Abstract
To address the proposition that familial B-cell chronic lymphocytic leukemia (CLL) may exhibit a more restricted phenotype than sporadic CLL with respect to immunoglobulin gene usage or ontogenic development, we compared immunoglobulin (Ig) heavy chain variable region (VH) gene usage and IgVH mutation status in 327 patients with CLL from 214 families with 724 patients with sporadic cases. The frequency of mutated CLL was higher in familial CLL (P < .001), and there was evidence of intrafamilial concordance in mutation status (P < .001). The repertoire and frequency of IgVH usage was, however, not significantly different between familial and sporadic CLL. Furthermore, IgVH usage was not correlated between affected members of the same family. These observations provide evidence that familial CLL is essentially indistinguishable from sporadic CLL, favoring a genetic basis to disease development in general rather than a simple environmental etiology.
Introduction
Evidence suggests the development of B-cell chronic lymphocytic leukemia (CLL) might be influenced by antigenic recognition or selection through the B-cell receptor (BCR). Hence, it is conceivable that familial disease is associated with a more restricted phenotype with respect to immunoglobulin gene usage or ontogenic development, reflected by presence or absence of somatic mutation
Initial reports based on two families, one American and one Israeli, provided evidence of a restriction in usage among CLL, compatible with the selection by a common super antigen.1 A subsequent study of 4 French families also revealed a restriction in heavy chain variable region (VH) usage.2 More recent findings from analyses of 8 Italian families3 and 11 American families,4 however, have failed to provide confirmatory data.
We have investigated immunoglobulin VH (IgVH) usage and mutation status in 327 individuals with familial CLL and compared findings with those in 724 patients with sporadic CLL, allowing us to address the shortcomings of previously published work, specifically the small sample size and failure to account for the frequency with which specific IgVH subtypes are used in CLL per se.
Methods
A total of 327 individuals with familial CLL from 214 families (2 or more per family) ascertained through hematologists in the United Kingdom (n = 236), Europe (n = 66), and Australia (n = 25) were studied. Vertical transmission of disease was apparent in 66 of the 214 families, and 15 of the families contained both affected offspring and sibships. A series of 724 unselected patients with CLL collected through the Leukemia Research CLL-4 trial5 (n = 336) and the Royal Marsden Hospital National Health Service (NHS) Trust (n = 388) acted as a source of patients with sporadic CLL. Diagnoses of B-cell CLL was established in accordance with World Health Organization (WHO) guidelines.6 The study was conducted with informed consent and ethical review board approval at each participating institution in accordance with the tenets of the Declaration of Helsinki.
Analysis of IgVH rearrangements was performed according to BIOMED-2 protocols as described previously7,8 using commercially available reagents (In VivoScribe Technologies, San Diego, CA). Clonality was assessed by size discrimination of polymerase chain reaction (PCR) products using semiautomated ABI3730xl sequencers in conjunction with Genescan software (Applied Biosystems, Foster City, CA). Sequence analysis was conducted using Chromas software version 2.23 (Applied Biosystems), and IgVH usage and mutational status were assigned using the international immunogenetics information system database.9 In accordance with published criteria, sequences with a germline homology of 98% or greater were classified as unmutated, and those displaying homology of less than 98% were classified as mutated.
The relationship between categorical variables was determined by the Fisher exact test. Concordance between traits was calculated by using the χ2 test statistic to test for differences between observed and expected numbers. A 2-sided P value of less than .05 was considered significant in all analyses.
Results and discussion
The proportion of females affected was higher in familial than sporadic CLL (1.4:1 compared with 2.3:1; P < .01). In both familial and sporadic CLL, 7% had either bi- or triclonal disease. Only in 3% of the sporadic and 4% of familial patients with monoclonal disease was it impossible to unambiguously define mutational status because sequence data recovered were of an insufficient quality despite repeated analysis. Our study findings are therefore unlikely to be biased as a consequence of issues relating to clonality or nonrecoverable mutational status.
In both sporadic and familial CLL, mutation status was unrelated to age at presentation (P = .90 and .21, respectively) or stage (P = .53). Mutated CLL, however, was significantly more common in familial than sporadic CLL (68% [190 of 297] compared with 47% [389 of 647]; P < .001). Although mutations were significantly more common in females with CLL, the association between mutation and familial disease remained statistically significant after adjusting for sex (frequency in female and male CLL, 0.58 and 0.43; 0.74 and 0.63 in sporadic and familial disease, respectively; P < .001).
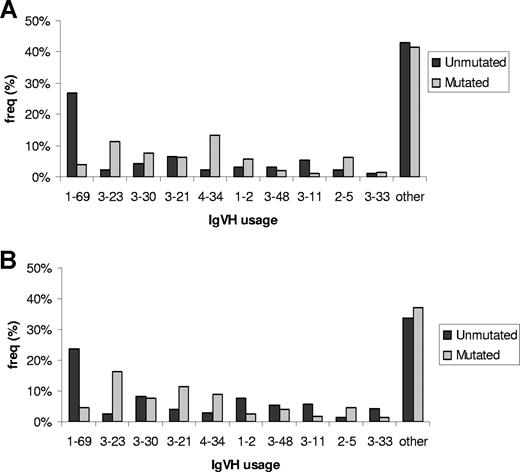
In both familial and sporadic CLL, IgVH usage was nonrandom, with VH3, VH1, and VH4 families expressed at the highest frequencies: 49%, 25%, and 18%, respectively (Figure 1). Globally, there was no difference in usage of specific VH subtypes between familial and sporadic CLL. Stratifying cases by sex showed no significant differences in the distribution of VH usage in either familial or sporadic CLL. In contrast, VH usage was significantly different between mutated and unmutated in both sporadic and familial CLL (Figure 1; P < .001). Restricting familial cases to United Kingdom cases did not affect study findings (data not shown).
IgVH gene segment usage in familial and sporadic CLL stratified by mutational status. (A) Familial CLL. (B) Sporadic CLL. Only the 10 most frequently used gene segments are individually presented. For sporadic CLL, “other” encompasses the subtypes: 3-7; 4-39; 3-9; 3-74; 4-59; 5-51; 1-3; 1-18; 3-15; 4-4; 4-30; 1-46; 3-53; 4-31; 1-8; 3-49; 6-1; 3-13; 3-72; 2-26; 4-61; 4-b; 5-a; 1-58; 3-64; 1-24; 3-20; 2-70; 3-22; 3-43; 3-66; 3-73; 3-h; 7-4; 7-81. For familial CLL, “other” encompasses the subtypes: 3-7; 4-59; 3-74; 4-39; 1-18; 3-13; 1-3; 4-b; 5-51; 6-1; 3-72; 4-31; 1-8; 3-15; 3-53; 4-4; 4-61; 5-a; 3-9; 3-20; 3-73; 1-46; 2-70; 3-64; 3-66; 7-4.
IgVH gene segment usage in familial and sporadic CLL stratified by mutational status. (A) Familial CLL. (B) Sporadic CLL. Only the 10 most frequently used gene segments are individually presented. For sporadic CLL, “other” encompasses the subtypes: 3-7; 4-39; 3-9; 3-74; 4-59; 5-51; 1-3; 1-18; 3-15; 4-4; 4-30; 1-46; 3-53; 4-31; 1-8; 3-49; 6-1; 3-13; 3-72; 2-26; 4-61; 4-b; 5-a; 1-58; 3-64; 1-24; 3-20; 2-70; 3-22; 3-43; 3-66; 3-73; 3-h; 7-4; 7-81. For familial CLL, “other” encompasses the subtypes: 3-7; 4-59; 3-74; 4-39; 1-18; 3-13; 1-3; 4-b; 5-51; 6-1; 3-72; 4-31; 1-8; 3-15; 3-53; 4-4; 4-61; 5-a; 3-9; 3-20; 3-73; 1-46; 2-70; 3-64; 3-66; 7-4.
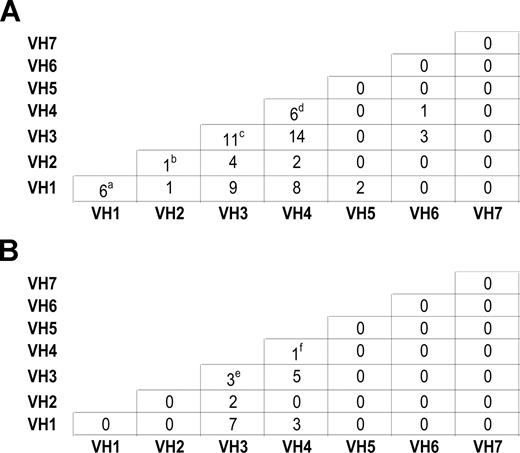
For 86 CLL families, samples were available from more than one affected individual within each family. Mutation status was correlated within families (P < .001). In contrast, there was no evidence of concordance in IgVH usage within families, in both parent-offspring pairs and sibships (Figure 2), thereby confirming previously published findings from Pritch et al3 and Sakai et al.4
Intrafamilial correlation of IgVH usage between individuals with CLL. (A): Within sibships. (a) IgVH1-69/IgVH1-69, IgVH1-69/IgVH1-69, IgVH1-69/IgVH1-2, IgVH1-69/IgVH1-2, IgVH1-69/IgVH1-2, IgVH1-69/IgVH1-18. (b) IgVH2-5/IgVH2-5 (c) IgVH3-21/IgVH3-21, IgVH3-23/IgVH3-74, IgVH3-23/IgVH3-1, IgVH3-33/IgVH3-7, IgVH3-15/IgVH3-30, IgVH3-53/IgVH3-7, IgVH3-21/IgVH3-7, IgVH3-23/IgVH3-48, IgVH3-23/IgVH3-7, IgVH3-9/IgVH3-74, IgVH3-72/IgVH3-74. (d) IgVH4-34/IgVH4-34, IgVH4-34/IgVH4-34, IgVH4-59/IgVH4-59, IgVH4-31/IgVH4-34, IgVH4-31/IgVH4-34, IgVH4-34/IgVH4-59. (B): Within parents and offspring. (e) IgVH3-53/IgVH3-21, IgVH3-48/IgVH3-23, IgVH3-7/IgVH3-23 (f) IgVH4-34/IgVH4-b.
Intrafamilial correlation of IgVH usage between individuals with CLL. (A): Within sibships. (a) IgVH1-69/IgVH1-69, IgVH1-69/IgVH1-69, IgVH1-69/IgVH1-2, IgVH1-69/IgVH1-2, IgVH1-69/IgVH1-2, IgVH1-69/IgVH1-18. (b) IgVH2-5/IgVH2-5 (c) IgVH3-21/IgVH3-21, IgVH3-23/IgVH3-74, IgVH3-23/IgVH3-1, IgVH3-33/IgVH3-7, IgVH3-15/IgVH3-30, IgVH3-53/IgVH3-7, IgVH3-21/IgVH3-7, IgVH3-23/IgVH3-48, IgVH3-23/IgVH3-7, IgVH3-9/IgVH3-74, IgVH3-72/IgVH3-74. (d) IgVH4-34/IgVH4-34, IgVH4-34/IgVH4-34, IgVH4-59/IgVH4-59, IgVH4-31/IgVH4-34, IgVH4-31/IgVH4-34, IgVH4-34/IgVH4-59. (B): Within parents and offspring. (e) IgVH3-53/IgVH3-21, IgVH3-48/IgVH3-23, IgVH3-7/IgVH3-23 (f) IgVH4-34/IgVH4-b.
The strongest evidence for BCR selection by chronic antigen stimulation in the development of CLL is provided by the nonrandom usage of variable domain elements. This hypothesis is provided by the subset of patients with CLL who, regardless of mutational status, use IgVH3-21, have homologous CDR3s, and primarily use V lambda 2-14.10,11 Because IgVH3-21 usage was limited in our cohort of familial CLL, it is unlikely that this plays a major role in familial disease. Moreover, our analysis of intrafamilial correlations showed that VH usage is generally discordant within families. Notably, the absence of concordance between affected sibships is especially relevant in view of the possibility that exposure to a common environmental risk factor early in life plays a role in etiology.
In our study, familial CLL appears to affect the sexes more equally than in sporadic disease. This observation is in keeping with data from the Utah registry.12 It is therefore conceivable that female patients with CLL are more likely to harbor a genetic predisposition than males a priori. Under such a model, females with CLL and their relatives would harbor the same strong inherited liability, thereby explaining the higher proportion of familial cases among female patients with CLL than among males.
In both familial and sporadic CLL, the presence of somatic mutations was higher in females with the disease. Although a number of other studies have also reported a sex effect on mutation status,13,14 it remains a possibility that this association may in part be a consequence of confounding from survivorship.
Regardless of sex differences, the prevalence of somatic mutations in familial CLL was higher than that in sporadic unrelated CLL. A number of published studies have also reported a higher frequency of mutations in familial disease observations, albeit based on small numbers. As mutational status is associated with a better prognosis of survivorship, the probability of phenotyping family members is a function of survivorship, thereby favoring mutated disease. Such an assertion does not, however, address the observation of a correlation in mutation status between affected family members. While concordance is not absolute, the higher frequency of mutated CLL in familial disease is intriguing, as monoclonal B-cell lymphocytosis (MBL) characterized by mutation-positive B cells is detectable at a high frequency in families and is likely to represent a marker of inherited predisposition.15
These data provide evidence that familial CLL can be subcategorized by mutation status but is essentially indistinguishable from sporadic CLL. This favors a genetic basis to the CLL in general rather than simple model susceptibility based on a single environmental etiologic risk factor.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We are grateful to all patients and their clinicians for their participation. We acknowledge useful discussions and advice from David Gonzalez De Castro (Section of Haemato-Oncology, Institute of Cancer Research).
This work was supported primarily by a grant from Leukemia Research. The Arbib Foundation and Cancer Research UK provided additional funding. S.F. and J.W. are supported by the Leukemia Foundation of Australia. D.C.-S. was in receipt of a Studentship from the Institute of Cancer Research.
Authorship
Contribution: D.C.-S. performed research and analyzed data; R.W. performed research; G.S. designed research; M.J.S.D., F.R.M., R.J.G.C. V.J., E.M., C.D., J.W., and S.F. performed sample acquisition; D.C. contributed to project development and obtained funding; R.S.H. designed research, obtained funding and wrote paper; and all authors contributed to the final paper
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Richard S. Houlston, 15 Cotswold Road, Sutton, United Kingdom SM2 5NG; e-mail: richard.houlston@icr.ac.uk.