Abstract
CD38 rules proliferation signals in chronic lymphocytic leukemia (CLL) cells, suggesting that the molecule is not merely a prognostic marker but also a key element in the pathogenetic network underlying the disease. CD38 has a genetic polymorphism, characterized by a C>G variation in the regulatory region of intron 1. The working hypothesis is that the presence of different alleles in CLL patients marks (or accounts for) some of the clinical heterogeneity. CD38 allele distribution in 248 Italian patients overlapped with that of the controls (n = 232), suggesting that susceptibility to CLL is not influenced by CD38 genotype. Stratification of patients according to markers of unfavorable prognosis constantly resulted in a significantly higher frequency of the rare G allele. Furthermore, analysis of clinical parameters showed that G allele is independently associated with nodal/splenic involvement. The highest G allele frequency was observed in the 16 patients of the cohort that developed Richter syndrome (RS). Five-year cumulative incidence of transformation was significantly higher in G allele carriers than in CC homozygotes. Multivariate analysis on a total of 30 RS patients confirmed that the probability of transformation is strongly associated with G allele, likely representing an independent risk factor for RS development.
Introduction
Chronic lymphocytic leukemia (CLL), the most common leukemia in Europe and North America, results from the neoplastic transformation of a population of B lymphocytes expressing surface CD5.1 Diagnosis is usually relatively simple, but predicting the clinical course can be difficult. Approximately one-third of CLL patients are affected by an indolent form of disease that does not require treatment or modify survival.2 Another third of patients present with a leukemia that will require iterative therapies, profoundly affecting their quality and length of life. A small fraction (2%-8%) of CLL patients develop Richter syndrome (RS), represented in most cases by diffuse large B-cell lymphoma (DLBCL) arising from the transformation of the original CLL clone or, less frequently, representing a new or secondary neoplasm.3,4 RS is a highly aggressive syndrome with a median overall survival of 5 to 8 months. Today, poor prognosis CLL may be identified at diagnosis based on a combination of clinical, morphologic, and molecular parameters. The most credited molecular indicators are the absence of mutations in the IgV genes5 and expression of CD386 and ZAP-70.7 Other acknowledged markers include genomic aberrations,8 such as deletions at chromosomes 17p9 and 11q.10
CLL was adopted by our group as a convenient human disease model for studying the role of CD38. Several lines of evidence indicate that CD38 may not be merely a prognostic marker but also a key element in the pathogenetic network underlying CLL.11 CD38 ligation in CLL cells results in robust proliferation/survival signals modulated through interactions with the CD31 ligand expressed by nurselike cells and by the stromal/endothelial components.12
CD38 expression by the leukemic clone ranges from negative to highly positive. Further, it is dependent on body district13 and can apparently vary over time in the same individual.14 The mechanisms controlling CD38 expression are complex and partly unknown. However, the microenvironment has been shown to provide soluble and membrane-bound signals that strongly up-regulate CD38 expression, likely potentiating its receptor functions.15,16 The CD38 gene, localized on the short arm of chromosome 4 (4p15),17,18 is more than 80 kb long, more than 98% of which is represented by intronic sequences. CD38 expression is tightly regulated at different levels. The first level of control lies in the 5′ flanking promoter region of the gene, which, with no TATA box and the presence of a CpG island, resembles many noninducible housekeeping genes.19 The second involves trans interactions with sequences that lie further upstream from the transcription start site, including T-cell transcription factor-1α (TCF-1α), nuclear factor for interleukin-6 (NF-IL-6), interferon-responsive element-1 (IRF-1), and binding sites for glucocorticoid hormones. The third level of control is located at the 5′ end of intron 1 and is involved in the up-regulation of CD38 expression by retinoids in myeloid cells.20 Within the same region is a well-characterized biallelic single nucleotide polymorphism (SNP). A restriction endonuclease PvuII site is located at the 5′ end of the first intron and marks a C>G variation at position 184 (hereafter referred to as 184 C>G).21 The gene frequencies among white populations are well known.21-23
The aim of this study was to investigate the distribution of CD38 SNP in CLL, in view of its potential contribution to the pathogenetic network underlying the disease. Therefore, we analyzed the distribution of CD38 alleles in (1) randomly collected patients and (2) in CLL subgroups selected on the basis of biologic biases. Results indicate that the G allele is significantly associated with molecular markers of unfavorable prognosis. Furthermore, the G allele is associated with clinical markers of nodal disease and represents a significant risk factor for RS transformation.
Methods
Patients and controls
The study was based on a series of 248 previously untreated CLL patients recruited from June 1991 to August 2007 in 3 referring institutions (University of Torino [Turin, Italy] and of Eastern Piedmont [Novara, Italy] and Fondazione Pascale [Naples, Italy]). Informed consent was obtained from all patients in accordance with institutional guidelines and the Declaration of Helsinki. The Ethical Committee of the University Hospital Molinette has reviewed and approved the research project on the role of CD38 in chronic lymphocytic leukemia in December 2005. Diagnosis of CLL was based on National Cancer Institute (NCI) Working Group criteria and confirmed by flow cytometry.24 Demographic (age, sex), clinical (disease stage, lymphocyte doubling time [LDT], therapy requirement and response, laboratory data at diagnosis) and molecular (IgV gene mutational status, CD38 and ZAP-70 expression, genomic aberrations) variables were collected for the whole cohort.
IgV mutations, CD38 expression, and ZAP-70 expression were determined as described,25 with cutoff values of 2% or more, 30% or more, and 20% or more, respectively. A CLL panel of multicolor probe sets (Vysis, Downers Grove, IL) was used for fluorescence in situ hybridization (FISH) analysis of chromosomes 11, 12, 13, and 17.
RS diagnosis was confirmed after histologic examination of lymph node or tissue biopsies showing transformation to DLCBL. Clinical, laboratory, and molecular data pertaining to RS patients were collected whenever available. The first series of RS (n = 16) was derived from the consecutive cohort of CLL patients described in the first paragraph of this section. A second series of RS, used for validation purposes, was represented by 14 consecutive RS patients referred to the Istituto Nazionale Tumori (Milan, Italy). The control group consisted of 232 blood donors (128 males and 104 females), randomly collected from the Molinette Blood Bank (Turin, Italy).
CD38 genotyping
Genomic DNA was isolated with the QIAamp DNA Mini Kit (Qiagen, Milan, Italy). Polymerase chain reaction (PCR) amplification was performed as reported23 using the primers 5′-CCGGGTGGTGCTGAGTAGGGAGTC-3′ (forward) and 5′-CTACGCAGCAGAGCCACCGAGCAG-3′ (reverse). The reaction was performed in a 25-μL volume with 1.5 mM MgCl2, 200 nM dNTPs, 1.5 pmol of each primer, and 2 U Taq polymerase (ReadyMix Taq PCR Reaction Mix; Sigma, Milan, Italy). Amplification conditions were as follows: denaturation at 95°C for 5 minutes, followed by 35 cycles of 95°C for 1 minute, 63°C for 1 minute, and 72°C for 1 minute, followed by an extension step of 72°C for 10 minutes on an Authorized Thermal Cycler (Eppendorf, Hamburg, Germany). The 128-bp amplicon was digested with 1 U PvuII (Roche Diagnostics, Milan, Italy) according to the manufacturer's instructions. The presence of the C allele resulted in digestion of the amplicon to 63- and 65-bp products. CD38 genotypes were identified following electrophoresis in a 4% NuSieve gel (Biospa, Milan, Italy). Genotyping results from 10 random samples representing the 3 genotypes (2 CC, 3 GC, and 5 GG) were confirmed by direct sequencing of PCR products.
In vitro cultures and cell surface staining
Peripheral blood mononuclear cells were cultured (106/mL) in RPMI 1640 medium (Sigma) with 10% fetal calf serum (Seromed, Berlin, Germany), 50 μg/mL gentamicin, 100 U/mL penicillin, and 100 μg/mL streptomycin (all from Sigma). Where indicated, IL-2 (100 IU/mL) was added at the beginning of cultures. After 72 hours, cells were harvested and stained for CD38-FITC (produced and conjugated in-house) and CD19-PE (eBioscience, San Diego, CA). Cells were analyzed with a FACSort flow cytometer (BD Biosciences, San Jose, CA), acquiring at least 5000 events per sample. Data were analyzed using the CellQuest software (BD Biosciences) and were expressed as mean fluorescence intensity (MFI).
Statistical analysis
The Student t test was used to determine the statistical significance of CD38 up-regulation upon IL-2 exposure in genotyped patients. The chi-square test was used to determine whether allele frequencies in the study were in Hardy-Weinberg equilibrium. The association between CD38 SNP and patient/disease-related variables, namely sex, age (≤ 65 years vs > 65 years), disease stage, LDT (≤ 12 months vs > 12 months), IgV gene mutational status, CD38 expression, ZAP-70 expression, number of affected lymph node areas (≤ 2 vs > 2), number of therapy regimens (≤ 2 vs > 2), LDH levels (≤ 450 U/L vs > 450 U/L), β2-microglobulin levels (≤ 2.5 vs > 2.5 mg/L), and genomic aberrations (good risk [no genomic aberrations or deletion 13] vs poor risk [deletion 11 and/or deletion 17]), was also assessed by chi-square test. Stratification of continuous clinical variables was based on the best predictive cutoff value or usual limit of normal. All statistical tests were 2-sided.
Multivariate analysis using probit or ordered probit regression model (95% confidence interval, CI) was used to explore the association between CD38 genotypes and variables previously described as predictors of prognosis (IgV mutations, CD38 and ZAP-70 expression, and genomic aberrations) and of RS (genomic aberrations, indicators of lymphoid mass, previous treatment regimens, and elevated LDH levels). Nonsignificant variables were stepwise removed from the model.
Date of transformation corresponds to the date of the biopsy showing RS transformation. Cumulative incidence of transformation was measured from date of CLL diagnosis to date of transformation, death, or last follow up. Survival analysis was performed by Kaplan-Meier method using log-rank to test for significant associations.
Statistical analyses were performed using SPSS 15.0 for Windows (SPSS, Chicago, IL) and Stata 9.0 (StataCorp, College Station, TX). Unadjusted P values less than .05 were considered significant in all statistical analyses.
Results
Allele frequencies in the healthy population
Analysis of 232 Italian individuals resulted in frequencies of 0.79 and 0.21 for the C and G allele, respectively (CC 62%, GC 34%, and GG 4%). These results are comparable with data derived from larger series from Spain22 or Ireland.23
Pooling together the information reported for 1056 white individuals resulted in a C allele frequency of 0.78 and a G allele frequency of 0.22 (CC 61%, GC 33%, and GG 6%; Table 1). The frequencies of the Italian control population serving as the reference for this study, namely C 0.79 and G 0.21, reflect the geographic composition of the population from which the clinical sample is derived.
CD38 genotypes and allele frequencies in different control populations
| Control population . | No. . | Genotype . | Allele frequency . | Reference . | |||
|---|---|---|---|---|---|---|---|
| CC, % . | GC, % . | GG, % . | C . | G . | |||
| Spain | 194 | 53 | 40 | 7 | 0.73 | 0.27 | 22 |
| Ireland | 630 | 64 | 30 | 6 | 0.79 | 0.21 | 23 |
| Italy | 232 | 62 | 34 | 4 | 0.79 | 0.21 | Present study |
| Theoretic cohort | 1056 | 61 | 33 | 6 | 0.78 | 0.22 | |
Allele frequencies in the CLL cohort
CD38 expression is a reliable and independent negative prognostic marker in CLL (reviewed in Matrai26 ). Further, CD38 is part of the pathogenetic circuit of the disease, performing as a cell surface receptor delivering proliferation/survival signals (reviewed in Deaglio et al11 ). The aim of this study was to analyze the distribution of the CD38 alleles in a sample of clinically and molecularly characterized CLL patients. In addition, we investigated whether the alleles segregate differently in CLL subgroups characterized by distinct molecular and clinical parameters. The characteristics of the CLL cohort are summarized in Table 2.
Clinical and molecular characteristics of the CLL cohort
| Variable . | No. (%) . |
|---|---|
| Patients | 248 |
| Median age at diagnosis, y | 65.7 |
| Sex, male | 143 (58) |
| Binet stage at diagnosis, n = 244 | |
| A | 172 (71) |
| B | 49 (20) |
| C | 23 (9) |
| CD38, n = 245 | |
| 30% or more | 102 (42) |
| ZAP-70, n = 190 | |
| 20% or more | 59 (31) |
| IgV gene mutation status, n = 236 | |
| UM, less than 2% | 87 (37) |
| Genomic aberrations, n = 212 | |
| Deletion 11 | 15 (7) |
| Deletion 17 | 28 (13) |
| Deletion 13 | 103 (49) |
| Trisomy 12 | 44 (21) |
| Normal | 58 (27) |
| CD38 genotype, n = 248 | |
| CC | 153 (62) |
| GC | 86 (34) |
| GG | 9 (4) |
| CD38 allele frequency, n = 248 | |
| C | 0.79 |
| G | 0.21 |
| Variable . | No. (%) . |
|---|---|
| Patients | 248 |
| Median age at diagnosis, y | 65.7 |
| Sex, male | 143 (58) |
| Binet stage at diagnosis, n = 244 | |
| A | 172 (71) |
| B | 49 (20) |
| C | 23 (9) |
| CD38, n = 245 | |
| 30% or more | 102 (42) |
| ZAP-70, n = 190 | |
| 20% or more | 59 (31) |
| IgV gene mutation status, n = 236 | |
| UM, less than 2% | 87 (37) |
| Genomic aberrations, n = 212 | |
| Deletion 11 | 15 (7) |
| Deletion 17 | 28 (13) |
| Deletion 13 | 103 (49) |
| Trisomy 12 | 44 (21) |
| Normal | 58 (27) |
| CD38 genotype, n = 248 | |
| CC | 153 (62) |
| GC | 86 (34) |
| GG | 9 (4) |
| CD38 allele frequency, n = 248 | |
| C | 0.79 |
| G | 0.21 |
Percentages refer to total number of events scored.
UM indicates unmutated.
Analysis of the CLL cohort resulted in a C allele frequency of 0.79 and a G allele frequency of 0.21, with a genotype distribution of CC 62%, GC 34%, and GG 4%. These results overlap with those obtained with the control population. The observed genotype frequencies defined by the CD38 SNP were in accordance with the Hardy-Weinberg law of equilibrium, providing no evidence of population stratification within the data set.
The G allele is associated with CLL patients carrying molecular markers of poor prognosis
The next step was to analyze the distribution of CD38 alleles in subgroups of patients with distinct clinical and molecular features. The cohort was therefore classified into subgroups according to molecular markers of unfavorable prognosis. The allele frequencies and genotype distributions obtained for each subgroup were then compared with those of the counterpart subgroup and with the whole CLL cohort.
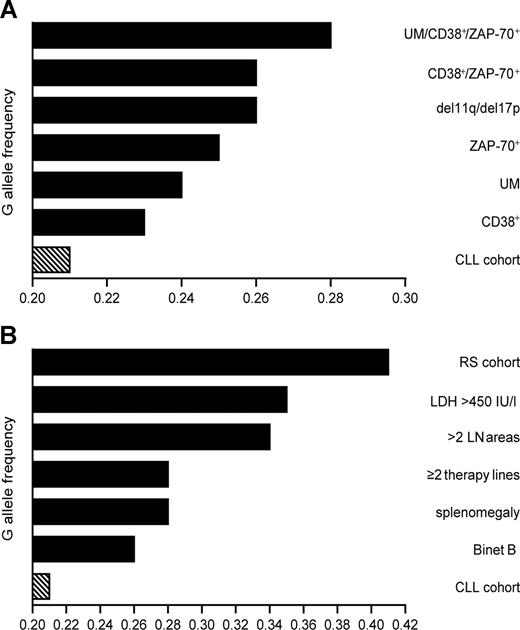
Grouping the patients on the basis of IgV gene mutational status (mutated [MUT] vs unmutated [UM]) or CD38 expression (CD38+ vs CD38−) revealed no significant differences in allele frequency. However, the rare G allele tended to be more frequent in the UM and CD38+ subgroups than in their corresponding counterparts (Table 3; Figure 1A). Genotype distribution showed a significant difference when comparing CD38+ to CD38− patients (P = .03), attributable mainly to an increased representation of GG homozygotes among CD38+ CLL (Table 4). Classification according to ZAP-70 expression yielded significant differences in allele frequencies (P = .03) and genotype distributions (P = .01).
Allele frequencies in molecularly distinct subgroups of the CLL cohort
| CLL subgroup . | Allele frequency . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|
| No. . | C . | G . | |||
| Total cohort | 248 | 0.79 | 0.21 | .77 (vs control) | |
| IgV mutational status | |||||
| MUT | 149 | 0.81 | 0.19 | .57 | .12 |
| UM | 87 | 0.76 | 0.24 | .59 | .24 |
| CD38 | |||||
| Less than 30% | 143 | 0.80 | 0.20 | .37 | .27 |
| 30% or more | 102 | 0.77 | 0.23 | .76 | .56 |
| ZAP-70 | |||||
| Less than 20% | 131 | 0.82 | 0.18 | .39 | .03 |
| 20% or more | 59 | 0.75 | 0.25 | .32 | .07 |
| CD38−/ZAP-70− | 88 | 0.83 | 0.17 | .29 | .027 |
| CD38+/ZAP-70+ | 35 | 0.74 | 0.26 | .28 | .07 |
| MUT/CD38−/ZAP-70− | 73 | 0.84 | 0.16 | .34 | .007 |
| UM/CD38+/ZAP-70+ | 25 | 0.72 | 0.28 | .29 | .05 |
| Normal/del 13q | 161 | 0.79 | 0.21 | .51 | .16 |
| del 11q/del 17p | 43 | 0.74 | 0.26 | .17 | .18 |
| CLL subgroup . | Allele frequency . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|
| No. . | C . | G . | |||
| Total cohort | 248 | 0.79 | 0.21 | .77 (vs control) | |
| IgV mutational status | |||||
| MUT | 149 | 0.81 | 0.19 | .57 | .12 |
| UM | 87 | 0.76 | 0.24 | .59 | .24 |
| CD38 | |||||
| Less than 30% | 143 | 0.80 | 0.20 | .37 | .27 |
| 30% or more | 102 | 0.77 | 0.23 | .76 | .56 |
| ZAP-70 | |||||
| Less than 20% | 131 | 0.82 | 0.18 | .39 | .03 |
| 20% or more | 59 | 0.75 | 0.25 | .32 | .07 |
| CD38−/ZAP-70− | 88 | 0.83 | 0.17 | .29 | .027 |
| CD38+/ZAP-70+ | 35 | 0.74 | 0.26 | .28 | .07 |
| MUT/CD38−/ZAP-70− | 73 | 0.84 | 0.16 | .34 | .007 |
| UM/CD38+/ZAP-70+ | 25 | 0.72 | 0.28 | .29 | .05 |
| Normal/del 13q | 161 | 0.79 | 0.21 | .51 | .16 |
| del 11q/del 17p | 43 | 0.74 | 0.26 | .17 | .18 |
Schematic representation of G allele frequencies in CLL subgroups. (A) G allele frequencies display a progressive increase when patients are grouped according to combination of negative prognostic markers. (B) G allele frequencies in patients grouped on the basis of clinical parameters of high tumor mass. RS patients present in the CLL cohort scored the highest G allele frequency. Sided bars in panels A,B indicate the frequency of the whole CLL cohort.
Schematic representation of G allele frequencies in CLL subgroups. (A) G allele frequencies display a progressive increase when patients are grouped according to combination of negative prognostic markers. (B) G allele frequencies in patients grouped on the basis of clinical parameters of high tumor mass. RS patients present in the CLL cohort scored the highest G allele frequency. Sided bars in panels A,B indicate the frequency of the whole CLL cohort.
Genotype distributions in molecularly distinct subgroups of the CLL cohort
| CLL subgroup . | No. . | Genotype . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|---|
| CC, % . | GC, % . | GG, % . | ||||
| Total cohort | 248 | 62 | 34 | 4 | .98 (vs control) | |
| IgV mutational status | ||||||
| MUT | 149 | 64 | 33 | 3 | .73 | .24 |
| UM | 87 | 57 | 37 | 6 | .54 | .09 |
| CD38 | ||||||
| Less than 30% | 143 | 62 | 35 | 2 | .65 | .27 |
| 30% or more | 102 | 61 | 33 | 6 | .58 | .03 |
| ZAP-70 | ||||||
| Less than 20% | 131 | 67 | 29 | 3 | .39 | .02 |
| 20% or more | 59 | 54 | 42 | 3 | .28 | .01 |
| CD38−/ZAP-70− | 88 | 67 | 32 | 1 | .25 | .007 |
| CD38+/ZAP-70+ | 35 | 51 | 46 | 3 | .07 | .002 |
| MUT/CD38−/ZAP-70− | 73 | 68 | 30 | 1 | .21 | <.001 |
| UM/CD38+/ZAP-70+ | 25 | 48 | 48 | 4 | .01 | <.001 |
| Normal/del 13q | 161 | 60 | 37 | 3 | .87 | .14 |
| del 11q/del 17p | 43 | 51 | 47 | 2 | .04 | .12 |
| CLL subgroup . | No. . | Genotype . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|---|
| CC, % . | GC, % . | GG, % . | ||||
| Total cohort | 248 | 62 | 34 | 4 | .98 (vs control) | |
| IgV mutational status | ||||||
| MUT | 149 | 64 | 33 | 3 | .73 | .24 |
| UM | 87 | 57 | 37 | 6 | .54 | .09 |
| CD38 | ||||||
| Less than 30% | 143 | 62 | 35 | 2 | .65 | .27 |
| 30% or more | 102 | 61 | 33 | 6 | .58 | .03 |
| ZAP-70 | ||||||
| Less than 20% | 131 | 67 | 29 | 3 | .39 | .02 |
| 20% or more | 59 | 54 | 42 | 3 | .28 | .01 |
| CD38−/ZAP-70− | 88 | 67 | 32 | 1 | .25 | .007 |
| CD38+/ZAP-70+ | 35 | 51 | 46 | 3 | .07 | .002 |
| MUT/CD38−/ZAP-70− | 73 | 68 | 30 | 1 | .21 | <.001 |
| UM/CD38+/ZAP-70+ | 25 | 48 | 48 | 4 | .01 | <.001 |
| Normal/del 13q | 161 | 60 | 37 | 3 | .87 | .14 |
| del 11q/del 17p | 43 | 51 | 47 | 2 | .04 | .12 |
Because of rounding, percentages do not always add up to 100.
MUT indicates mutated; UM, unmutated; and del, deletion.
Recent studies indicate that the combination of molecular markers can identify patients with unfavorable prognosis more accurately than the use of one marker alone.27,28 CD38+/ZAP-70+ patients displayed a slightly higher G allele frequency than patients expressing only 1 of the 2 molecules (Table 3). CD38+/ZAP-70+ patients showed a significantly different genotype distribution compared with CD38−/ZAP-70− patients (P = .002; Table 4). Factoring in the IgV gene mutational status and comparing UM/CD38+/ZAP-70+ with MUT/CD38−/ZAP-70− patients rendered the difference highly significant, both in terms of allele frequencies (P = .05) and of genotype distributions (P < .001). The genotype distribution of UM/CD38+/ZAP-70+ patients was also significantly different from the CLL cohort as a whole (P = .01) and from the control population (P = .002). The UM/CD38+/ZAP-70+ group displayed the highest G allele frequency, contained more heterozygotes than any of the other subgroups, and had the highest percentage of G-carrying patients. Increased G allele frequencies were also observed when considering patients harboring genetic aberrations correlated with poor prognosis (Table 3; Figure 1A).
Subsequent multivariate analysis showed no association between the G or C allele and CD38 surface expression. CD38 expression was strongly correlated with the absence of mutations (P = .008), confirming the original observation.6 The presence of the G allele displayed a statistically significant association with the expression of ZAP-70 20% or more (P = .01; Table 5). High ZAP-70 expression also correlated with the absence of mutations in IgV genes, again in agreement with published data.7
Probit or ordered probit regression model for multivariate analysis
| . | Variable . | Marginal effect . | P . |
|---|---|---|---|
| CD38 30% or more, n = 102 | Mutation | −0.022 | .008 |
| Male | 0.168 | .017 | |
| Age at diagnosis | 0.006 | .026 | |
| GC | 0.027 | .531 | |
| GG | 0.037 | .666 | |
| ZAP-70 20% or more, n = 59 | Mutation | −0.048 | <.001 |
| GC + GG | 0.194 | .010 | |
| Trisomy 12 | 0.230 | .010 | |
| Splenomegaly, n = 51 | Deletion 11 | 0.420 | .004 |
| GG | 0.531 | .005 | |
| Male | 0.140 | .015 | |
| Trisomy 12 | 0.224 | .022 | |
| Deletion 17 | 0.197 | .065 | |
| Richter syndrome, n = 30 | GC | 0.124 | .007 |
| GG | 0.306 | .040 | |
| Lines of treatment, n = 67 | Trisomy 12 | 0.078 | .004 |
| Deletion 11 | 0.148 | .001 | |
| Deletion 17 | 0.112 | .005 | |
| Mutation | −0.007 | .014 |
| . | Variable . | Marginal effect . | P . |
|---|---|---|---|
| CD38 30% or more, n = 102 | Mutation | −0.022 | .008 |
| Male | 0.168 | .017 | |
| Age at diagnosis | 0.006 | .026 | |
| GC | 0.027 | .531 | |
| GG | 0.037 | .666 | |
| ZAP-70 20% or more, n = 59 | Mutation | −0.048 | <.001 |
| GC + GG | 0.194 | .010 | |
| Trisomy 12 | 0.230 | .010 | |
| Splenomegaly, n = 51 | Deletion 11 | 0.420 | .004 |
| GG | 0.531 | .005 | |
| Male | 0.140 | .015 | |
| Trisomy 12 | 0.224 | .022 | |
| Deletion 17 | 0.197 | .065 | |
| Richter syndrome, n = 30 | GC | 0.124 | .007 |
| GG | 0.306 | .040 | |
| Lines of treatment, n = 67 | Trisomy 12 | 0.078 | .004 |
| Deletion 11 | 0.148 | .001 | |
| Deletion 17 | 0.112 | .005 | |
| Mutation | −0.007 | .014 |
Marginal effect measures the variation in probability generated by a unit increase in the considered variable.
The G allele is associated with CLL cases showing clinical and laboratory markers of high tumor mass
Next, patients were grouped on the basis of clinical parameters. Age, β2-microglobulin level, LDT, and blood counts showed no preferential association with the CD38 SNP. Male patients were characterized by a significantly higher G allele frequency (P = .04; Table 6) and different genotype distribution (P = .03; Table 7) compared with female patients. No significant difference was apparent compared with the healthy male and/or female population. This result likely reflects the fact that male CLL patients in our cohort display molecular and clinical markers of more aggressive disease (Tables 6,7).
Allele frequencies in clinically distinct subgroups of the CLL cohort
| CLL subgroup . | Allele frequency . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|
| No. . | C . | G . | |||
| Sex | |||||
| Female | 105 | 0.82 | 0.18 | .38 | .12 |
| Male | 143 | 0.77 | 0.23 | .56 | .04 |
| Binet stage | |||||
| A+C | 195 | 0.80 | 0.20 | .43 | .03 |
| B | 49 | 0.74 | 0.26 | .40 | .27 |
| Affected LN areas | |||||
| 2 or less | 188 | 0.81 | 0.19 | .50 | <.001 |
| More than 2 | 50 | 0.66 | 0.34 | .14 | .051 |
| Spleen size | |||||
| Normal | 191 | 0.81 | 0.19 | .29 | <.001 |
| Enlarged | 51 | 0.72 | 0.28 | .11 | .027 |
| LDH | |||||
| 450 IU/L or less | 192 | 0.81 | 0.19 | .01 | .003 |
| More than 450 IU/L | 34 | 0.65 | 0.35 | .64 | <.001 |
| Lines of therapy | |||||
| Less than 2 | 170 | 0.81 | 0.19 | .41 | <.001 |
| 2 or more | 67 | 0.72 | 0.28 | .11 | .018 |
| CLL subgroup . | Allele frequency . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|
| No. . | C . | G . | |||
| Sex | |||||
| Female | 105 | 0.82 | 0.18 | .38 | .12 |
| Male | 143 | 0.77 | 0.23 | .56 | .04 |
| Binet stage | |||||
| A+C | 195 | 0.80 | 0.20 | .43 | .03 |
| B | 49 | 0.74 | 0.26 | .40 | .27 |
| Affected LN areas | |||||
| 2 or less | 188 | 0.81 | 0.19 | .50 | <.001 |
| More than 2 | 50 | 0.66 | 0.34 | .14 | .051 |
| Spleen size | |||||
| Normal | 191 | 0.81 | 0.19 | .29 | <.001 |
| Enlarged | 51 | 0.72 | 0.28 | .11 | .027 |
| LDH | |||||
| 450 IU/L or less | 192 | 0.81 | 0.19 | .01 | .003 |
| More than 450 IU/L | 34 | 0.65 | 0.35 | .64 | <.001 |
| Lines of therapy | |||||
| Less than 2 | 170 | 0.81 | 0.19 | .41 | <.001 |
| 2 or more | 67 | 0.72 | 0.28 | .11 | .018 |
Genotype distributions in clinically distinct subgroups of the CLL cohort
| CLL subgroup . | No. . | Genotype . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|---|
| CC, % . | GC, % . | GG, % . | ||||
| Sex | ||||||
| Female | 105 | 67 | 31 | 2 | .26 | .14 |
| Male | 143 | 58 | 37 | 5 | .55 | .03 |
| Binet stage | ||||||
| A+C | 195 | 63 | 35 | 3 | .79 | .11 |
| B | 49 | 57 | 35 | 8 | .08 | .002 |
| Affected LN areas | ||||||
| 2 or less | 188 | 64 | 34 | 3 | .76 | .01 |
| More than 2 | 50 | 50 | 42 | 8 | .019 | <.001 |
| Spleen size | ||||||
| Normal | 191 | 64 | 34 | 2 | .62 | .014 |
| Enlarged | 51 | 53 | 37 | 10 | .006 | <.001 |
| LDH | ||||||
| 450 IU/L or less | 192 | 65 | 32 | 3 | .78 | <.001 |
| More than 450 IU/L | 34 | 38 | 53 | 9 | <.001 | <.001 |
| Lines of therapy | ||||||
| Less than 2 | 170 | 64 | 33 | 4 | .88 | .002 |
| 2 or more | 67 | 46 | 49 | 5 | .008 | .001 |
| CLL subgroup . | No. . | Genotype . | P vs CLL . | P vs counterpart . | ||
|---|---|---|---|---|---|---|
| CC, % . | GC, % . | GG, % . | ||||
| Sex | ||||||
| Female | 105 | 67 | 31 | 2 | .26 | .14 |
| Male | 143 | 58 | 37 | 5 | .55 | .03 |
| Binet stage | ||||||
| A+C | 195 | 63 | 35 | 3 | .79 | .11 |
| B | 49 | 57 | 35 | 8 | .08 | .002 |
| Affected LN areas | ||||||
| 2 or less | 188 | 64 | 34 | 3 | .76 | .01 |
| More than 2 | 50 | 50 | 42 | 8 | .019 | <.001 |
| Spleen size | ||||||
| Normal | 191 | 64 | 34 | 2 | .62 | .014 |
| Enlarged | 51 | 53 | 37 | 10 | .006 | <.001 |
| LDH | ||||||
| 450 IU/L or less | 192 | 65 | 32 | 3 | .78 | <.001 |
| More than 450 IU/L | 34 | 38 | 53 | 9 | <.001 | <.001 |
| Lines of therapy | ||||||
| Less than 2 | 170 | 64 | 33 | 4 | .88 | .002 |
| 2 or more | 67 | 46 | 49 | 5 | .008 | .001 |
Because of rounding, percentages do not always add up to 100.
Stratification according to the Binet staging system showed that stage B patients presented with the highest G allele frequency and the highest number of GG homozygotes. These results were statistically significant compared with Binet A patients (P < .001) or the rest of the CLL cohort (P = .002; Table 6). This finding provided an initial association between G allele and high tumor mass. We therefore investigated the distribution of the CD38 SNP in patients with extensive nodal and splenic involvement. The patient subgroup with 2 or fewer lymph node areas affected showed a lower G allele frequency than the subgroup with more than 2 lymph nodes areas involved (P < .001; Table 6). Genotype distribution in these patients accounted for G allele carriers in 50% of the sample (P < .001), statistically different from the whole CLL cohort as well as the counterpart subgroup (Table 7; Figure 1B). Supporting evidence came from the examination of patients divided on the basis of splenomegaly: the allele frequency and genotype distribution of this subgroup differed from the counterpart lacking spleen enlargement as well as from the whole CLL cohort. According to chi-square test, a significantly high G allele frequency was observed in patients with an enlarged spleen, compared with patients without splenomegaly (P = .027). The same group also displayed a significant difference in their genotype distribution in comparison with the counterpart group or with the whole CLL cohort (P < .001 and P = .006, respectively).
High serum levels of LDH at diagnosis are an indirect indicator for high tumor mass. Elevated LDH levels in the present cohort were associated with a G allele frequency of 0.35, highly significant with all comparisons (counterpart subgroup, whole CLL cohort, and control population). G allele carriers represented the majority of this subgroup (62% vs 39% in the whole cohort).
In summary, the statistically significant variables (ie, Binet stage B, number of affected lymph node areas, splenomegaly, and elevated LDH levels) suggest that the G allele is associated with nodal and splenic involvement (Tables 6,7; Figure 1B).
Multivariate analysis confirmed the statistically significant association of the GG genotype with splenomegaly (P = .005). GG patients displayed a 53.1% increase in probability of presenting with splenomegaly. Other variables associated with splenomegaly were deletions in chromosomes 11 and 17 and male sex, again in agreement with independent studies29-31 (Table 5).
RS patients display the highest G allele frequency
CLL patients with a high tumor mass have a significant risk of developing RS (ie, transformation of CLL into aggressive DLCBL).4 Sixteen patients of our cohort (6.5%) developed RS, in line with general expectations.32 The clinical and molecular characteristics are presented in Table 8. Eleven of these patients bore at least one G allele (9 GC [56.4%] and 2 GG [12.5%]), and 5 patients were homozygous for C (31%). The resulting frequencies were 0.59 for C and 0.41 for G alleles (Figure 1B).
Clinical and molecular characteristics of Richter syndrome patients
| Variable . | Cohort RS, no. (%) . | Collected RS, no. (%) . |
|---|---|---|
| Patients | 16 | 14 |
| Median age at diagnosis, y, n = 16, n = 10* | 64 | 59 |
| Sex (male) | 11 (69) | 10 (71) |
| Binet stage at diagnosis, n = 16, n = 9* | ||
| A | 6 (38) | 3 (33) |
| B | 6 (38) | 3 (33) |
| C | 4 (25) | 3 (33) |
| CD38, n = 14, n = 8* | ||
| 30% or more | 11 (79) | 6 (75) |
| ZAP-70, n = 8, n = 2* | ||
| 20% or more | 1 (12.5) | 1 (50) |
| IgV gene mutation status, n = 16, n = 3* | ||
| UM, less than 2% | 12 (75) | 0 (0) |
| Genomic aberrations, n = 14, n = 3* | ||
| Deletion 11 | 2 (14) | 0 (0) |
| Deletion 17 | 3 (21) | 3 (100) |
| Deletion 13 | 3 (21) | 1 (33) |
| Trisomy 12 | 8 (57) | 0 (0) |
| Normal | 3 (21) | 0 (0) |
| CD38 genotypes, n = 16, n = 14* | ||
| CC | 5 (31) | 5 (36) |
| GC | 9 (56) | 7 (50) |
| GG | 2 (13) | 2 (14) |
| CD38 allele frequencies, n = 16, n = 14* | ||
| C | 0.59 | 0.61 |
| G | 0.41 | 0.39 |
| Variable . | Cohort RS, no. (%) . | Collected RS, no. (%) . |
|---|---|---|
| Patients | 16 | 14 |
| Median age at diagnosis, y, n = 16, n = 10* | 64 | 59 |
| Sex (male) | 11 (69) | 10 (71) |
| Binet stage at diagnosis, n = 16, n = 9* | ||
| A | 6 (38) | 3 (33) |
| B | 6 (38) | 3 (33) |
| C | 4 (25) | 3 (33) |
| CD38, n = 14, n = 8* | ||
| 30% or more | 11 (79) | 6 (75) |
| ZAP-70, n = 8, n = 2* | ||
| 20% or more | 1 (12.5) | 1 (50) |
| IgV gene mutation status, n = 16, n = 3* | ||
| UM, less than 2% | 12 (75) | 0 (0) |
| Genomic aberrations, n = 14, n = 3* | ||
| Deletion 11 | 2 (14) | 0 (0) |
| Deletion 17 | 3 (21) | 3 (100) |
| Deletion 13 | 3 (21) | 1 (33) |
| Trisomy 12 | 8 (57) | 0 (0) |
| Normal | 3 (21) | 0 (0) |
| CD38 genotypes, n = 16, n = 14* | ||
| CC | 5 (31) | 5 (36) |
| GC | 9 (56) | 7 (50) |
| GG | 2 (13) | 2 (14) |
| CD38 allele frequencies, n = 16, n = 14* | ||
| C | 0.59 | 0.61 |
| G | 0.41 | 0.39 |
Percentages refer to total number of events scored.
UM indicates unmutated.
Number of events scored in the collected RS cohort.
These results implicate a strong correlation between the G allele and RS. To validate this observation in an independent series, another 14 consecutive cases of RS were collected from the Istituto Nazionale Tumori (Milan, Italy). Five were CC (36%), 7 were GC (50%), and 2 were GG (14%), scoring allele frequencies of 0.61 for C and 0.39 for G. Statistical analysis on the basis of allele frequencies as well as genotype distribution yielded highly significant differences in all comparisons (the whole CLL cohort, the CLL cohort that did not undergo transformation, or the control population).
Results from multivariate analysis support the hypothesis that the probability of transformation is strongly associated with the presence of a G allele. Compared with CC homozygotes, GG patients had a 30.6% increase in the relative risk of developing RS, whereas GC heterozygotes showed an intermediate probability of 12.4% (P = .040 and P = .007, respectively; Table 5).
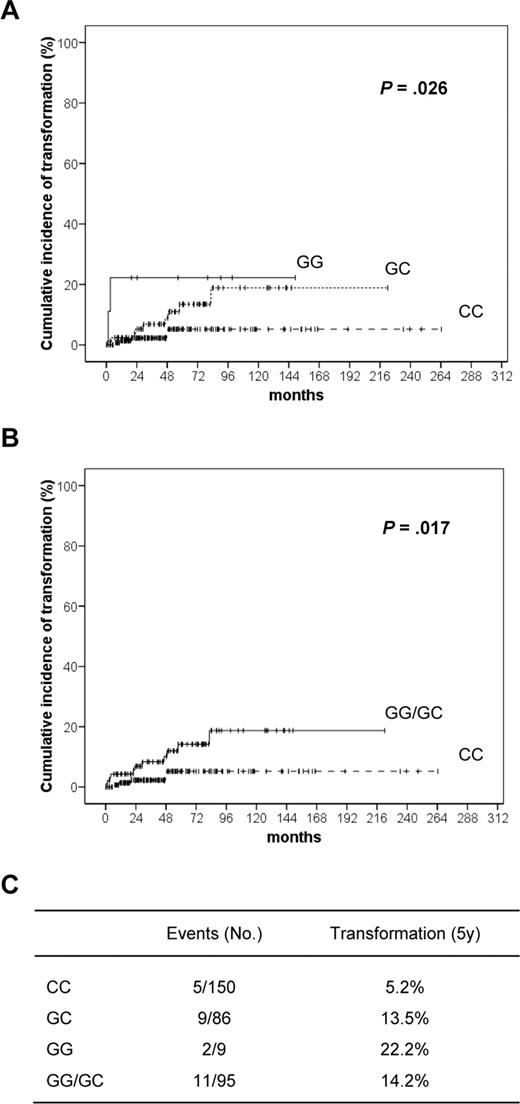
To confirm the relevance of 184C>G polymorphism in predicting RS transformation, an actuarial analysis was performed on the CLL cohort (n = 245). Univariate log-rank analysis identified G allele as a risk factor for RS transformation. Indeed, the 5-year risk of RS transformation was 22.2% in the GG homozygotes, as opposed to 13.5% in the GC heterozygotes and 5.2% in the CC homozygotes (P = .026, Figure 2A). This comparison remained significant even when all G allele carriers (GG/GC) were grouped and compared with CC homozygotes. In this instance, the 5-year risk of transformation was 14.2% in G carriers, as opposed to 5.2% in CC (P = .017; Figure 2B).
Kaplan-Meier curves showing time to RS transformation according to genotype. Cumulative incidence of RS transformation at 5 years performed on the CLL cohort (n = 245), divided on the basis of CD38 genotype. Univariate log-rank analysis identifies G allele as a risk factor for RS transformation (A). The result remains significant even when all G allele carriers are grouped (GG/GC) and compared with CC homozygotes (B). (C) Table showing the 5-year risk of RS transformation according to CD38 genotype.
Kaplan-Meier curves showing time to RS transformation according to genotype. Cumulative incidence of RS transformation at 5 years performed on the CLL cohort (n = 245), divided on the basis of CD38 genotype. Univariate log-rank analysis identifies G allele as a risk factor for RS transformation (A). The result remains significant even when all G allele carriers are grouped (GG/GC) and compared with CC homozygotes (B). (C) Table showing the 5-year risk of RS transformation according to CD38 genotype.
The presence of the G allele correlates with an increased CD38 up-regulation in response to IL-2
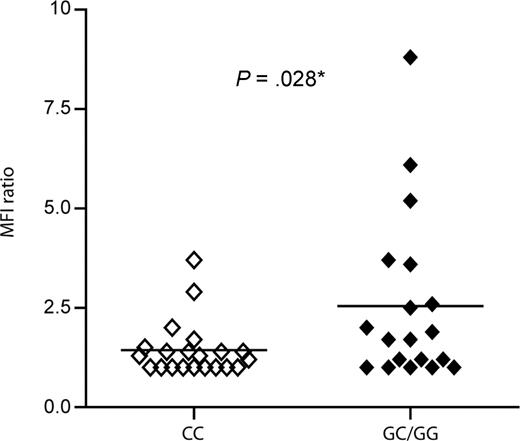
Once determined that the G allele is significantly associated with high tumor mass and with transformation of CLL to RS, we tried to determine the molecular reasons behind this observation. No significant differences were observed in basal peripheral blood CD38 expression between CC patients and G allele carriers, either when considering percentage of positive cells or MFI (data not shown). It is known that CD38 expression varies according to body district, being higher in the lymph nodes and spleen compared with peripheral blood.13,33 However, lymph node biopsies in our cohort were too few to make comparisons possible. To overcome this limitation, we took advantage of in vitro observations suggesting that CD38 expression is highly sensitive to exogenous IL-2.15 Peripheral blood CLL lymphocytes of 40 genotyped patients (21 CC, 17 GC, and 2 GG) were treated with IL-2 for 72 hours before checking CD38 MFI levels. A MFI ratio was determined as follows: MFI levels after IL-2 exposure/MFI levels after culture in complete medium. G allele carriers showed a significantly higher up-regulation of CD38 in response to IL-2, compared with CC homozygotes (P = .028, Figure 3). This preliminary result suggests that the presence of the G allele influences transcription of the CD38 gene in response to external signals.
Up-regulation of CD38 expression upon IL-2 exposure according to genotype. Peripheral blood mononuclear cells from 40 CLL patients (21 CC, 17 GC, and 2 GG) were cultured for 72 hours in the presence or absence of IL-2 (100 IU/mL) before staining for surface CD38 and CD19. A MFI ratio was determined as follows: MFI levels after IL-2 exposure/MFI levels after culture in complete medium. G allele carriers showed a significantly higher surface CD38 expression in response to IL-2, compared with CC homozygotes (P = .028).
Up-regulation of CD38 expression upon IL-2 exposure according to genotype. Peripheral blood mononuclear cells from 40 CLL patients (21 CC, 17 GC, and 2 GG) were cultured for 72 hours in the presence or absence of IL-2 (100 IU/mL) before staining for surface CD38 and CD19. A MFI ratio was determined as follows: MFI levels after IL-2 exposure/MFI levels after culture in complete medium. G allele carriers showed a significantly higher surface CD38 expression in response to IL-2, compared with CC homozygotes (P = .028).
Discussion
The clinical behavior of CLL is highly heterogeneous, ranging from silent nonsymptomatic accumulation of cells to a chronic leukemia, which gradually compromises bone marrow function and significantly shortens life expectancy. In some CLL patients, the disease develops into Richter syndrome, an aggressive lymphoma with a rapidly fatal outcome. Because of this variety in clinical course, it is important to be able to determine the risk category of patients at diagnosis, so as to define the best treatment options.34 Thanks to extensive research, a number of clinical parameters, morphologic features, and molecular markers are now available as tools for identifying patients with an unfavorable prognosis.
One of these tools is CD38, a reliable and independent negative prognostic marker,26 which is also involved in the delivery of growth and survival signals to the neoplastic cells.11 The percentage of CD38+ cells in the peripheral blood fluctuates widely, and its surface levels are sensitive to changes in environment, stage of disease, and therapy administered. Further, it is becoming clear that CD38+ cells within the same neoplastic clone are the ones that express markers of proliferation, such as Ki-67, and have shorter telomeres.35-37 In vitro experiments have identified soluble and membrane-bound signals that can up-modulate CD38 expression.15,16 The time lag in response suggests that de novo gene transcription occurs.
Our research interest lies in exploitation of the CLL model as a suitable tool for gathering information on the functions of CD38, while at the same time supporting the working hypothesis that CD38 is part of the pathogenetic network of CLL and thus constitutes a valid therapeutic target.38,39 Starting from the premise that CD38 is genetically polymorphic, we asked whether the presence of different alleles in CLL patients might mark and/or account for some of the clinical and biologic heterogeneity described. In this context, the analysis of CD38 polymorphism in CLL patients may offer insights into the regulation of the molecule's expression, and it may provide clinicians with a novel diagnostic/prognostic marker.
The CD38 gene lacks a conventional TATA box and a regulatory region is partially contained within the first intron. Consensus sequences for several nuclear factors have been hypothesized, including one for E2A (reviewed in Malavasi et al40 ). A SNP resulting in a C>G nucleotide substitution is located inside the putative E2A consensus sequence. The allele frequencies for this SNP have been characterized in Spanish, Italian, and Irish populations, the latter comprising only females. So far, no clear-cut association between this SNP and a clinical setting has been determined.
CD38 allele distribution in 248 Italian CLL patients did not differ from the allele distributions of the control cohort, suggesting that susceptibility to CLL is not influenced by the CD38 genotype. However, stratification of patients on the basis of molecular markers of unfavorable prognosis constantly resulted in a higher frequency of the rare G allele. Multivariate analysis confirmed the independent association between the G allele and ZAP-70 expression 20% or more. This observation is interesting in light of previous reports showing that CD38 and ZAP-70 are functionally linked in the same growth pathway.25 Furthermore, the G allele is increased in the subset of patients presenting a high tumor mass, as measured by splenomegaly, lymph node involvement, and elevated LDH levels.
The fraction of patients that developed RS within this cohort, characterized by increased lymphoid masses, showed the highest G allele frequency (0.41). Univariate analysis confirmed that the 5-year cumulative incidence of transformation is significantly higher in G allele carriers than in CC homozygotes. The frequency of GG homozygotes is approximately twice that of the heterozygotes, which implies that the G allele exerts a quantitative effect. The validity of this observation was confirmed on the basis of data from an additional series of 14 consecutive cases of RS. This validation subset displayed gene frequencies similar to RS included in the first data set. The 2 RS series were then pooled together and analyzed by multivariate analysis based on the probit system, measuring the variation in probability of developing RS based on a given parameter and without considering time. This test selectively highlighted the G allele as an independent risk factor for transformation. In accordance with the univariate test, the probability of developing RS for GG homozygotes was approximately twice that of the heterozygotes.
How does the CD38 SNP impact on this difference? Knowing that enhanced CD38 expression predicts aggressiveness of CLL, we surmised that the G allele might influence CD38 expression. No association between percentage or MFI levels of peripheral blood CD38 expression and the G allele could be highlighted. However, in vitro studies performed on 40 genotyped CLL patients indicated that G allele carriers are significantly more sensitive to the effects exerted by IL-2, with a resulting higher surface CD38 expression. This observation is in line with a recent report showing that the cells with the highest levels of CD38 are nested in lymph nodes and spleen, surrounded by CD4+/CD25+/foxp3− cells.33 The latter population likely represents activated effector T cells, the highest producers of IL-2 in vivo.
The results of our study may be read from 2 perspectives. From a biologic point of view, they suggest that CD38 polymorphism plays a role in the complex network of interactions taking place in vivo in open and closed systems. Further, they prompt a reinterpretation of an old observation obtained in unselected CLL samples, where antibody-driven CD38 signaling induced the appearance of a population of plasmablast-like cells, reminiscent of RS transformation.15 The marked heterogeneity observed in that study could now be attributed to the quantitative presence or absence of the G allele. The number of possible combinations and their pathophysiological implications are further compounded by the highly polymorphic nature of the CD31 ligand.41
A reading of these results from a clinical standpoint suggests that the G allele may represent an independent risk factor for RS development with potential relevance as a prognostic tool.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
Thanks to Dr F. Castagno (Molinette Hospital Blood Bank, Turin, Italy) for providing blood samples and to Dr M. Magni (Istituto Nazionale Tumori, Milan, Italy) for providing clinical data for some of the RS patients. The excellent technical help of Ms F. Cottino is gratefully acknowledged.
This work was supported by grants from the Chronic Lymphocytic Leukemia Global Research Foundation (CLL-GRF, Houston, TX; S.D.), Associazione Italiana Ricerca Cancro (AIRC, Milan, Italy; S.D. and F.M.), Programmi di ricerca di Rilevante Interesse Nazionale (PRIN, Rome, Italy; S.D., F.M., and G.G.), Fondi di Ateneno, University of Turin, Turin, Italy; (S.D. and F.M.), and Regione Piemonte (F.M. and G.G.). Fondazione Internazionale Ricerche Medicina Sperimentale (FIRMS, Turin, Italy), Fondazione CRT, Fondazione “A. Scorza” (Cosenza, Italy) and Associazione Italiana Leucemie/Linfomi of Novara (Novara-AIL; Italy) provided financial contributions.
S.A. is a student in the PhD Course on Advanced Techniques in Tumor Localization, University of Torino, and is sponsored by Dr Werner Jackstädt-Stiftung.
Authorship
Contribution: S.A. performed experiments and wrote the paper; D.R. provided patient samples and performed statistical analyses; L. Bergui and G.D. provided patient samples; E.F. set up experimental PCR conditions; L.B. performed IgV mutation studies; P.O. performed ZAP-70 staining and FISH analyses; D.N. provided RS patient samples, F. Morabito provided patient samples; A.C. provided RS patient samples; G.G. provided patient samples and helped in discussion and interpretation of results; F. Malavasi designed experiments and wrote the paper; S.D. designed experiments and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Silvia Deaglio, Department of Genetics, Biology and Biochemistry, Via Santena 19, 10126 Torino, Italy; e-mail: silvia.deaglio@unito.it; or Fabio Malavasi, Department of Genetics, Biology and Biochemistry, Via Santena 19, 10126 Torino, Italy; e-mail: fabio.malavasi@unito.it.