Mantle cell lymphoma (MCL) is defined pathologically by the detection of CD20, CD5, and most importantly cyclin D1 (CCND1). Its distinction from other lymphomas is important for prognosis and appropriate therapy, but occasional cases may fail to express CCND1 and morphologic simulators may express CD20 and CD5 but not CD23. In this study, we show that the transcription factor Sox11 is specifically expressed in the nucleus of MCL compared with other lymphomas and benign lymphoid tissue. Although the role of Sox11 presently is not known in lymphocyte ontogeny, it is normally expressed in the developing central nervous system in the embryo and shows sequence homology with Sox4, a transcription factor crucial for B lymphopoiesis. Sox11 mRNA is increased in gliomas compared with healthy brain tissue, suggesting a role in malignant transformation and/or cell survival. Our novel finding of specific overexpression of Sox11 mRNA and nuclear protein in both cyclin D1–positive and – negative MCL may be useful for the diagnosis of MCL as a complement to cyclin D1 and also suggests a functional role for Sox11 in MCL.
Introduction
B-cell lymphomas (BCLs) are divided into several subgroups depending on their morphologic and phenotypic properties,1 and accurate diagnosis is necessary for prognosis and targeted therapy. Mantle cell lymphoma (MCL) accounts for 5% to 10% of BCLs and is characterized by overexpression of cyclin D1 (CCND1) due to the specific translocation t(11;14)(q13;q32), which brings the cyclin D1 (BCL1/CCND1) gene under the control of the immunoglobulin heavy chain enhancer. MCL may be distinguished from potential morphologic mimics, including chronic lymphocytic leukemia/small cell lymphoma (CLL), follicular lymphoma (FL), and marginal zone lymphoma (MZL) based on immunohistochemical staining (IHC) for CD5, CD23, and CD10. MCL and CLL both express CD5 but MCL, in contrast to CLL, generally lacks CD23 by immunohistochemistry. FL lacks both CD5 and CD23 but most often expresses CD10, whereas MZL is typically negative for all 3 antigens. Nevertheless, cases of CD23− CLL occur and rare instances of CD5+ MZL have been described. Follicular dendritic cell networks are characteristically expanded in MCL, but this occurs also in FL and MZL. CCND1 overexpression is thus the hallmark of MCL even though approximately 10% of MCLs lack CCND1 expression and may be misdiagnosed by overreliance on CCND1 IHC.2,3 For these reasons, fluorescence in situ hybridization (FISH) targeting the specific translocation probably affords the most sensitive detection of MCL, although it is not widely performed and the results may be more easily affected by technical factors than modern IHC. Thus, specific markers that can aid in stratification and diagnosis of lymphomas in general and MCL in particular would be of major interest
In this study, we demonstrate that the transcription factor Sox11 is specifically up-regulated in MCL and distinguishes MCL from other B-cell lymphomas. Sox11 belongs to the Sox gene family and is mapped to chromosome 2p25.3.4 Proteins are grouped into this family if they contain a DNA-binding high-mobility group (HMG) domain with strong amino acid similarity (usually > 50%) to the HMG domain of Sry, a sex-determining protein.5 More than 20 Sox gene family members have been identified in different species, and Sox genes are divided into 7 subgroups according to the degree of homology within and outside the HMG domain. Human Sox11 belongs to the subgroup C and is homologous to Sox4 with 55% amino acid identity within the C-terminal transactivation domain and 86% identity for the HMG domain.5 Sox4 is a prominent transcription factor in lymphocytes of both the B- and T-cell lineage5,6 and is crucial for B lymphopoiesis.7 In contrast, Sox11 has no known lymphopoietic function and is not expressed in B cells.
The present finding of the specific nuclear expression of Sox11 in MCL compared with other B-cell lymphomas and various benign tissues is based on previous work, in which gene expression profiling was followed by high-throughput screening of the corresponding proteins8,–10 using protein epitope signature tag (PrEST)-specific antibodies.11,,,,–16 Nuclear Sox11 may not only be a valuable marker for diagnosing cyclin D1–positive and – negative MCL but as discussed within may also be expected to have a functional role in the development and/or survival of the tumor cells.
Methods
mRNA analysis of Sox11 and cyclin D1
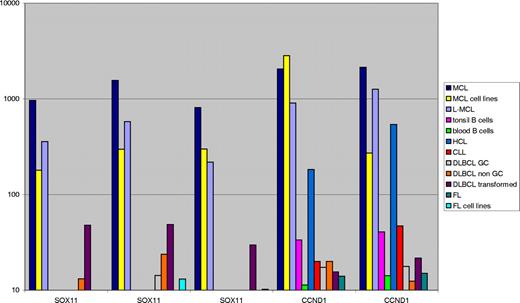
All B-cell lymphomas and reference samples were purified using flow cytometry–assisted cell sorting, and isolated RNA was run on Affymetrix U133 Plus 2.0 (Santa Clara, CA), as previously described.17 The probe sets available for the SOX11 genes are 204913_s_at, 204914_s_at, and 204915_s_at. Cyclin D1 is represented by 208711_s_at and 208712_at. The value for the individual probe sets is displayed in Figure 1 in the order given.
Diagnostic material, tissue microarrays, and immunohistochemical analysis
All material was reviewed by a histopathologist (M.D.) using recent World Health Organization criteria, including a standard diagnostic panel of antibodies (CD20, Bcl-6, CD3, CD5, CD10, CD23, CD21, cyclin D1).1 Separate formalin-fixed paraffin TMAs were constructed for normal adult tissues, 3 benign tonsillar controls, and mature B-cell lymphomas, which included 10 cases of MCL, 27 cases of chronic lymphatic leukemia/small lymphocytic lymphoma (CLL), 27 cases of follicular lymphoma (FL), and 30 cases of diffuse large B-cell lymphoma (DLBCL), as previously described.18 Representative, nonnecrotic tumor areas were marked on H&E-stained sections and 2 1.0-mm tissue cores were then removed from each corresponding paraffin block and assembled in a new recipient block using an automated device (ATA-27; Beecher Instruments, Sun Prairie, WI). For analysis, an Olympus BX45 equipped with 40×/0.75 UPlanFl dry objective and an Olympus U-CMAD3 digital camera was used. Images were captured and processed in Adobe Photoshop CS2 version 9.0.2 (Adobe Systems, San Jose, CA).
Sox11.
Protein expression was studied in both types of tissue microarrays (TMAs) (Tables 2,3) and additionally in whole 2- to 4-μm–thick sections (Tables 1,2; blocks supplied by the Departments of Pathology in Lund and Malmö). After deparaffination, sections were immersed in BORGdecloaker (Biocare, Concord, CA) at pH 9.0 and placed in an electric decloaking chamber for antigen retrieval, then stained with the primary rabbit antihuman Sox11 antibody (1:100) at room temperature for 25 minutes. This polyclonal antibody targeted the following protein sequence, FMVWSKIERRKIMEQSPDMHNAEISKRLGKRWKMLKDSE-KIPFIREAERLRLKHMADYPDYKYRPRKKPKMDPSAKPSASQSP-EKSAAGGGGGSAGGGAGGAKTSKGSSKK, and was raised as part of the HPR project, as previously described.8,11 Signal was detected, using Dako REAL Detection system (Glostrup, Denmark) containing the secondary biotinylated goat antirabbit/mouse antibody, the streptavidin/horseradish peroxidase complex, and 3,3′-diaminobenzidine, according to the manufacturer's protocol. Slides were counterstained with Mayer hematoxylin (Sigma-Aldrich, St Louis, MO).
IHC staining of Sox11 and cyclin D1 on whole MCL tissue sections
| Entity . | Case ID . | Sox11 . | Description . |
|---|---|---|---|
| MCL | 1 | Positive | Nuclear staining |
| MCL-BV | 2 | Positive | Strong nuclear staining |
| MCL | 3 | Positive | Nuclear staining and localized cytoplasmic staining |
| MCL | 4 | Few positive nuclei | Nuclear staining in a proportion of the tumor cells and cytoplasmic staining |
| MCL | 5 | Positive | Nuclear staining |
| MCL-BV | 6 | Negative | Negative, some background |
| MCL | 7 | Positive | Strong nuclear staining |
| MCL | 8 | Few positive nuclei | Granular nuclear staining in a proportion of the tumor cells and cytoplasmic staining |
| MCL | 9 | Positive | Strong nuclear staining |
| MCL | 10 | Positive | Nuclear and localized cytoplasmic staining |
| MCL | 11 | Positive | Strong nuclear staining |
| MCL | 12 | Positive | Nuclear staining |
| MCL | 13 | Positive | Strong nuclear staining |
| MCL | 14 | Positive | Nuclear staining |
| MCL | 15 | Positive | Nuclear staining |
| MCL | 16 | Positive | Strong nuclear staining |
| MCL | 17 | Positive | Nuclear staining |
| MCL | 18 | Few positive nuclei | Granular nuclear staining in a proportion of the tumor cells and cytoplasmic staining |
| Entity . | Case ID . | Sox11 . | Description . |
|---|---|---|---|
| MCL | 1 | Positive | Nuclear staining |
| MCL-BV | 2 | Positive | Strong nuclear staining |
| MCL | 3 | Positive | Nuclear staining and localized cytoplasmic staining |
| MCL | 4 | Few positive nuclei | Nuclear staining in a proportion of the tumor cells and cytoplasmic staining |
| MCL | 5 | Positive | Nuclear staining |
| MCL-BV | 6 | Negative | Negative, some background |
| MCL | 7 | Positive | Strong nuclear staining |
| MCL | 8 | Few positive nuclei | Granular nuclear staining in a proportion of the tumor cells and cytoplasmic staining |
| MCL | 9 | Positive | Strong nuclear staining |
| MCL | 10 | Positive | Nuclear and localized cytoplasmic staining |
| MCL | 11 | Positive | Strong nuclear staining |
| MCL | 12 | Positive | Nuclear staining |
| MCL | 13 | Positive | Strong nuclear staining |
| MCL | 14 | Positive | Nuclear staining |
| MCL | 15 | Positive | Nuclear staining |
| MCL | 16 | Positive | Strong nuclear staining |
| MCL | 17 | Positive | Nuclear staining |
| MCL | 18 | Few positive nuclei | Granular nuclear staining in a proportion of the tumor cells and cytoplasmic staining |
All entities were positive for cyclin D1.
MCL-BV indicates MCL blastoid variant; and ND, not determined.
Sox11 expression in whole tissue sections of B-cell lymphomas and benign tonsils
| Entity . | WTS or TMA . | Cases, no. . | Nuclear staining,* no. . |
|---|---|---|---|
| MCL | WTS | 18 | 17 |
| CLL | WTS | 20 | 0 |
| FL | WTS | 20 | 0 |
| Tonsil, GC | WTS | 4 | 0 |
| MCL | TMA | 10 | 9 (strong) |
| CLL | TMA | 27 | 0 |
| FL | TMA | 27 | 0 |
| DLBCL | TMA | 30 | 2 (weak to moderate) |
| Tonsil | TMA | 3 | 0 |
| Entity . | WTS or TMA . | Cases, no. . | Nuclear staining,* no. . |
|---|---|---|---|
| MCL | WTS | 18 | 17 |
| CLL | WTS | 20 | 0 |
| FL | WTS | 20 | 0 |
| Tonsil, GC | WTS | 4 | 0 |
| MCL | TMA | 10 | 9 (strong) |
| CLL | TMA | 27 | 0 |
| FL | TMA | 27 | 0 |
| DLBCL | TMA | 30 | 2 (weak to moderate) |
| Tonsil | TMA | 3 | 0 |
WTS indicates whole tissue section.
Six cases of MCL, 10 CLL, 10 FL, 11 DLBCL, and 2 tonsils showed cytoplasmic staining.
Sox11 expression in TMA of adult tissues
| Cell type . | Organ . | IHC staining . | |
|---|---|---|---|
| Nuclear . | Cytoplasmic . | ||
| Schwann | Peripheral nerve | Positive | Negative |
| Keratinocyte | Skin | Positive | Positive |
| Squamous | Tonsil | Positive | Negative |
| Epithelium | Bronchus | Scattered positive | Positive |
| Follicular epithelium | Thyroidea | Occ. weak positive | Positive |
| Enterocyte | Appendix | Negative | Positive |
| Enterocyte | Colon | Negative | Positive |
| Hematopoietic | Bone marrow | Negative | Negative |
| Cerebral cortex | Adult brain | Negative | Negative |
| Acinar epithelium | Breast | Negative | Negative |
| Acinar epithelium | Pancreas | Negative | Positive |
| Acinar epithelium | Prostate | Negative | Positive |
| Hepatocyte | Liver | Negative | Positive |
| Renal tubule | Kidney | Negative | Positive |
| Glomeruli | Kidney | Negative | Negative |
| Melanoma | Skin | Negative | Positive |
| Skeletal muscle | Muscle | Negative | Negative |
| Trophoblast | Placenta | Negative | Negative |
| Smooth muscle | Uterus | Negative | Positive |
| Cell type . | Organ . | IHC staining . | |
|---|---|---|---|
| Nuclear . | Cytoplasmic . | ||
| Schwann | Peripheral nerve | Positive | Negative |
| Keratinocyte | Skin | Positive | Positive |
| Squamous | Tonsil | Positive | Negative |
| Epithelium | Bronchus | Scattered positive | Positive |
| Follicular epithelium | Thyroidea | Occ. weak positive | Positive |
| Enterocyte | Appendix | Negative | Positive |
| Enterocyte | Colon | Negative | Positive |
| Hematopoietic | Bone marrow | Negative | Negative |
| Cerebral cortex | Adult brain | Negative | Negative |
| Acinar epithelium | Breast | Negative | Negative |
| Acinar epithelium | Pancreas | Negative | Positive |
| Acinar epithelium | Prostate | Negative | Positive |
| Hepatocyte | Liver | Negative | Positive |
| Renal tubule | Kidney | Negative | Positive |
| Glomeruli | Kidney | Negative | Negative |
| Melanoma | Skin | Negative | Positive |
| Skeletal muscle | Muscle | Negative | Negative |
| Trophoblast | Placenta | Negative | Negative |
| Smooth muscle | Uterus | Negative | Positive |
Occ. indicates occasional.
Cyclin D1.
For IHC determination of cyclin D1 (CCND1), sections were microwaved in EDTA buffer for antigen retrieval and immunostained as described for Sox11 but using a highly sensitive rabbit monoclonal anti-CCND1 antibody (NeoMarkers, Fremont, CA) at a dilution of 1:10019 for whole sections and mouse monoclonal anti-CCND1 antibody diluted 1:50 (Clone DSC-6; Dako) for tissue microarray sections.
Sox11 in CCND1− MCL
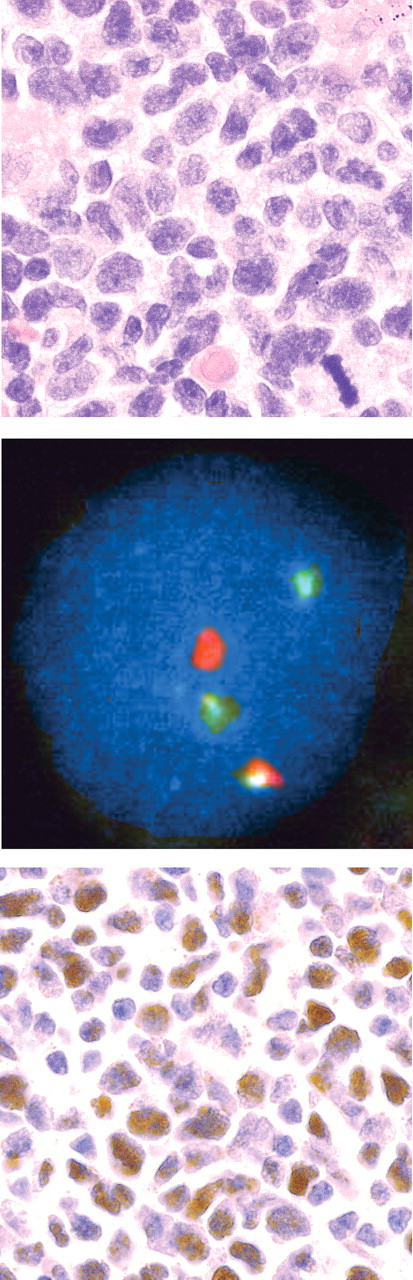
As a further test of the discriminating power of Sox11, one case of small B-cell lymphoma expressing CD20, CD5, and surface IgM but not CD23 or CCND1 was analyzed with fluorescence in situ hybridization (FISH) for the specific MCL translocation using LSI IGH/CCND1 XT dual color, dual fusion translocation probe (Vysis, Downers Grove, IL) and a cutoff value of 6% based on fusion counts in 350 total nuclei prior to staining for Sox11. For FISH analysis, an Olympus BX40 with 10×/1.30 UPlanFl oil objective and a Pixera Pro 150ES digital camera was used. Images were captured and processed in Adobe Photoshop CS2 version 9.0.2 (Adobe Systems).
Results
mRNA levels of Sox11 and CCND1/BCL1 in lymphomas
Sox11 and CCND1/BCL1 mRNA levels were assessed in various B-cell lymphomas and reference samples on Affymetrix U133 Plus 2.0 arrays (Figure 1). The transcriptional level of Sox11 was 10 to 100 times higher for the MCL samples compared with most other lymphomas (Figure 1). Primary MCL as well as leukemic MCL (L-MCL) and MCL cell lines all showed high transcript levels of Sox11. A few samples of DLBCL also showed elevated levels of Sox11 mRNA, reflected both in an increased mean average value and a variable cytoplasmic IHC signal. CCND1/BCL1 transcript levels were high in all types of MCL samples, including L-MCL and, as expected, also in samples of hairy cell leukemia (HCL). Importantly, HCL failed to show elevated Sox11 expression.
Sox11 mRNA in lymphomas and reference material. The mean average transcriptional level (scaled raw values) of Sox11 and cyclin D1 for different lymphoma specimens, MCL and FL cell lines, and benign tonsil and blood reference material is illustrated. The genes are represented by 2 different cyclin D1 or 3 Sox11 probes on the array as described in “mRNA analysis of Sox11 and cyclin D1.” The expression level of Sox11 is on average 10 to 100 times higher for MCL, L-MCL, and MCL cell lines than for the other categories, as is the expression of cyclin D1, with the exception of HCL, which also expresses cyclin D1.
Sox11 mRNA in lymphomas and reference material. The mean average transcriptional level (scaled raw values) of Sox11 and cyclin D1 for different lymphoma specimens, MCL and FL cell lines, and benign tonsil and blood reference material is illustrated. The genes are represented by 2 different cyclin D1 or 3 Sox11 probes on the array as described in “mRNA analysis of Sox11 and cyclin D1.” The expression level of Sox11 is on average 10 to 100 times higher for MCL, L-MCL, and MCL cell lines than for the other categories, as is the expression of cyclin D1, with the exception of HCL, which also expresses cyclin D1.
Immunohistochemistry
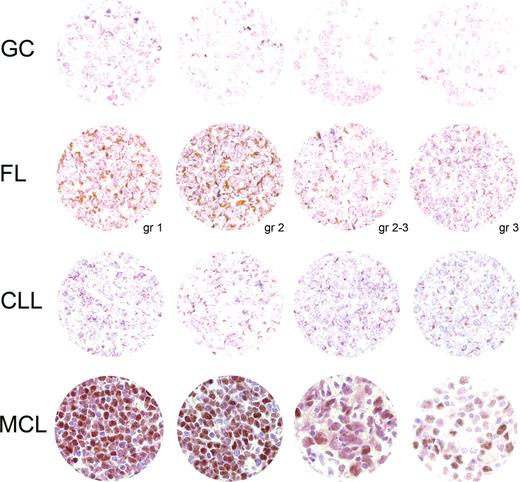
Sox11 was investigated as a potential antigen in the differential diagnosis of B-cell lymphomas. The distribution of Sox11 was assessed in formalin-fixed normal adult tissues, B-cell lymphoma, and benign lymphoid tissue. Interestingly, both whole MCL tissue sections (17/18, Tables 1,2) and TMA of MCL (9/10, Table 2) showed nuclear staining for Sox11, while other lymphomas generally were negative or showed cytoplasmic staining only. Representative IHC images are shown in Figure 2.
Sox11 IHC analysis. A TMA montage of representative Sox11 IHC signals in each category tested includes rows of benign tonsillar germinal centers (GCs), follicular lymphoma (FL, with tumor grade at the bottom right), chronic lymphatic leukemia/small lymphocytic lymphoma (CLL), and mantle cell lymphoma (including blastoid variant in the third image from the left). Each section in the first 3 rows displays granular cytoplasmic staining. A single positive nucleus in the second FL section is probably endothelial. In contrast, most MCL nuclei express Sox11 with varying intensity. Hematoxylin counterstain; magnification, 185×.
Sox11 IHC analysis. A TMA montage of representative Sox11 IHC signals in each category tested includes rows of benign tonsillar germinal centers (GCs), follicular lymphoma (FL, with tumor grade at the bottom right), chronic lymphatic leukemia/small lymphocytic lymphoma (CLL), and mantle cell lymphoma (including blastoid variant in the third image from the left). Each section in the first 3 rows displays granular cytoplasmic staining. A single positive nucleus in the second FL section is probably endothelial. In contrast, most MCL nuclei express Sox11 with varying intensity. Hematoxylin counterstain; magnification, 185×.
Sox11 staining of whole tissue sections.
Nuclear staining was present in 17 of 18 cases (Tables 1,2). In most cases, the staining was intense and present in the majority of the tumor cells (14/17), while 3 cases showed fewer positive nuclei (Figure 2) but were still easy to differentiate from non-MCL tissues. For FL, none of 20 cases showed nuclear positivity (5 cases showed cytoplasmic staining only), whereas CLL failed to stain in the nuclei of 20 cases (2 positive only in the cytoplasm). Nuclear and cytoplasmic signals were readily distinguishable in all cases. Benign tonsil showed no nuclear signal in the lymphoid compartment but consistently positive nuclei in the squamous epithelium.
Sox11 staining of TMA.
Tissue microarrays of MCL (CCND1+), CLL, FL, DLBCL, and benign tonsil were also surveyed (Table 2; Figure 2). MCL showed positive nuclear staining of Sox11 in 9 of 10 cases, whereas CLL expressed no nuclear Sox11 in 27 cases. FL showed similar results where 22 of 27 cases were completely devoid of nuclear signal and the remaining 5 cases had staining limited to the cytoplasm. Of 30 DLBCLs, 28 lacked the nuclear transcription factor but the remaining 2 cases showed only weak diffuse nuclear staining in contrast to the intense nuclear staining in MCL. In the 3 tonsillar controls, no lymphoid cells expressed Sox11.
Sox11 staining in cyclin D1− MCL.
Staining of Sox11 was analyzed in a case of cyclin D1− MCL-like lymphoma in which specific FISH translocation signals were present in 22% of nuclei (Figure 3). Intense Sox11 nuclear staining was noted, consistent with our hypothesis that Sox11 is a nuclear marker for MCL and is expressed independently of cyclin D1.
CCND1− MCL. The top panel displays a lymphoma histologically consistent with MCL (H&E stain). The diagnosis was confirmed by FISH, which in the middle panel shows probe hybridization to IGH (green) and CCND1 (red) loci in a nucleus with a single yellow fusion signal. In the bottom panel, tumor nuclei strongly express Sox11.
CCND1− MCL. The top panel displays a lymphoma histologically consistent with MCL (H&E stain). The diagnosis was confirmed by FISH, which in the middle panel shows probe hybridization to IGH (green) and CCND1 (red) loci in a nucleus with a single yellow fusion signal. In the bottom panel, tumor nuclei strongly express Sox11.
Sox11 adult tissue survey.
We checked the expression of Sox11 in various adult human tissues on a TMA (Table 3). Nuclear staining was found in Schwann cells, keratinocytes, and other squamous cells, while all other tissue, including bone marrow and brain, failed to express nuclear Sox11.
Discussion
We present here the novel findings that the non–B-cell lineage transcription factor Sox11 is specifically expressed in the nucleus of MCL compared with other B-cell lymphomas and nonmalignant lymphoid tissue (Tables 1,Table 2–3,Figures 1,2). Sox11 is normally expressed throughout the central nervous system in the human embryo and may thus play a role in the developing nervous system.20 Quantitative reverse-transcription–polymerase chain reaction (Q-RT-PCR) analysis has not only confirmed expression in fetal brain but also showed Sox11 overexpression in malignant gliomas compared with normal adult brain and other organs.21 In contrast, it has been shown that many adult tissues, for example, spleen, thymus, prostate, testis, ovary, small intestine, colon, peripheral blood leukocytes, heart, brain, placenta, lung, liver, skeletal muscle, kidney, and pancreas lack Sox11 mRNA.22 The lack of expression in adult brain and leukocytes is corroborated by our findings, but the results of the mRNA study notwithstanding, we detected cytoplasmic but not nuclear signal for the Sox11 protein in kidney, liver, lung, and pancreas (Table 3).
The function of Sox11 and many other members of the Sox HMG box superfamily is not clear.5 Sox HMG domains have been found to bind to the consensus sequence 5′-(A/T)(A/T)CAA(A/T)G-3′ in the minor groove of DNA23 and thereby induce a large conformational change.24 It has been suggested that the function of the Sox proteins is at least partly architectural, allowing other transcription factors to bind the major groove and/or bring together regulatory elements, thereby facilitating the formation of protein complexes.25 There is growing support for a model in which the HMG domain serves 2 functions, (1) DNA binding and (2) partner selection, which may permit selective recruitment of individual Sox proteins to specific genes. The function of Sox11 and Sox2 and their interaction with 2 Pit-Oct-Unc (POU) proteins, Oct-3 and Brn-2, have been studied in humans. It was shown that both proteins cooperated with Oct-3, while only Sox11 could partner with Brn-2 to activate transcription.26 Brn-2 is involved in the proliferation of melanoma cells and can, like cyclin D1, be induced by the WNT/Beta-catenin pathway.27
In the murine system, it has been shown in primary and continuous cell cultures that neuron survival and neurite growth are dependent on Sox11.28 Its role in tissue remodeling has also been studied in a mouse knockout model showing various malformations, indicating that Sox11 is important for tissue remodeling and that mutated Sox11 in humans can potentially be associated with malformation syndromes.29 Furthermore, using siRNA knockdown in a neuroblastoma cell line and in cultured mouse dorsal root ganglia cells, it was shown that knockdown of Sox11 modulated the level of mRNAs for several proteins related to cell survival and death. For example, increased expression of the proapoptotic gene BNIP3 and decreased expression of the antiapoptotic gene TANK (TNF receptor–associated factor family member–associated NFκB activator) were demonstrated.28
Thus, although Sox11 function in humans is not clear, it can be hypothesized that the overexpression of Sox11 in MCL leads to aberrant regulation of genes involved in cell survival and/or death. This is further supported by the fact that many of the Sox genes have been shown to be involved in different types of cancer, as reviewed by Dong et al25 Sox4, which is highly homologous to Sox11, has been identified as a potential regulator of the human breast cancer oncogene, HER2/neu (c-ErbB2)30 and both Sox4 and Sox11 are strongly expressed in most medulloblastomas, anaplastic oligodendrogliomas, and glioblastomas.
In this study, we show that Sox11 transcription is specifically up-regulated and the protein overexpressed in MCL cell nuclei in comparison with both benign tissue and other B-cell lymphomas, including a case of CCND1− MCL. Transcript analysis of the different malignancies confirmed, as expected, high overexpression of cyclin D1 in MCL and HCL, but more surprisingly more than 100 × overexpression of Sox11 only in MCL. The overexpression of Sox11 was not only quantitative but also qualitative as most non-MCL cases produced no transcript. Furthermore, Sox11 was not restricted to primary nodular MCL but was also characteristic of L-MCL and MCL cell lines, which expressed the transcript at high levels.
The majority of cyclin D1–positive MCLs showed nuclear staining for Sox11, which was usually intense, while a few cases showed somewhat weaker and less frequent nuclear staining. No nuclear staining was found in FL, CLL, or in the tonsillar controls.
The cytoplasmic localization of Sox11 detected in some lymphomas and normal tissues cannot be readily explained, and the lack of nuclear staining correlates with the absence of Sox11 mRNA in FL, CLL, and most DLBCL. Nuclear localization signals are conserved among all Sox proteins,31,32 and although a nuclear export signal has been found within the group E Sox proteins (Sox8-10),33,34 this has not yet been shown for Sox11. Interestingly, proteins with both nuclear import and export sequences will continuously shuttle between the cytoplasm and the nucleus,35 and for Sox10 this has been shown to be essential for the function of the protein.34
To further test the diagnostic potential of Sox11 for MCLs lacking the characteristic cyclin D1 protein expression, one case of cyclin D1–negative MCL with FISH-confirmed t(11;14) was analyzed and shown to express the transcription factor. This indicates that Sox11 may be useful in routine IHC to identify both cyclin D1–positive and – negative MCLs, and may thus reduce the need for a more time-consuming analysis such as FISH. This correlates with our results showing that there is no general coregulation of cyclin D1 and Sox11 mRNA, since HCLs that express cyclin D1 lack expression of Sox11. However, HCLs express cyclin D1 independent of chromosomal rearrangements,36 which might suggest that the nuclear overexpression of Sox11 in MCL is in some way dependent on the t(11;14) translocation.
In summary, we present the novel finding that the Sox11 transcription factor is specifically expressed in MCL, while other B-cell lymphomas lack nuclear Sox11. Thus, not only does Sox11 offer a potential marker for the diagnosis of MCL regardless of the presence or absence of cyclin D1, but the biology of this transcription factor also suggests an important functional role in the survival and transformation of tumor cells in which it is expressed.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Maria Lindström and Kristina Lövgren at the Department of Oncology, Lund University Hospital, for their excellent work with the immunohistochemical stainings.
This work was supported by a grant from the Leukemia and Lymphoma Society (USA, grant no. 6085-06) and from CREATE Health, a Strategic Center for Translational Cancer Research (http://www.createhealth.lth.se).
Authorship
Contribution: S.E. designed and performed the study and wrote the paper; M.D. evaluated the diagnostic material and the results of the immunohistochemical analysis and participated in the writing of the paper; K.J. collected and evaluated the diagnostic material used for the tissue microarrays; M.J. identified and collected the diagnostic material and participated in the evaluation of the immunohistochemical analysis; and C.A.K.B. participated in the planning of the study, the evaluation of the results, and the writing of the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Sara Ek, Department of Immunotechnology, Lund University, BMC D13, SE-221 84 Lund, Sweden; e-mail: sara.ek@immun.lth.se.