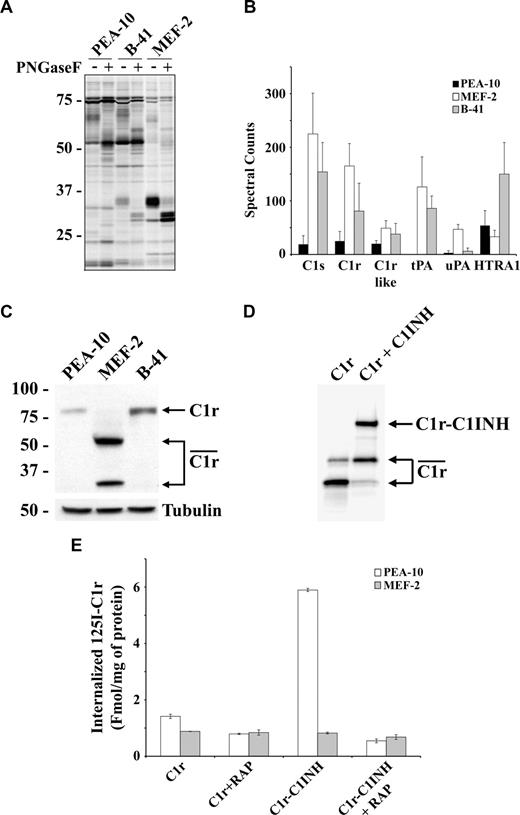
When the LRP-1 gene is deleted in macrophages in vivo, increased levels of inflammatory mediators collect in atherosclerotic plaques.20 To identify novel serine hydrolases that are regulated by LRP-1 and may function as mediators of the inflammatory response, we analyzed CM from PEA-10, MEF-2, and B-41 cells by ABPP. To begin, active serine hydrolases were labeled with rhodamine-coupled fluorophosphonate and imaged in gels. Figure 1A shows that the pattern of identified serine hydrolases was different in CM from LRP-1-deficient MEF-2 cells, compared with LRP-1-expressing PEA-10 cells. Expression of human LRP-1 in MEF-2 cells (the B-41 cell line) restored a pattern that more closely resembled that observed in PEA-10 cells.
Active proteases are increased in conditioned medium (CM) from LRP-1–deficient cells. (A,B) CM from PEA-10, MEF-2, and B-41 cells was analyzed by ABPP. The probes used are specific for active serine hydrolases. (C) CM from PEA-10, MEF-2, and B-41 cells was analyzed by immunoblot using C1r-specific antibody (top panel). The corresponding cell extracts were immunoblotted using antibody specific for tubulin to confirm that the CM was collected from an equivalent number of cells (bottom panel). (D) 125I-labeled active C1r was incubated for 1 hour with or without C1-INH. Active C1r is shown by the connected arrows. The complex of the C1r light chain with C1-INH (C1r-C1INH) migrates as the low mobility band. (E) 125I-C1r or 125I-C1r-C1-INH was incubated for 3 hours at 37°C with PEA-10 or MEF-2 cells. Receptor-associated protein (RAP; 0.5 μM) was added to some cells during the incubation. Cells were washed and treated with pronase to remove cell surface-associated 125I-C1r. Radioactivity associated with the cell pellets was determined. Error bars represent SD.
Active proteases are increased in conditioned medium (CM) from LRP-1–deficient cells. (A,B) CM from PEA-10, MEF-2, and B-41 cells was analyzed by ABPP. The probes used are specific for active serine hydrolases. (C) CM from PEA-10, MEF-2, and B-41 cells was analyzed by immunoblot using C1r-specific antibody (top panel). The corresponding cell extracts were immunoblotted using antibody specific for tubulin to confirm that the CM was collected from an equivalent number of cells (bottom panel). (D) 125I-labeled active C1r was incubated for 1 hour with or without C1-INH. Active C1r is shown by the connected arrows. The complex of the C1r light chain with C1-INH (C1r-C1INH) migrates as the low mobility band. (E) 125I-C1r or 125I-C1r-C1-INH was incubated for 3 hours at 37°C with PEA-10 or MEF-2 cells. Receptor-associated protein (RAP; 0.5 μM) was added to some cells during the incubation. Cells were washed and treated with pronase to remove cell surface-associated 125I-C1r. Radioactivity associated with the cell pellets was determined. Error bars represent SD.
Next, CM samples were treated with FP-PB, which covalently modifies the serine hydrolase active site for subsequent purification and analysis by mass spectrometry.31,37 Figure 1B shows that the complement proteases, C1s and C1r, C1r-like protease, uPA, and tPA were significantly increased (P < .05) in CM from LRP-1–deficient MEF-2 cells compared with PEA-10 cells. The increases in tPA and uPA were anticipated because LRP-1 functions as an endocytic receptor for these proteases.38,39 C1s is known to be internalized by LRP-1 after complex formation with C1-INH.40 Expression of human LRP-1, in B-41 cells, rescued these cells with regard to their ability to regulate uPA. Human LRP-1 less completely reversed the effects of LRP-1 deficiency on the other serine proteases, perhaps reflecting partial species specificity in the activity of LRP-1. The level of cell-surface human LRP-1 in B-41 cells is equivalent to the level of murine LRP-1 in PEA-10 cells.29
Regulation of C1r by LRP-1
To validate our ABPP results, CM samples from PEA-10 and MEF-2 cells were subjected to immunoblot analysis to detect C1r (Figure 1C). C1r was present in CM from PEA-10 and B-41 cells, principally in the single-chain 90-kDa form, which is known to be the zymogen.43 In MEF-2 cells, the 90-kDa band was largely absent. Instead, substantial amounts of the 2-chain form were present. This is known to be the active species, formed by auto-activation44 and detected by ABPP. Considering the variousC1r bands together, the total level of C1r was substantially increased in MEF-2 cell CM.
On activation, C1r and C1s form covalent complexes with C1-INH.45 LRP-1 is necessary for the clearance of the C1s-C1-INH complex.40 To determine whether LRP-1 mediates the clearance of C1r-C1-INH complex, 125I-C1r was incubated with purified C1-INH. A high molecular mass band, detected by autoradiography, corresponded to the mass of the 125I-C1r light chain bound to C1-INH (Figure 1D). 125I-C1r and 125I-C1r-C1-INH complex were incubated with PEA-10 and MEF-2 cells. As shown in Figure 1E, 125I-C1r internalization was promoted by C1-INH only in LRP-1–expressing cells. Internalization of 125I-C1r-C1-INH complex was inhibited by receptor-associated protein (RAP), confirming the role of LRP-1.
Expression of complement components is increased in LRP-1–deficient cells
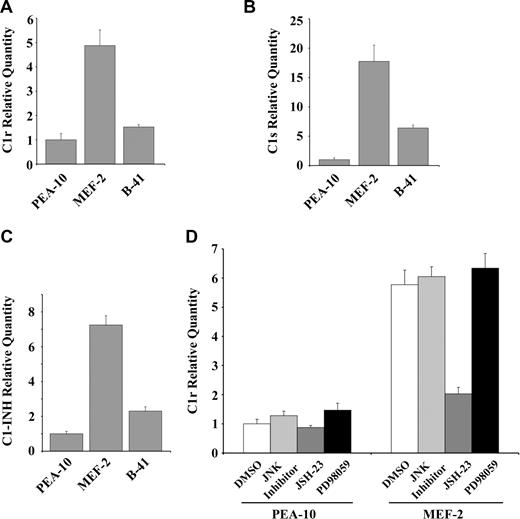
In addition to its role as an endocytic receptor, LRP-1 functions in cell signaling and regulates gene transcription.18,46 To determine whether LRP-1 regulates expression of complement components, we determined mRNA levels for C1r, C1s, and C1-INH in PEA-10, MEF-2, and B-41 cells. As shown in Figure 2A-C, expression of all 3 mRNAs was significantly increased in LRP-1–deficient MEF-2 cells. This effect was at least partially rescued by expression of human LRP-1 in B-41 cells.
LRP-1 regulates expression of complement components. PEA-10, MEF-2, and B-41 cells were grown for 18 hours in SFM and then analyzed by qPCR to determine expression of (A) C1r, (B) C1s, and (C) C1INH. (D) PEA-10 and MEF-2 cells were grown in SFM for 18 hours in the presence of JSH-23 (10 μM), JNK inhibitor (10 μM), PD98059 (40 μM), or vehicle (DMSO). C1r expression was determined by qPCR. Error bars represent SD.
LRP-1 regulates expression of complement components. PEA-10, MEF-2, and B-41 cells were grown for 18 hours in SFM and then analyzed by qPCR to determine expression of (A) C1r, (B) C1s, and (C) C1INH. (D) PEA-10 and MEF-2 cells were grown in SFM for 18 hours in the presence of JSH-23 (10 μM), JNK inhibitor (10 μM), PD98059 (40 μM), or vehicle (DMSO). C1r expression was determined by qPCR. Error bars represent SD.
To identify candidate signaling cascades involved in the regulation of complement component expression by LRP-1, we examined C1r gene expression in PEA-10 and MEF-2 cells, after treatment with pharmacologic inhibitors of several cell-signaling enzymes (Figure 2D). Specific JNK inhibitor and the MEK-1 inhibitor, PD98059, did not regulate C1r mRNA expression. By contrast, JSH-23, which inhibits nuclear translocation of NF-κB,47 decreased C1r expression selectively in MEF-2 cells.
NF-κB functions as a major regulator of inflammation, cell survival, and as a mediator of the response to TNF-α.25 To test whether the basal level of NF-κB activity is increased in LRP-1–deficient MEFs, nuclear extracts were prepared and subjected to EMSA, using an NF-κB–specific biotin-labeled probe. As shown in Figure 3A (representative experiment, n = 3), active NF-κB was substantially increased in MEF-2 cells, compared with PEA-10 and B-41 cells. Excess unlabeled NF-κB probe (cold probe) blocked the specific band in all 3 cell lines. Similar results were obtained when we compared CL-16 cancer cells in which LRP-1 was silenced (shLRP-1 cells) with control CL-16 cells in which LRP-1 is expressed (Figure 3B). Again, loss of LRP-1 was associated with increased NF-κB activity.
LRP-1 inhibits NF-κB activation. (A) Nuclear extracts from PEA-10, MEF-2, and B-41 cells were analyzed by EMSA using an NF-κB–specific biotin-labeled probe, in the presence or absence of an excess concentration of the cold probe. (B) Nuclear extracts from CL-16 cells that express LRP-1 and from shLRP-1 cells, which are CL16 cells in which LRP-1 has been silenced with shRNA, were analyzed by EMSA for NF-κB activity. (C-E) RAW 264.7 cells were transiently transfected with NTC or LRP-1-specific siRNA. (C) Expression of LRP-1 was determined by qPCR 36 hours later. (D) Expression of C1r was determined 36 hours later. Error bars represent SD. (E) EMSA assay for active NF-κB using nuclear extracts from RAW 264.7 cells transfected with NTC or LRP-1-specific siRNA.
LRP-1 inhibits NF-κB activation. (A) Nuclear extracts from PEA-10, MEF-2, and B-41 cells were analyzed by EMSA using an NF-κB–specific biotin-labeled probe, in the presence or absence of an excess concentration of the cold probe. (B) Nuclear extracts from CL-16 cells that express LRP-1 and from shLRP-1 cells, which are CL16 cells in which LRP-1 has been silenced with shRNA, were analyzed by EMSA for NF-κB activity. (C-E) RAW 264.7 cells were transiently transfected with NTC or LRP-1-specific siRNA. (C) Expression of LRP-1 was determined by qPCR 36 hours later. (D) Expression of C1r was determined 36 hours later. Error bars represent SD. (E) EMSA assay for active NF-κB using nuclear extracts from RAW 264.7 cells transfected with NTC or LRP-1-specific siRNA.
Next, we silenced LRP-1 expression in RAW 264.7 macrophage-like cells and studied the effects on C1r expression and NF-κB activity. Figure 3C shows that, 36 hours after transfection, LRP-1 mRNA was decreased by more than 40%. The transfection efficiency was approximately 50%, as determined by green fluorescent protein expression, suggesting that the degree of LRP-1–silencing in the transfected cell subpopulation was probably much greater than 40%. Figure 3D shows that C1r mRNA expression was significantly increased in RAW 264.7 cells in which LRP-1 was silenced. NF-κB activity also was increased in LRP-1–silenced RAW 264.7 cells, as determined by EMSA (Figure 3E).
The IKK–NF-κB pathway is activated in LRP-1–deficient cells
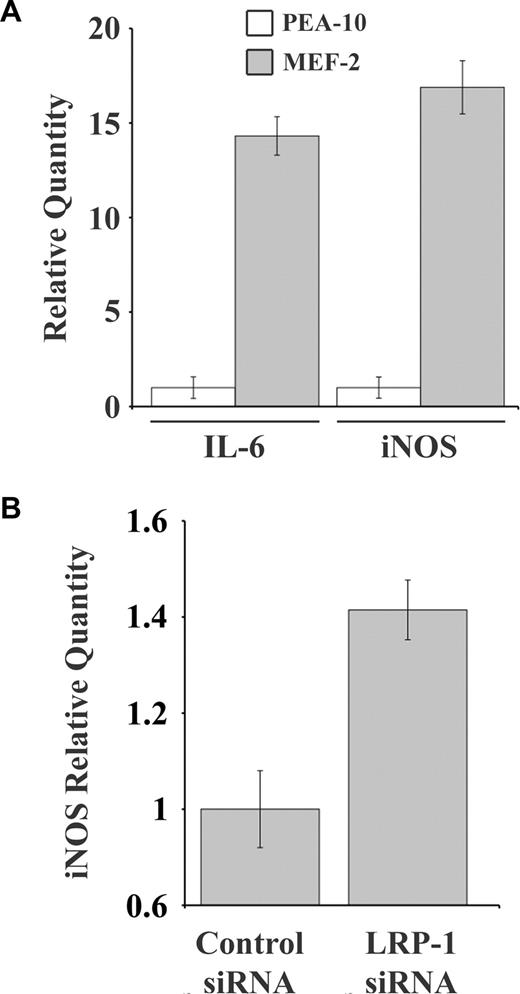
Nuclear NF-κB promotes transcription of various genes, including inducible nitric oxide synthase (iNOS) and interleukin-6 (IL-6).26,27 Figure 4A shows that the mRNA levels for iNOS and IL-6 are increased in MEF-2 cells, compared with PEA-10 cells, providing further evidence for activation of NF-κB in LRP-1–deficient cells. iNOS mRNA also was significantly increased (P < .05) in RAW 264.7 cells that were transfected with LRP-1–specific siRNA (Figure 4B).
IL-6 and iNOS expression is increased in LRP-1–deficient cells. (A) PEA-10 and MEF-2 cells were grown for 18 hours in SFM. Expression of IL-6 and iNOS was analyzed by qPCR. (B) RAW 264.7 cells were transiently transfected with NTC or LRP-1–specific siRNA. Expression of iNOS was analyzed by qPCR 36 hours after transfection. Error bars represent SD.
IL-6 and iNOS expression is increased in LRP-1–deficient cells. (A) PEA-10 and MEF-2 cells were grown for 18 hours in SFM. Expression of IL-6 and iNOS was analyzed by qPCR. (B) RAW 264.7 cells were transiently transfected with NTC or LRP-1–specific siRNA. Expression of iNOS was analyzed by qPCR 36 hours after transfection. Error bars represent SD.
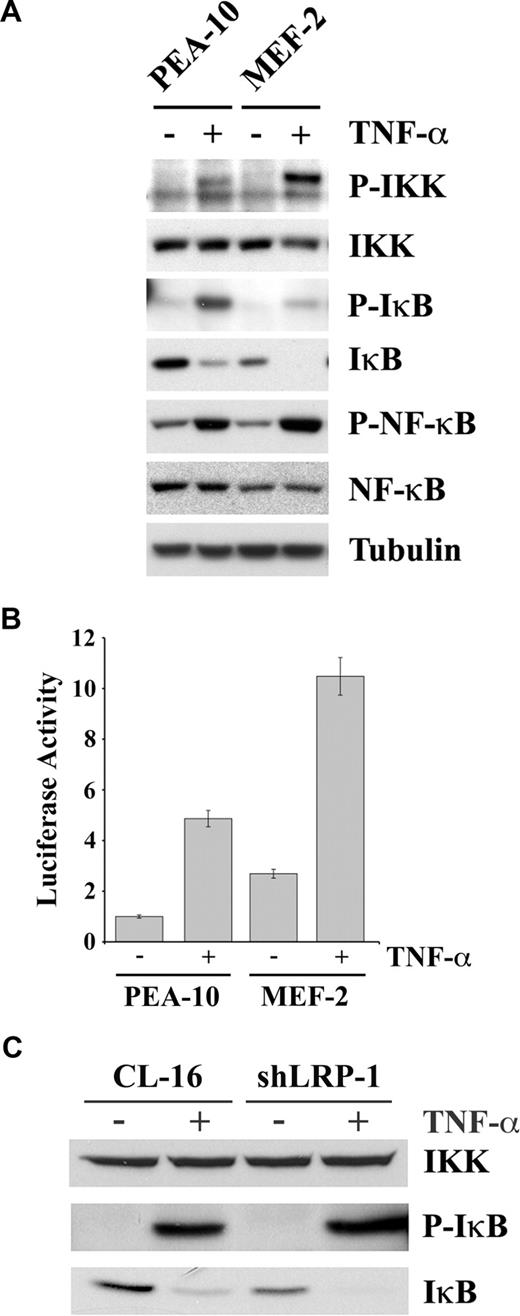
To determine the mechanism by which LRP-1 deficiency may be associated with activation of NF-κB, we studied cell signaling under basal conditions and in response to TNF-α in MEFs. Figure 5A shows that, in response to TNF-α, phosphorylation of IKK was substantially increased in MEF-2 cells. The β subunit of IKK, which is the slower migrating of the IKK bands, seemed to be preferentially phosphorylated. The NF-κB inhibitory protein, IκB, was present at a substantially decreased level in MEF-2 cells, before treatment with TNF-α. IκB binds NF-κB, retaining it in the cytoplasm.25 IKK-induced phosphorylation of IκB promotes IκB degradation and release of activated NF-κB.48,49 In response to TNF-α, more IκB phosphorylation was observed in PEA-10 cells, compared with MEF-2 cells, probably reflecting the higher substrate concentration. However, after TNF-α treatment, IκB was undetectable in MEF-2 cells. The p65/RelA form of NF-κB was phosphorylated in response to TNF-α in both PEA-10 and MEF-2 cells. This can occur downstream of IKK or other kinases.50 The total level of p65/RelA was slightly decreased in MEF-2 cells, despite the increase in NF-κB activity.
The IKK-NF-κB pathway is activated in LRP-1-deficient cells. (A) PEA-10 and MEF-2 cells were treated with TNF-α (50 ng/mL) or with vehicle for 10 minutes. Equal amounts of cell extract were subjected to immunoblot analysis for phosphorylated IKK, total IKK, phosphorylated IκB, total IκB, phosphorylated NF-κB, total NF-κB, and tubulin. (B) PEA-10 and MEF-2 cells were transfected with pNF-κB-Luc to measure NF-κB activity. Cells were treated with TNF-α (50 ng/mL) or vehicle. Luciferase activity was measured 6 hours later. Error bars represent SD. (C) CL-16 and shLRP-1 cells were treated with TNF-α (50 ng/mL) or with vehicle for 10 minutes. Equal amounts of cell extract were subjected to immunoblot analysis for total IKK, phosphorylated IκB, and total IκB.
The IKK-NF-κB pathway is activated in LRP-1-deficient cells. (A) PEA-10 and MEF-2 cells were treated with TNF-α (50 ng/mL) or with vehicle for 10 minutes. Equal amounts of cell extract were subjected to immunoblot analysis for phosphorylated IKK, total IKK, phosphorylated IκB, total IκB, phosphorylated NF-κB, total NF-κB, and tubulin. (B) PEA-10 and MEF-2 cells were transfected with pNF-κB-Luc to measure NF-κB activity. Cells were treated with TNF-α (50 ng/mL) or vehicle. Luciferase activity was measured 6 hours later. Error bars represent SD. (C) CL-16 and shLRP-1 cells were treated with TNF-α (50 ng/mL) or with vehicle for 10 minutes. Equal amounts of cell extract were subjected to immunoblot analysis for total IKK, phosphorylated IκB, and total IκB.
We hypothesized that the decrease in the basal level of IκB in MEF-2 cells was the result of activation of autocrine signaling through the IKK/NF-κB pathway, in the absence of exogenously added agents. To test this hypothesis, first we measured IκB mRNA expression by qPCR and found that IκB expression is higher in MEF-2 cells compared with PEA-10 cells (results not shown). Next, we measured NF-κB activity before and after TNF-α treatment, in PEA-10 and MEF-2 cells, using a quantitative method. The cells were transfected with pNF-κB-Luc, a promoter-reporter construct with 3 NF-κB response elements, which reports NF-κB activity and with pRL-TK as a control. Figure 5B demonstrates that, in the absence of TNF-α, luciferase expression was increased 3-fold in MEF-2 cells, compared with PEA-10 cells. Thus, NF-κB is indeed activated under basal conditions in MEF-2 cells, in the absence of exogenously added agents, consistent with the loss of IκB and the results of our EMSA assays. In response to TNF-α, NF-κB activity was increased in both cell lines; however, NF-κB activity remained more elevated in the MEF-2 cells.
To confirm our results regarding LRP-1 and IκB in a second model system, we examined expression of IκB protein in CL-16 cells. As shown in Figure 5C, the level of IKK was unchanged when LRP-1 was silenced in CL-16 cells; however, the basal level of IκB was decreased (bottom panel), confirming the results obtained with MEFs.
Cell-surface TNFR1 is increased in LRP-1–deficient cells
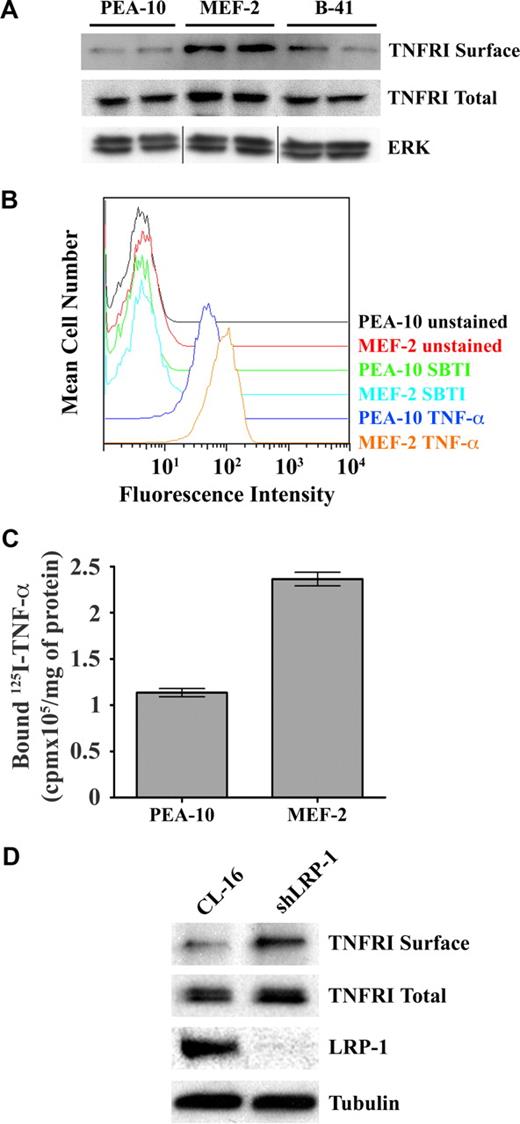
To further probe the mechanism by which LRP-1 controls NF-κB activity, we examined TNFR1 in MEFs. Figure 6A shows that the total level of TNFR1 was slightly increased in MEF-2 cells, compared with PEA-10 and B-41 cells; however, cell-surface TNFR1 was substantially increased in MEF-2 cells. The increase in cell-surface TNFR1 in MEF-2 cells was confirmed by FACS analysis of the binding of human TNF-α to MEFs (Figure 6B). We also examined the binding of 125I-TNF-α to MEFs at 4°C. Figure 6C shows that 125I-TNF-α-binding to MEF-2 cells was increased approximately 2-fold compared with PEA-10 cells (P < .05). Because our binding assay measured total as opposed to specific binding, the actual difference in specific binding was probably greater. Figure 6D shows that, in a second model system (CL-16 cancer cells), LRP-1-silencing is associated with a modest increase in total cellular TNFR1 and with a more substantial increase in cell-surface TNFR1, confirming our results in MEFs.
LRP-1 regulates cell-surface TNFR1. (A) PEA-10, MEF-2, and B-41 cells were grown for 18 hours in SFM. Cell-surface proteins then were biotin-labeled. The biotin-labeled proteins were purified by streptavidin-Sepharose affinity chromatography and then subjected to immunoblot analysis for TNFR1 (TNFR1 surface). Total cell extracts were also subjected to immunoblot analysis for TNFR1 (TNFR1 total) and for total ERK/MAP kinase as a control for protein load (vertical lines have been inserted to represent a repositioned gel lane). (B) PEA-10 and MEF-2 cells were incubated with biotin-labeled human TNF-α, followed by fluorescein isothiocyanate-labeled streptavidin. The cells then were subjected to FACS analysis. Control preparations were incubated with biotin-labeled soybean trypsin inhibitor (SBTI), which does not bind to cell-surface proteins. (C) 125I-labeled TNF-α was incubated with PEA-10 or MEF-2 cells for 4 hours at 4°C. The cells were washed and lysed in 0.1 M NaOH, 1% SDS. Radioactivity associated with the cell lysates was determined using a γ-counter. Error bars represent SD. (D) Cell-surface proteins from CL-16 cells and shLRP-1 cells were labeled with biotin and purified by streptavidin-Sepharose chromatography. The purified proteins were subjected to immunoblot analysis for TNFR1 (TNFR1 surface) or LRP-1. Total cell extracts also were subjected to immunoblot analysis for TNFR1 or tubulin as a measure of protein load.
LRP-1 regulates cell-surface TNFR1. (A) PEA-10, MEF-2, and B-41 cells were grown for 18 hours in SFM. Cell-surface proteins then were biotin-labeled. The biotin-labeled proteins were purified by streptavidin-Sepharose affinity chromatography and then subjected to immunoblot analysis for TNFR1 (TNFR1 surface). Total cell extracts were also subjected to immunoblot analysis for TNFR1 (TNFR1 total) and for total ERK/MAP kinase as a control for protein load (vertical lines have been inserted to represent a repositioned gel lane). (B) PEA-10 and MEF-2 cells were incubated with biotin-labeled human TNF-α, followed by fluorescein isothiocyanate-labeled streptavidin. The cells then were subjected to FACS analysis. Control preparations were incubated with biotin-labeled soybean trypsin inhibitor (SBTI), which does not bind to cell-surface proteins. (C) 125I-labeled TNF-α was incubated with PEA-10 or MEF-2 cells for 4 hours at 4°C. The cells were washed and lysed in 0.1 M NaOH, 1% SDS. Radioactivity associated with the cell lysates was determined using a γ-counter. Error bars represent SD. (D) Cell-surface proteins from CL-16 cells and shLRP-1 cells were labeled with biotin and purified by streptavidin-Sepharose chromatography. The purified proteins were subjected to immunoblot analysis for TNFR1 (TNFR1 surface) or LRP-1. Total cell extracts also were subjected to immunoblot analysis for TNFR1 or tubulin as a measure of protein load.
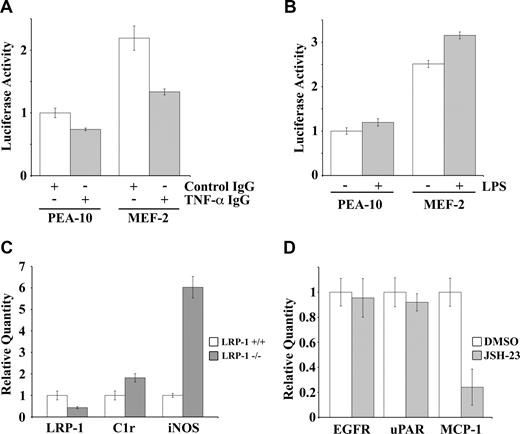
We hypothesized that the ability of LRP-1 to regulate cell-surface TNFR1 and TNFR1-dependent cell signaling accounts for the increase in NF-κB activity in LRP-1–deficient cells. To test this hypothesis, first we examined NF-κB activity in cells that were cultured in the presence of TNF-α-neutralizing antibody or control immunoglobulin G for 18 hours. As shown in Figure 7A, TNF-α-neutralizing antibody decreased NF-κB activity by 40% in MEF-2 cells. A more modest effect was observed in PEA-10 cells.
LRP-1 deletion in macrophages in vivo increases expression of C1r and iNOS. (A) PEA-10 and MEF-2 cells were transfected with pNF-κB-Luc to measure NF-κB activity. Cells were treated with TNF-α-neutralizing antibody or control IgG for 18 hours. Luciferase activity then was measured. (B) PEA-10 and MEF-2 cells were transfected with pNF-κB-Luc. Cells were treated with LPS (100 ng/mL) or vehicle. Luciferase activity was measured 6 hours later. (C) Macrophages were harvested from the IP space of mice in which LRP-1 was conditionally deleted in macrophages and neutrophils and from control wild-type mice in the same genetic background. RNA was isolated from peritoneal cells 3 days after thioglycolate stimulation. Expression of LRP-1, C1r, and iNOS was determined by qPCR. (D) Peritoneal macrophages were cultured for 12 hours in the presence of JSH-23 (10 μM) or vehicle. Expression of EGFr, uPAR, and MCP-1 was analyzed by qPCR. Error bars represent SD.
LRP-1 deletion in macrophages in vivo increases expression of C1r and iNOS. (A) PEA-10 and MEF-2 cells were transfected with pNF-κB-Luc to measure NF-κB activity. Cells were treated with TNF-α-neutralizing antibody or control IgG for 18 hours. Luciferase activity then was measured. (B) PEA-10 and MEF-2 cells were transfected with pNF-κB-Luc. Cells were treated with LPS (100 ng/mL) or vehicle. Luciferase activity was measured 6 hours later. (C) Macrophages were harvested from the IP space of mice in which LRP-1 was conditionally deleted in macrophages and neutrophils and from control wild-type mice in the same genetic background. RNA was isolated from peritoneal cells 3 days after thioglycolate stimulation. Expression of LRP-1, C1r, and iNOS was determined by qPCR. (D) Peritoneal macrophages were cultured for 12 hours in the presence of JSH-23 (10 μM) or vehicle. Expression of EGFr, uPAR, and MCP-1 was analyzed by qPCR. Error bars represent SD.
Our model also predicts that LRP-1–deficient cells may be selectively responsive to TNF-α because of the increase in TNFR1. This prediction is supported by our results showing greatly increased IKK phosphorylation in response to TNF-α in MEF-2 cells (Figure 5A). To further test this prediction, we treated MEFs with LPS, a known activator of the NF-κB pathway. LPS had only a minimal effect of NF-κB activity in PEA-10 and MEF-2 cells; selective enhancement of the response to LPS in MEF-2 cells was not observed (Figure 7B).
LRP-1 regulates NF-κB in vivo
Next, we sought to determine whether NF-κB activation is responsible for the increase in accumulation of inflammatory mediators in atherosclerotic plaques, in mice in which LRP-1 is deleted in macrophages.20 To address this question, we harvested macrophages after intraperitoneal thioglycolate injection, from mice in which LRP-1 is conditionally deleted and from wild-type mice in the equivalent genetic background. RNA was isolated from the cells without culturing in vitro, and levels of mRNA for iNOS and C1r were determined (Figure 7C). To control for the presence of other cells in the peritoneal wash, we assessed LRP-1 mRNA expression and observed a 60% decrease in the cells harvested from mice in which LRP-1 was conditionally deleted. The same cells demonstrated an 80% increase in C1r mRNA and a 6-fold increase in iNOS mRNA.
Overton et al20 demonstrated that MCP-1 expression is greatly increased in LRP-1–deficient macrophages isolated by peritoneal wash. To test whether MCP-1 expression is regulated by NF-κB, we examined MCP-1 expression in LRP-1–deficient macrophages that were treated with JSH-23 or vehicle for 12 hours. JSH-23 decreased MCP-1 expression by 75%. As a control, we showed that expression of uPAR and epidermal growth factor receptor were unchanged (Figure 7D). Thus, MCP-1 expression is selectively regulated downstream of NF-κB in LRP-1–deficient macrophages.
Discussion
LRP-1 controls cell signaling by direct pathways, in which LRP-1 functions alone or as part of a multireceptor cell signaling complex, and by indirect pathways, in which the principal role of LRP-1 is to regulate the level of another receptor or its ligands. Direct pathways are activated when specific ligands bind to LRP-1, including tPA,11 activated α2M,10 apolipoprotein E,12 and connective tissue growth factor.51 Although the mechanism by which LRP-1 ligands activate cell signaling remains unclear, binding of signaling adaptor proteins to LRP-1 may be involved.13-15 By contrast, LRP-1 indirectly regulates cell signaling by mediating the endocytosis of uPAR52 and uPA.38 By this pathway, LRP-1 controls the activation state of Ras, ERK/MAP kinase, and Rac1.5,6
In this study, ABPP experiments identified the complement proteases, C1r and C1s, as gene products that are regulated at the mRNA level by LRP-1. Regulation of C1r expression by LRP-1 was explained by the ability of LRP-1 to suppress the basal activity of the IKK/NF-κB pathway. We obtained evidence for regulation of NF-κB by LRP-1 in multiple cell culture model systems, including MEFs, CL-16 cancer cells, and RAW 264.7 cells. We demonstrated that regulation of NF-κB impacts on expression of inflammatory mediators beyond the complement cascade, including iNOS and IL-6. Furthermore, we showed that LRP-1–deficient macrophages, harvested from mice, demonstrate increased expression of iNOS and C1r. The previously demonstrated increase in expression of MCP-1, by LRP-1–deficient macrophages,20 was attributed to NF-κB. We propose that the ability of LRP-1 to regulate the activity of the IKK/NF-κB pathway provides a mechanistic explanation for the antiinflammatory activity of LRP-1, which is evident in mouse models of atherosclerosis.20
When LRP-1 is absent, cell-surface TNFR1 is increased, IκB is decreased, and NF-κB activity is increased. TNF-α–neutralizing antibody inhibits NF-κB activity in LRP-1–deficient cells. We interpret these data as consistent with a model in which LRP-1 deficiency derepresses autocrine TNFR1-initiated cell signaling to NF-κB. Although at least part of the response represents sensitization to endogenously produced TNF-α, we cannot rule out a role for ligand-independent TNFR1-initiated cell signaling.53 In response to TNF-α, IKK phosphorylation is increased in LRP-1-deficient MEFs. Based on our studies, examining cells under basal conditions and in response to TNF-α, we propose that TNFR1 may be grouped with uPAR, among receptors that are indirectly regulated by LRP-1. Increased expression of inflammatory mediators, including C1r, C1s, iNOS, and IL-6, occurs downstream of TNFR1 when LRP-1 is deficient or silenced.
The precise mechanism by which LRP-1 regulates cell-surface TNFR1 remains to be determined; however, it is reasonable to assume that the importance of this process in controlling the activity of the IKK/NF-κB pathway may be context dependent. It is also plausible that LRP-1 ligands may signal directly through LRP-1 to regulate NF-κB. Platelet factor 4 has been reported to induce endothelial E-selectin expression by a mechanism that involves binding to LRP-1 and activation of NF-κB.54 Furthermore, tPA binding to LRP-1 has been implicated in the induction of NF-κB activity that accompanies cerebral ischemia secondary to arterial occlusion.55 Taken together, our work and these other studies suggest that the overall activity of LRP-1, in the regulation of the IKK/NF-κB pathway, may represent the integration of responses occurring directly downstream of LRP-1 and downstream of receptors, such as TNFR1, that are regulated by LRP-1.
Our studies reveal evidence for cross-talk between cell signaling pathways regulated by LRP-1. LRP-1 silencing in Schwann cells and deficiency in MEFs increase susceptibility to cell death in response to stressful conditions.17 This result may be explained by the positive effect of LRP-1 on Akt activation; however, the increase in cell death in response to TNF-α, in LRP-1–deficient cells, may also be explained by the effects of LRP-1 on TNFR1 shown here. On the other hand, the increase in NF-κB activity, in LRP-1–deficient cells, might be expected to favor cell survival.56 We hypothesize that the effects of LRP-1 on Akt and NF-κB may oppose one another in controlling cell survival. Because NF-κB may be activated downstream of Akt,57 understanding how LRP-1 regulates cross-talk between these 2 important pathways emerges as an important future goal.
Precise control of the complement cascade is critical in inflammation.58 In the C1 complex, when C1q binds to antibodies or pathogens, it undergoes conformational change, which induces activation of C1r and subsequent activation of C1s.59 These proteases target C2 and C4 to form C3 convertase.60 Previous studies have shown that catabolism of C1s and C3 is mediated by LRP-1.40,61 We have shown that LRP-1 mediates the catabolism of C1r. Furthermore, LRP-1 regulates expression of C1r, C1s, and C1-INH at the mRNA level. Thus, LRP-1 emerges as a key regulator of the complement cascade at multiple levels.
We previously demonstrated that interferon-γ represses LRP-1 expression in macrophages.62,63 We now know that down-regulation of LRP-1 expression, as an isolated event, is sufficient to induce expression of complement components via NF-κB activation. Interferon-γ is known to induce expression of C1r and C1-INH.64,65 Thus, it will be interesting to determine whether the ability of interferon-γ to control LRP-1 expression is integral to the mechanism by which it regulates expression of C1r and C1-INH.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by National Institutes of Health grant R01 HL-60551.
National Institutes of Health
Authorship
Contributions: A.G. and S.A. contributed equally to this work, designing and executing the majority of the experiments and interpreting the data; S.N. and C.D.O. performed select experiments and interpreted the resulting data; M.F.L., S.F., W.M.C., and B.F.C. participated in the design of experiments and in the interpretation of data; S.L.G. designed the experiments, interpreted data and, together with A.G., wrote the paper; all authors contributed to the editing of the final draft.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Steven L. Gonias, University of California–San Diego, 9500 Gilman Drive, La Jolla, CA 92093-0612; e-mail: sgonias@ucsd.edu.
References
Author notes
A.G. and S.A. contributed equally to this work.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal