CD49d/α4-integrin is variably expressed in chronic lymphocytic leukemia (CLL). We evaluated its relevance as independent prognosticator for overall survival and time to treatment (TTT) in a series of 303 (232 for TTT) CLLs, in comparison with other biologic or clinical prognosticators (CD38, ZAP-70, immunoglobulin variable heavy chain (IGHV) gene status, cytogenetic abnormalities, soluble CD23, β2-microglobulin, Rai staging). Flow cytometric detection of CD49d was stable and reproducible, and the chosen cut-off (30% CLL cells) easily discriminated CD49dlow from CD49dhigh cases. CD49d, whose expression was strongly associated with that of CD38 (P < .001) and ZAP-70 (P < .001), or with IGHV mutations (P < .001), was independent prognosticator for overall survival along with IGHV mutational status (CD49d hazard ratio, HRCD49d = 3.52, P = .02; HRIGHV = 6.53, P < .001) or, if this parameter was omitted, with ZAP-70 (HRCD49d = 3.72, P = .002; HRZAP-70 = 3.32, P = .009). CD49d was also a prognosticator for TTT (HR = 1.74, P = .007) and refined the impact of all the other factors. Notably, a CD49dhigh phenotype, although not changing the outcome of good prognosis (ZAP-70low, mutated IGHV) CLL, was necessary to correctly prognosticate the shorter TTT of ZAP-70high (HR = 3.12; P = .023) or unmutated IGHV (HR = 2.95; P = .002) cases. These findings support the introduction of CD49d detection in routine prognostic assessment of CLL patients, and suggest both pathogenetic and therapeutic implications for CD49d expression in CLL.
Introduction
B-cell chronic lymphocytic leukemia (CLL) is a heterogeneous disease with highly variable clinical courses.1 Two major clinical staging systems, mainly based on tumor load, were developed to estimate prognosis in CLL.2,3 Both these systems, however, are unable to prospectively discriminate the rapidly evolving patients from those destined to remain with a stable disease for decades.1
In recent years, several publications reported the prognostic significance of specific immunoglobulin variable heavy chain (IGHV) gene features in CLL, including the amount of IGHV mutations,4,,,,–9 as well as the expression of particular IGHV genes, including IGHV3-21, mostly associated with a bad outcome,10,,–13 or IGHV3-72, identifying CLL with stable clinical courses.14,15 Because IGHV gene sequencing is a technique usually not available in diagnostic laboratories, many studies focused on the identification of alternative markers with similar prognostic value, and whose expression could be easily investigated (eg, by flow cytometry). As an example, expression of CD38 has been shown to correlate with unfavorable prognosis and with the presence of unmutated IGHV genes,4 although this latter relationship was then questioned.6,16,17
The techniques of gene expression profiling (GEP) have been used for the identification of additional molecules to be used as potential prognosticators in CLL.18,,–21 Among them, the gene encoding for the T cell specific zeta-associated protein-70 (ZAP-70) was demonstrated to have both prognostic relevance and predictive power as surrogate for IGHV gene mutations when its intracytoplasmic expression is investigated by flow cytometry,20,22,,–25 although a common standardized protocol for its detection is still to be defined.26,27
In analogy with GEP, an approach analyzing the simultaneous and coordinated expression of surface antigens has been recently proposed by us and named surface-antigen expression profiling.28,–30 According to this procedure, antigen expression values, as measured by flow cytometry in a series of CLL with known clinical courses, were analyzed with data mining tools identical to those used in GEP, with the aim of identifying the immunophenotypic signatures associated with different prognosis.28,–30 A striking finding of these studies was the simultaneous overexpression of CD38 and CD49d as part of the signature characterizing the subgroup with the worst outcome.28,–30 The CD49d/α4 integrin is a molecule functionally acting as adhesion structure for extracellular matrix components or mediating cell-cell interactions through the binding with fibronectin or vascular cell adhesion molecule-1, respectively.31,32 In this regard, CD49d-expressing CLL cells were shown to have a high propensity to adhere to fibronectin substrates, and an increased CD49d protein expression was demonstrated in CLL cells from advanced Rai stage patients.33 Consistently, preliminary studies by our group suggested that high CD49d, alone or in combination with high CD38, were both prognostic factors for short overall survival (OS) in CLL,34 although the independent prognostic relevance of CD49d was not tested by appropriate multivariate analyses.34
By taking advantage of a wide cohort of CLL patients, the aims of the present study were: (1) to use a reproducible flow cytometry method for testing CD49d expression in peripheral blood (PB) CLL samples for prognostic purposes; (2) to investigate the relationship between CD49d and other well-established biologic prognosticators (CD38 and ZAP-70 expression, IGHV gene mutational status, presence of specific genomic aberrations), or markers of tumor burden (clinical stage, β2-microglobulin, soluble CD23); and (3) to test the impact of CD49d as an independent prognostic factor for OS and disease progression, this latter evaluated as time to treatment (TTT).35
Methods
Patients
This is a retrospective study that includes PB samples from 303 patients affected by CLL and recruited in a single center during the period 1988 to 2006. Approval of the study was obtained from the institutional review board of the Chair of Hematology, University of Tor Vergata (Rome, Italy). Informed consent was obtained in accordance with the Declaration of Helsinki. Inclusion criteria for patients were as follows: (1) untreated disease at time of referral; and (2) diagnosis made less than 12 months before, according to the current criteria.36 The median age at diagnosis was 63.5 years (range, 32-97 years) and the male/female ratio was 1.2 (164 males, 139 females). The distribution of clinical stages at diagnosis according to the modified Rai staging system (mod-Rai),3,35 available for 292 of 303 patients, was as follows: 105 patients, stage I (low-risk category); 181 patients, stage II (intermediate-risk category); 6 patients, stage III (high-risk category). Median follow-up was 6.2 years with 48 deaths and 255 censored patients. In 232 of 303 patients, treatments were established after the National Cancer Institute Working Group criteria.35 In this cohort of patients, chosen to evaluate the relevance of CD49d as prognostic factor for progressive disease that needs therapy, median age at diagnosis was 63.5 years (range, 32-87 years), the male/female ratio was 1.1 (121 male patients, 111 female patients), and the distribution of mod-Rai stages at diagnosis was 86 patients, stage I; 142 patients, stage II; 4 patients, stage III. The median follow-up for this subset was 6.1 years; 123 patients were censored without event, and 109 experienced a progressive disease requiring therapy. Among them, 27 patients were intermittently treated with a combination of chlorambucil at conventional doses and prednisone, whereas 82 cases received fludarabine alone or fludarabine-containing regimens.23,37
In all cases, immunophenotypic, molecular, serologic, and fluorescence in situ hybridization (FISH) analyses were carried out either at diagnosis or before start protocol therapies. A total of 107 of 303 cases were already reported in a preliminary study of ours.34 Fifty-six of 303 cases were available as cryopreserved samples, frozen at diagnosis, and kept in liquid nitrogen until use.
Immunophenotypic analyses
All the flow cytometric analyses of the present study were performed on a FACScalibur flow cytometer (Becton Dickinson, San Jose, CA). Throughout the course of the present study, the control of instrument calibration and the stability of instrument setup were made on a daily basis by immunophenotyping leukocyte subpopulations from normal PB fresh and/or commercially available stabilized blood samples; further compensation controls were run on a weekly basis using reference fluorospheres (Calibrite beads; Becton Dickinson).38,39 Expression of CD38 and CD49d was analyzed by 3-color immunofluorescence, as described,29,40 by combining phycoerythrin (PE)–conjugated anti-CD38 or anti-CD49d monoclonal antibodies (mAbs) with peridinin-chlorophyll-protein-cyanine-5.5 (PerCP-Cy5.5)–conjugated anti-CD19 mAbs and fluorescein isothiocyanate–conjugated anti-CD5 mAbs. Irrelevant isotype-matched antibodies (Becton Dickinson) were used to determine background fluorescence. Expression data were reported as percentage of CD5+CD19+ CLL cells displaying specific fluorescence intensity greater than the 98% to 99% of the same cell population stained with isotype– and fluorochrome–matched control immunoglobulins. Flow cytometry detection of ZAP-70 was performed as reported,23,27,29,30 using a combination of PerCP-Cy5.5–conjugated anti-CD19, allophycocyanin–conjugated anti-CD5, PE–conjugated anti-CD3/anti-CD56 (Becton Dickinson), and Alexa-488–conjugated anti-ZAP-70 mAbs (Caltag, Burlingame, CA). ZAP-70 expression was reported as percentage of CD19+ CLL cells expressing the protein above a marker set to include as positive all autologous T cells (ie, at the left tail of ZAP-70 specific fluorescence distribution, as detected on T cells).22,23 According to previous studies, cut-offs of 30% and 20% of positive cells were chosen to discriminate CD38low and CD38high, or ZAP-70low and ZAP-70high CLLs, respectively.4,22,,–25,40
Estimation of CD49d expression level yielding the best separation of 2 subgroups with different OS and/or TTT probabilities was made by applying various methods, including the maximally selected log-rank statistics, Youden's index, receiver-operating characteristic analysis and using the percentage of positive cells as measure for antigen expression level.30,41 All these tests and previous observations,30,34 concordantly indicated to use the cut-off of 30% of positive cells to discriminate CD49dhigh and CD49dlow cases when testing the prognostic impact of CD49d on both OS and TTT (data not shown).
To test the reproducibility of CD49d expression, as investigated by the flow cytometric approach reported above (this section, first paragraph), expression of CD49d was compared: (1) in fresh versus frozen PB samples (15 cases); (2) in fresh PB samples delivered by overnight courier and separately analyzed in 2 different laboratories (ie, located at the University of Tor Vergata in Rome and at the Clinical and Experimental Onco-Hematology Unit of the Centro di Riferimento Oncologico in Aviano; 44 cases). As summarized in Figure S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article), these sets of experiments yielded almost superimposable results in all cases. Finally, the stability of CD49d expression during the disease course was demonstrated in selected CD49dlow and CD49dhigh CLL cases in which frozen PB samples covering a period of 1 to 14.0 years from diagnosis were available (Figure S1).42,43
Identification of IGHV gene mutational status
Total RNAs were extracted, reverse-transcribed, checked for first-strand synthesis,6,44 and amplified in the presence of sense primers annealing to either VH1-VH6 leader sequences or 5′-ends of VH1-VH6 FR1, used in conjunction with JH- or Cμ-specific antisense primers.4,–6,44 Purified amplicons were cloned and at least 10 to 20 colonies for each case were sequenced; CLL specific IGHV transcripts, identified by sharing the same CDR3 in multiple clones, were analyzed for percent mutation as previously reported.6,44 Mutated (M) and unmutated (UM) CLL cases were identified when percent mismatch was above or below the 2% cut-off, respectively.4,–6
Soluble CD23 and β2-microglobulin detection
Analysis of cytogenetic aberrations
Cytogenetic abnormalities were detected by interphase FISH carried out for loci on chromosome 11, 12, 13, and 17. For chromosomes 11, 13, and 17, the 3 locus specific probes LSI-ATM, LSI-D13S319, and LSI-p53, directly labeled with SpectrumGreen (LSI-ATM) or SpectrumOrange (LSI-D13S319 and LSI-p53), were used, respectively (Vysis, London, United Kingdom). An alpha satellite DNA probe CEP12, directly labeled with SpectrumGreen, was used to detect aneuploidy of chromosome 12 (Vysis). FISH procedures were as reported previously23,37,45 ; in all cases, at least 200 interphase cells with well-delineated fluorescent spots were examined.
Statistical analysis
All statistical analyses were performed using the R statistical package (http://www.r-project.org/). Median follow-up was computed using the OS database and applying the inverted censoring method. The primary endpoints were OS (data available for 303 patients) and TTT (data available for 232 patients), defined as time from diagnosis to death (event) or end of follow-up (censored observation), or time from diagnosis to treatment (event) or end of follow-up (censored observation), respectively. For OS analyses, all events were considered as CLL–related, ie, all deaths were considered as events whatever the cause. In the cohort of cases used for TTT analysis, no patients died before disease progression. OS and TTT were estimated using the Kaplan-Meier plots, and comparisons between groups were made by log-rank test. The Cox proportional hazards regression model was chosen to assess the independent effect of covariables, treated as dichotomous, on OS or TTT, with a stepwise procedure for selecting significant variables.
The association between CD49d expression and other variables (age, mod-Rai, CD38, ZAP-70, IGHV gene mutations, FISH, sCD23, and β2M) was calculated using the χ2 test and by testing the Spearman's rank correlation.
Results
Expression of CD49d and association with other prognostic factors
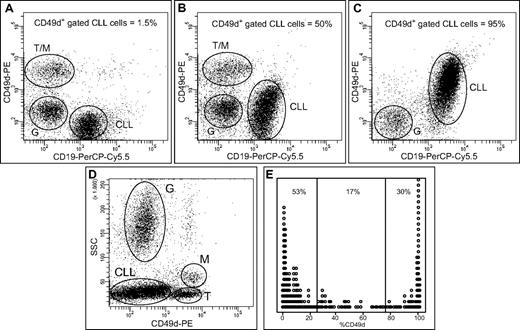
Expression of CD49d, as detected by 3-color flow cytometry on CD19+CD5+ CLL cells, revealed variable patterns of fluorescence intensity, corresponding to different percentages of positive CLL cells (Figure 1A-C). In all cases, expression of CD49d on CLL cells was clearly distinguishable to that of residual normal T cells and monocytes, expressing CD49d at brighter intensity, or neutrophils, usually lacking CD49d expression (Figure 1A-D). When CD49d expression levels, reported as percentage of positive CLL cells, were ordered in a stacked strip-plot, 83% of cases expressed CD49d at either very high (> 75% of positive cells) or negligible (< 25% of positive cells) levels. Conversely, as few as 17% of CLLs expressed CD49d at intermediate (25%-75% of positive cells) levels (Figure 1E).
Characteristics of CD49d expression. (A-C) Flow cytometric dot-plots of CD19-PerCP-Cy5.5 vs CD49d-PE expression on PB samples of 3 representative CLL cases; the reported percent values of CD49d-expressing cells are calculated on the gated CD5+CD19+ CLL population. T cells (T), monocytes (M), granulocytes (G), and CLL cells (CLL) are indicated in dot-plots. (D) Flow cytometric dot-plot of the physical parameter side-scatter (SSC) vs CD49d-PE expression; T cells, monocytes, granulocytes, and CLL cells are indicated as above; the dot-plot corresponds to CLL reported in panel B. (E) Stacked strip-plot of ordered CD49d percent expression values. Reported percent values refer to the amount of cases expressing low (< 25%), intermediate (25%-75%), or high (> 75%) percentages of CD49d.
Characteristics of CD49d expression. (A-C) Flow cytometric dot-plots of CD19-PerCP-Cy5.5 vs CD49d-PE expression on PB samples of 3 representative CLL cases; the reported percent values of CD49d-expressing cells are calculated on the gated CD5+CD19+ CLL population. T cells (T), monocytes (M), granulocytes (G), and CLL cells (CLL) are indicated in dot-plots. (D) Flow cytometric dot-plot of the physical parameter side-scatter (SSC) vs CD49d-PE expression; T cells, monocytes, granulocytes, and CLL cells are indicated as above; the dot-plot corresponds to CLL reported in panel B. (E) Stacked strip-plot of ordered CD49d percent expression values. Reported percent values refer to the amount of cases expressing low (< 25%), intermediate (25%-75%), or high (> 75%) percentages of CD49d.
As reported here and previously,30,34 a cut-off of 30% of positive cells was chosen to discriminate CD49dhigh and CD49dlow cases when testing the prognostic impact of CD49d on both OS and TTT. Table 1 shows the association of CD49d, used as categorical variable, with other prognosticators for CLL, including biologic markers (CD38 and ZAP-70 expression, the IGHV gene mutational status), markers of tumor burden (β2M, sCD23, and mod-Rai), the presence of specific chromosomal aberrations, and age.1,,,,,,–8,23,35,37,40,45,46 All the reported factors were proven to be significant prognosticators for OS and /or TTT (OS alone for age) also in our CLL series (Table 1). As previously described,29,30,34 the strongest statistically significant associations were between CD49d and CD38 (P < .001; Spearman's rank coefficient ρ = .59), CD49d and IGHV mutations (P < .001; ρ = .34) or CD49d and ZAP-70 (P < .001; ρ = .26), with overall concordances of 78%, 68%, and 64%, respectively. Lower but still significant associations were found between CD49d expression and serum sCD23 levels or mod-Rai. Finally, no significant associations were found with the presence of specific chromosomal aberrations, serum levels for β2M, and age (Table 1).
Distribution of prognostic factors in CLL cases according to CD49d expression
| Parameter, category* . | CD49d, no. . | P(χ2)† . | ρ‡ . | n§ . | 10-year survival, % . | 10-year untreated, % . | |
|---|---|---|---|---|---|---|---|
| Less than 30% . | More than 30% . | ||||||
| CD38 | |||||||
| Less than 30% | 153 | 45 | < .001 | 0.59 | 303 | 88 | 40 |
| More than or equal to 30% | 20 | 85 | — | — | — | 65¶ | 15¶ |
| IGHV | |||||||
| M | 98 | 45 | < .001 | 0.34 | 210 | 87 | 41 |
| UM | 22 | 45 | — | — | — | 43¶ | 9¶ |
| ZAP-70 | |||||||
| Less than 20% | 107 | 49 | < .001 | 0.26 | 270 | 95 | 44 |
| More than or equal to 20% | 49 | 65 | — | — | — | 79¶ | 17¶ |
| sCD23 | |||||||
| Less than 70 IU | 89 | 49 | .001 | 0.22 | 206 | 98 | 46 |
| More than or equal to 70 IU | 28 | 40 | — | — | — | 84¶ | 7¶ |
| mod-Rai | |||||||
| I | 73 | 32 | .002 | 0.18 | 292 | 97 | 53 |
| II-III | 96 | 91 | — | — | — | 91 | 19¶ |
| FISH | |||||||
| Normal‖ | 30 | 45 | .18 | 0.11 | 156 | NT | 46 |
| Abnormal | 41 | 40 | — | — | — | NT | 15¶ |
| β2M | |||||||
| Less than 2.2 g/L | 84 | 51 | .10 | 0.11 | 232 | 97 | 45 |
| More than or equal to 2.2 g/L | 50 | 47 | — | — | — | 88¶ | 14¶ |
| Age | |||||||
| Less than 65 years | 94 | 65 | .49 | 0.04 | 302 | 86 | 28 |
| More than or equal to 65 years | 79 | 64 | — | — | — | 73¶ | 35 |
| Parameter, category* . | CD49d, no. . | P(χ2)† . | ρ‡ . | n§ . | 10-year survival, % . | 10-year untreated, % . | |
|---|---|---|---|---|---|---|---|
| Less than 30% . | More than 30% . | ||||||
| CD38 | |||||||
| Less than 30% | 153 | 45 | < .001 | 0.59 | 303 | 88 | 40 |
| More than or equal to 30% | 20 | 85 | — | — | — | 65¶ | 15¶ |
| IGHV | |||||||
| M | 98 | 45 | < .001 | 0.34 | 210 | 87 | 41 |
| UM | 22 | 45 | — | — | — | 43¶ | 9¶ |
| ZAP-70 | |||||||
| Less than 20% | 107 | 49 | < .001 | 0.26 | 270 | 95 | 44 |
| More than or equal to 20% | 49 | 65 | — | — | — | 79¶ | 17¶ |
| sCD23 | |||||||
| Less than 70 IU | 89 | 49 | .001 | 0.22 | 206 | 98 | 46 |
| More than or equal to 70 IU | 28 | 40 | — | — | — | 84¶ | 7¶ |
| mod-Rai | |||||||
| I | 73 | 32 | .002 | 0.18 | 292 | 97 | 53 |
| II-III | 96 | 91 | — | — | — | 91 | 19¶ |
| FISH | |||||||
| Normal‖ | 30 | 45 | .18 | 0.11 | 156 | NT | 46 |
| Abnormal | 41 | 40 | — | — | — | NT | 15¶ |
| β2M | |||||||
| Less than 2.2 g/L | 84 | 51 | .10 | 0.11 | 232 | 97 | 45 |
| More than or equal to 2.2 g/L | 50 | 47 | — | — | — | 88¶ | 14¶ |
| Age | |||||||
| Less than 65 years | 94 | 65 | .49 | 0.04 | 302 | 86 | 28 |
| More than or equal to 65 years | 79 | 64 | — | — | — | 73¶ | 35 |
CLL cases with low (<30%) or high (>30%) CD49d expression were characterized for the distribution of several factors (CD38 and ZAP-70 expression, IGHV mutational status, modified Rai stage, sCD23 and β2M serum levels, normal and abnormal FISH, age), which in our series had prognostic relevance for OS and/or TTT.
IGHV indicates immunoglobulin heavy variable chain; ZAP-70, zeta-chain-associated protein 70 kDa; sCD23, soluble CD23; mod-Rai, modified Rai staging system; FISH, fluorescence in situ hybridization; β2M, β2-microglobulin; NT, not tested; and —, not applicable.
For each variable, the selected cutoff points or the compared categories are indicated.
χ2 tests were performed to evaluate the association between CD49d expression (below or above the established threshold set at 30% of positive CLL cells) and other prognostic factors.
Spearman ρ was calculated to show the degree of rank correlation.
Values refer to the number of cases analyzed for a given feature.
Normal FISH category included cases with normal karyotypes, that is, lacking 11q−, 13q−, 17p−, and chromosome +12; abnormal FISH included cases with 11q− (16 cases), 13q− (35 cases), 17p− (5 cases), and +12 (25 cases); 10-years untreated was computed by comparing cases with normal karyotypes with cases with 11q−, 17p−, and +12.
P < .05 (log-rank test).
Relevance of CD49d expression as prognostic factor for OS
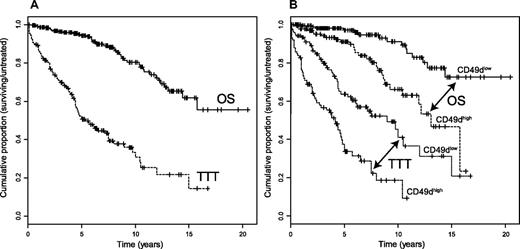
OS data were available for 303 CLL patients; altogether, the percentage survival was 80% at 10 years, with a “not reached” median survival (Figure 2A). When testing the prognostic relevance of CD49d expression in this cohort of patients, a significantly shorter survival was found in CD49dhigh compared with CD49dlow CLL patients (P < .001; Figure 2B), not dissimilar to that obtained by splitting patients according to the other biologic prognosticators (Table 1; Figure S2).
OS and TTT in CLL patients. (A) Kaplan-Meier estimates of OS in 303 CLL patients (median OS, not reached; solid line) and TTT in 232 patients (median TTT, 5.3 years; dashed line). (B) Prognostic impact of CD49d on OS and TTT. Kaplan-Meier curves obtained by comparing OS of CLL patients according to CD49dlow (173 patients) and CD49dhigh (130 patients) expression are depicted in solid and dashed lines, respectively (P < .001, log-rank test); Kaplan-Meier curves obtained by comparing TTT of CLL patients according to CD49dlow (134 patients) and CD49dhigh (98 patients) expression are depicted in dotted and dot-dashed lines, respectively (P < .001, log-rank test).
OS and TTT in CLL patients. (A) Kaplan-Meier estimates of OS in 303 CLL patients (median OS, not reached; solid line) and TTT in 232 patients (median TTT, 5.3 years; dashed line). (B) Prognostic impact of CD49d on OS and TTT. Kaplan-Meier curves obtained by comparing OS of CLL patients according to CD49dlow (173 patients) and CD49dhigh (130 patients) expression are depicted in solid and dashed lines, respectively (P < .001, log-rank test); Kaplan-Meier curves obtained by comparing TTT of CLL patients according to CD49dlow (134 patients) and CD49dhigh (98 patients) expression are depicted in dotted and dot-dashed lines, respectively (P < .001, log-rank test).
The value of CD49d as independent prognostic factor for OS was checked by multivariate Cox proportional hazards analyses applied to models that included the prognosticators proven to significantly correlate with OS in univariate analyses (Table 1). In particular, 263 cases were selected in which information regarding the biologic prognosticators CD49d, ZAP-70, and CD38 was simultaneously available, along with age; in this cohort of patients, multivariate analysis selected CD49d (P = .002) and ZAP-70 (P = .009) as significant independent biologic prognosticators (Table 2). In a further model adjusting also for the IGHV status, the cohort of patients decreased to 175 cases; also in this cohort of patients, multivariate analysis identified CD49d (P = .02), along with IGHV gene mutational status (P < .001) but not ZAP-70 and CD38, as independent biologic prognosticators for OS (Table 2). Finally, when adjusting for all the biologic prognosticators and the 2 tumor burden markers sCD23 and β2M (Table 1), again CD49d was an independent prognostic factor for shorter OS (P = .006), along with β2M (P = .013), although in the context of a small cohort of patients (131 cases; not shown). A significant impact of advanced age as prognosticator for shorter OS was also observed in all multivariate analyses (Table 2 and data not shown), as expected.47
Multivariate Cox regression analyses of OS
| Parameter . | 263 patients . | 175 patients . | P . | |
|---|---|---|---|---|
| HR (95% CI)* . | P . | HR (95% CI)* . | ||
| CD49d less than 30% | 3.72 (1.62-8.56) | .002 | 3.52 (1.22-10.2) | .02 |
| ZAP-70 more than 20% | 3.32 (1.46-7.51) | .009 | — | — |
| Age more than 65 years | 3.73 (1.63-8.51) | .002 | 6.85 (2.54-18.5) | .001 |
| IGHV UM | NT | NT | 6.53 (2.32-18.4) | .001 |
| Parameter . | 263 patients . | 175 patients . | P . | |
|---|---|---|---|---|
| HR (95% CI)* . | P . | HR (95% CI)* . | ||
| CD49d less than 30% | 3.72 (1.62-8.56) | .002 | 3.52 (1.22-10.2) | .02 |
| ZAP-70 more than 20% | 3.32 (1.46-7.51) | .009 | — | — |
| Age more than 65 years | 3.73 (1.63-8.51) | .002 | 6.85 (2.54-18.5) | .001 |
| IGHV UM | NT | NT | 6.53 (2.32-18.4) | .001 |
Multivariate Cox regression analyses of OS were performed by including the following covariates: CD49d, CD38, ZAP-70, age (263 patients) or CD49d, CD38, ZAP-70, age, and IGHV (175 patients). For each variable, the selected cutoff points or the compared categories are indicated.
OS indicates overall survival; HR, hazard ratio; CI, confidence interval; NT, not tested; other abbreviations are as in Table 1.
Based on the final model after stepwise selection of covariates.
Altogether, these data concordantly indicated the relevance of CD49d as independent prognosticator for OS in CLL.
Relevance of CD49d expression as prognostic factor for progressive disease
The relevance of CD49d as prognosticator for progressive disease, evaluated as TTT, was compared with CD38, ZAP-70, IGHV mutational status, β2M, sCD23, mod-Rai, and the presence of specific genomic aberrations. As shown in Figure 2A, the selected cohort of cases had a median TTT of 5.3 years.
Relationship of CD49d with CD38, ZAP-70 and IGHV mutational status.
Median TTT was shorter in CD49dhigh cases (4.2 years), compared with the CD49dlow subgroup of patients (9.0 years; Figure 2B). Similar trends were observed when investigating the impact on TTT duration of the 2 immunophenotypic prognosticators CD38 and ZAP-70, or of IGHV gene mutations (Table 1; Figure S2).
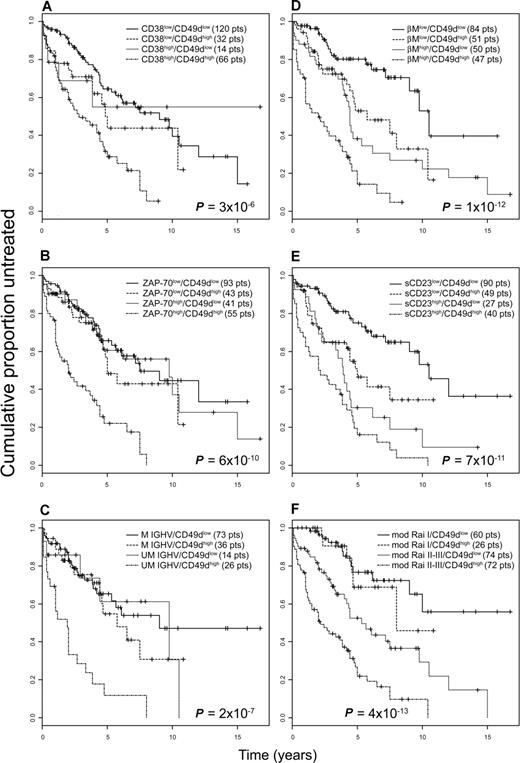
A better refinement in the prognostic assessment of TTT was possible by combining CD49d expression with that of CD38 and ZAP-70, or with the presence of a specific IGHV gene mutational status (Figure 3A-C). In particular, although the expression of CD49d only slightly affected the clinical outcome of good prognosis CD38low, ZAP-70low, or M-IGHV CLL patients, information on CD49d expression levels turned out to be necessary to correctly prognosticate TTT duration in the context of bad prognosis patients. Indeed, only cases in which a high CD38 (n = 66) or ZAP-70 (n = 55) expression, or a UM-IGHV gene configuration (n = 26), was associated with a CD49dhigh phenotype experienced a disease with significantly shorter TTT than those with CD38low, ZAP-70low, or M-IGHV phenotypes. Conversely, expression of CD49d below the threshold of 30% positive cells identified discrete subsets of cases, expressing the CD49dlow/CD38high (14 cases), CD49dlow/ZAP-70high (41 cases), or CD49dlow/UM-IGHV (14 cases) phenotypes, whose TTT intervals were not dissimilar to those of good prognosis CLLs (Figure 3A-C).
Prognostic relevance for TTT of CD49d in combination with other biologic prognosticators or markers of tumor burden. Kaplan-Meier curves obtained by associating CD49d expression to 3 biologic prognosticators, ie, CD38 (A), ZAP-70 (B), and IGHV gene mutational status (C), or to 3 tumor burden markers, ie, β2M (D) and sCD23 (E) serum levels and mod-Rai risk groups (F). For each Kaplan-Meier analysis, 4 groups were compared: patients lacking both negative prognosticators, patients with presence of both negative prognosticators, patients with presence of either one unfavorable prognosticator. The numbers of patients (pts) included in each group are reported in parentheses; the reported P values refer to the log-rank test.
Prognostic relevance for TTT of CD49d in combination with other biologic prognosticators or markers of tumor burden. Kaplan-Meier curves obtained by associating CD49d expression to 3 biologic prognosticators, ie, CD38 (A), ZAP-70 (B), and IGHV gene mutational status (C), or to 3 tumor burden markers, ie, β2M (D) and sCD23 (E) serum levels and mod-Rai risk groups (F). For each Kaplan-Meier analysis, 4 groups were compared: patients lacking both negative prognosticators, patients with presence of both negative prognosticators, patients with presence of either one unfavorable prognosticator. The numbers of patients (pts) included in each group are reported in parentheses; the reported P values refer to the log-rank test.
Relationship of CD49d with markers of disease burden (β2M, sCD23, and mod-Rai).
Serum levels for β2M or sCD23 and mod-Rai are additional variables with proven prognostic relevance in CLL, overall indicating disease burden.1,3,35,37,45 Using cut-offs set at 2.2 g/L or 70 U/mL for β2M and sCD23, respectively,23,40 2 subgroups with significantly different risk for treatment were identified; a similar prognostic impact was observed by comparing patients belonging to the mod-Rai low-risk group with those of the intermediate-/high-risk categories (Table 1; Figure S3).35
For all these variables, expression of CD49d had true additive properties as prognostic factor for TTT (Figure 3D-F). Indeed, expression of CD49d over the established threshold, when associated with high serum levels for β2M or sCD23 or with a mod-Rai high-/intermediate-risk group, identified the subsets of patients with the shortest TTT. Conversely, a CD49dlow phenotype, if paralleled with low serum levels for β2M or sCD23, or in the context of a mod-Rai low-risk group, always identified the subgroups with the longest TTT. Of note, patients who were discordant for CD49d expression and at least one of these markers displayed intermediate TTT; these groups were identified by the following combinations: 1) a favorable CD49dlow phenotype associated with prognostically unfavorable categories for β2M (50 patients), sCD23 (27 patients), or mod-Rai (high-/intermediate-risk group, 74 patients); and 2) an unfavorable CD49dhigh phenotype associated with prognostically favorable categories for β2M (51 patients), sCD23 (49 patients), or mod-Rai (low-risk group, 26 patients).
Relationship of CD49d with cytogenetic abnormalities.
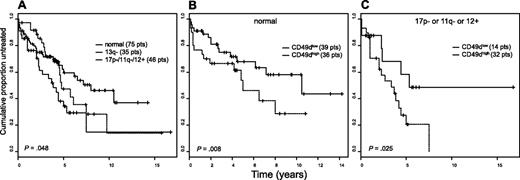
Significantly shorter TTT intervals were observed in patients with prognostically unfavorable chromosomal aberrations, including 17p−, 11q−, and trisomy 12, which altogether identified the poor risk cytogenetic subset (46 patients), compared with normal karyotype patients (75 cases; median TTT 3.8 vs 8.0 years; Figure 4A).23,48,49 As previously shown,23,50 patients with 13q- aberrations (35 cases) showed an intermediate outcome (Figure 4A). To explore the clinical impact of CD49d among different cytogenetic groups, we investigated its expression within the normal karyotype and the poor-risk cytogenetic subsets. As shown in Figure 4B and C, the CD49dhigh phenotype identified a group of patients with shorter TTT both in the context of CLL without chromosomal aberrations (median TTT, 5.0 years vs not reached; P = .008), and within the poor-risk subset (median TTT, 3.3 years vs not reached; P = .025).
Prognostic impact on TTT risk of cytogenetic abnormalities alone and in combination with CD49d. (A) Patients were split according to their karyotype in 3 groups and compared for TTT: normal karyotype (n = 75), presence of the unfavorable genomic aberrations 17p−, 11q−, and trisomy 12 (n = 46), presence of 13q− chromosomal aberration (n = 35). (B,C) Kaplan-Meier curves comparing TTT according to CD49d expression in the group with a normal karyotype (B) and in the group with presence of the unfavorable genomic aberrations (C). The numbers of patients (pts) included in each group are reported in parentheses; the reported P values refer to the log-rank test.
Prognostic impact on TTT risk of cytogenetic abnormalities alone and in combination with CD49d. (A) Patients were split according to their karyotype in 3 groups and compared for TTT: normal karyotype (n = 75), presence of the unfavorable genomic aberrations 17p−, 11q−, and trisomy 12 (n = 46), presence of 13q− chromosomal aberration (n = 35). (B,C) Kaplan-Meier curves comparing TTT according to CD49d expression in the group with a normal karyotype (B) and in the group with presence of the unfavorable genomic aberrations (C). The numbers of patients (pts) included in each group are reported in parentheses; the reported P values refer to the log-rank test.
Multivariate analyses.
CD49d, CD38, ZAP-70, β2M, sCD23, and mod-Rai were included in a proportional hazard regression model of Cox to test their strength as independent prognostic factors for TTT. In a cohort of 206 patients in which all the prognostic information was simultaneously available, CD49d seemed to be the sole independent prognosticator for TTT among the biologic risk factors, along with all the 3 markers of tumor burden (Table 3). On the other hand, if we included the IGHV gene mutational status to the analysis, the multivariate model selected this covariate as independent prognosticator (P = .021) in the context of a cohort of 131 cases (not shown). Because of the differences in the prognostic relevance of CD49d expression in ZAP-70low versus ZAP-70high CLL, or in CLL with M vs UM-IGHV gene status, as emerged by the trend of TTT curves (Figure 3B,C), the possibility of interactions between CD49d expression and these variables was considered and first assessed by bivariate Cox proportional hazards models. According to these analyses, CD49dhigh expression in CLL with ZAP-70high or UM-IGHV gene status was associated with a hazard ratio (HR) for progressive disease of 2.72 (95% confidence interval (CI), 1.21-6.11; P = .015) or 3.14 (95% CI, 1.02-9.71; P = .005), respectively, whereas no additional prognostic information was provided when CD49dhigh expression was tested in the context of CLL expressing a ZAP-70low (HR = 1.36; 95% CI, 0.77-2.41; P = .29), or a M-IGHV phenotype (HR = 1.30; 95% CI, 0.68-2.48; P = .43), or, conversely, ZAP-70high (HR = 1.11; 95% CI, 0.62-1.99; P = .73) or UM-IGHV (HR = 1.21; 95% CI, 0.49-2.96; P = .68) were tested in CD49dlow CLL. Finally, despite the trend of TTT graphs (Figure 3A), no statistically significant interaction was observed between CD49dhigh and CD38high expression (HR = 1.77; 95% CI, 0.59-5.35; P = .31).
Multivariate Cox regression analyses of TTT
| Parameter . | HR (95% CI)* . | P . |
|---|---|---|
| CD49d more than or equal to 30% | 1.74 (1.16-2.62) | .007 |
| β2M more than or equal to 2.2 g/L | 1.67 (1.07-2.60) | .02 |
| sCD23 more than or equal to 70 IU | 2.07 (1.35-3.17) | <.001 |
| Mod-Rai II-III | 2.87 (1.70-4.83) | <.001 |
| Parameter . | HR (95% CI)* . | P . |
|---|---|---|
| CD49d more than or equal to 30% | 1.74 (1.16-2.62) | .007 |
| β2M more than or equal to 2.2 g/L | 1.67 (1.07-2.60) | .02 |
| sCD23 more than or equal to 70 IU | 2.07 (1.35-3.17) | <.001 |
| Mod-Rai II-III | 2.87 (1.70-4.83) | <.001 |
Multivariate Cox regression analyses of TTT were performed by including the following covariates: CD49d, CD38, ZAP-70, β2M, sCD23, and mod-Rai (206 patients). For each variable, the selected cutoff points or the compared categories are indicated.
TTT indicates time to treatment; RA, hazard ratio; CI, confidence interval; other abbreviations are as in Tables 1 and 2.
Based on the final model after stepwise selection of covariates.
To take into account interactions between CD49d and IGHV gene mutations or CD49d and ZAP-70, and to evaluate their independent prognostic value for TTT, these combinations were incorporated in a further multivariate analysis along with all the biologic or tumor load markers tested in our study. As summarized in Table 4, CLLs expressing a CD49dhigh/ZAP-70high or a CD49dhigh/UM-IGHV phenotype were associated with an increased and independent risk of progressive disease (HRs of 3.12 or 2.95, respectively), whereas CLLs expressing CD49dlow with either ZAP-70high or a UM-IGHV mutational status, or, conversely, CLL expressing CD49dhigh with either ZAP-70low or a M-IGHV mutational status, failed to maintain their prognostic relevance. Of the other markers investigated, only 2 tumor load markers (ie, mod-Rai and β2M) maintained an independent relevance as TTT prognosticators (Table 4).
Multivariate Cox regression analyses of TTT with interactions between biological prognostic markers
| Parameter . | HR (95% CI)* . | P . |
|---|---|---|
| ZAP-70 more than 20% and CD49d more than 30% | 3.12 (1.17-8.32) | .023 |
| IGVH UM and CD49d more than 30% | 2.95 (1.46-5.96) | .002 |
| Mod-Rai II-III | 3.36 (1.59-7.13) | .002 |
| β2M more than 2.2 g/L | 2.08 (1.20-3.60) | .009 |
| Parameter . | HR (95% CI)* . | P . |
|---|---|---|
| ZAP-70 more than 20% and CD49d more than 30% | 3.12 (1.17-8.32) | .023 |
| IGVH UM and CD49d more than 30% | 2.95 (1.46-5.96) | .002 |
| Mod-Rai II-III | 3.36 (1.59-7.13) | .002 |
| β2M more than 2.2 g/L | 2.08 (1.20-3.60) | .009 |
Multivariate Cox regression analyses of TTT were performed by including the following covariates: CD49d, CD38, ZAP-70, IGHV, β2M, sCD23, mod-Rai, and interactions between ZAP-70/CD49d and IGHV/CD49d (131 patients). For each variable, the selected cutoff points or the compared categories are indicated.
Based on the final model after stepwise selection of covariates.
Discussion
The aim of the present study was to evaluate, in a large cohort of patients, the clinical value of CD49d as independent prognostic factor for CLL, in relationship with other well-established prognosticators of biologic relevance (CD38, ZAP-70, IGHV mutations, cytogenetic aberrations), or indicators of CLL tumor burden (clinical stage, β2M, sCD23).4,–6,23,40
Expression of CD49d was performed by a direct 3-color immunofluorescence strategy, and data were reported as percent of CD5+CD19+ CLL cells expressing the antigen, in analogy with the current methodologies for CD38 and ZAP-70 detection.4,22,,–25,40 The optimal cut-off for CD49d expression yielding the best separation of CLL patients into 2 subgroups with different prognosis was fixed at 30% of positive cells by applying the most commonly used methods, including the maximally selected log-rank statistics, Youden's index and receiver-operating characteristic analysis.8,30,44 Of note, the great majority of cases of our series either completely lacked expression of CD49d or expressed CD49d at very high levels. This fact, by minimizing the number of cases expressing borderline levels of the molecule, turned out to be a convenient feature making CD49d a more suitable prognosticator compared with other markers whose expression values are usually distributed around the cut-off. For this aspect, CD49d was similar to CD3840 and clearly different from ZAP-70,51 whose expression levels are frequently in the close proximity of the 20% of positive cells standard cut-off (P.B., unpublished observation, April 2007).22,,–25 In the perspective of using CD49d for prognostic purposes, we also tested the interlaboratory reproducibility, the variation in fresh vs frozen samples, and the stability over time of its expression in CLL.23,25,42,43 All these studies concordantly indicate that CD49d is a surface marker whose expression can be easily investigated, is highly reproducible, and is not subjected to variation in fresh vs frozen samples or over time.
CD49d protein expression was an independent risk factor for OS of CLL patients and was necessary to correctly prognosticate the status of active/progressive disease in the context of selected subsets of CLL patients.25,35,52 In particular, a refinement of prognosis for TTT was always observed by combining CD49d with the markers of tumor burden (β2M, sCD23, mod-Rai) or in the context of the 2 main cytogenetic subsets (normal karyotypes or bearing the 17p−/11q−/+12 chromosomal aberrations). Moreover, high CD49d expression levels, although not changing the outcome of good prognosis CLL, as identified by low levels of CD38 and ZAP-70 or by a M-IGHV gene status, turned out to be necessary to correctly identify groups of patients with bad clinical courses in the context of CLL with a ZAP-70high phenotype or a UM-IGHV gene status. In other terms, the more rapid disease progression of ZAP-70high CLL or of UM-IGHV CLL only holds true if associated with high CD49d expression levels. Notably, no statistical differences in terms of TTT were observed by comparing ZAP-70high/CD49dlow CLL or UM-IGHV/ CD49dlow CLL with cases with ZAP-70low or M-IGHV phenotypes, irrespective of CD49d expression. Although not reaching a statistical significance in our series, similar interactions allegedly occurred also between CD49d and CD38, as suggested by the trend of TTT curves and the HR reported in the present study. As shown by our multivariate analyses, CD49d had also an independent prognostic value regarding OS along with other biologic prognosticators, ie, ZAP-70 and IGHV gene status. Therefore, the possibility of interactions between CD49d and these prognosticators also in term of OS was hypothesized and tested by additional multivariate analyses, which, however, lacked statistical significance supposedly because of the low number of events (48 of 303 cases) in our OS series (P.B., unpublished observation, July 2007). The additional clinical value of CD49d, indicated by the present study and suggested in a recent review on CLL prognostic factors,53 has to be validated in more homogeneous and larger datasets of well-characterized CLL patients, eg, prospectively collected by combining the efforts of several Institutions, as well as in independent sets of patients.1
Relationships among prognosticators similar to those described in the present study have been already described between CD38 and ZAP-70,23,54,55 or ZAP-70 and IGHV mutations,23,25 or, more recently, between IGHV mutations and the CLL-specific gene CLLU1.46 These interactions, besides providing a more precise prognostic assessment of patients than that relying on a single marker, may have potential biologic implications. As an example, ZAP-70 and CD38 have been shown to synergize in sustaining the downstream signaling on surface Ig triggering.56,–58 By considering the high correlation between CD38 and CD49d expression in CLL, as shown here and previously,29,30,34 and the known propensity of CD38 to laterally associate with several molecules in the context of membrane lipid rafts,59,60 it is conceivable that specific CD49d/CD38 interactions might also occur in CLL. If so, the resulting molecular complexes might have pathogenetic implications in determining the aggressive clinical course of CD49d-expressing CLLs. Notably, in preliminary experiments of ours, the simultaneous engagement of CD38 and CD49d had a role in the protection of CLL cells against apoptotic stimuli (A.Z., unpublished observation, November 2006).59,61
Prospectively, the present and future studies exploring the role of CD49d in CLL may have also therapeutic implications, envisioning the use for CLL patients of natalizumab (Tysabri; Biogen Idec, Cambridge, MA and Elan Pharmaceuticals, South San Francisco, CA), a humanized anti-CD49d monoclonal antibody already available and currently used in multiple sclerosis.62
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by Ministero della Salute (Ricerca Finalizzata IRCCS and Alleanza Contro il Cancro), Associazione Italiana contro le leucemie linfomi e mielomi (A.I.L.), Venezia Section, Pramaggiore group (fellowship to M.D.B.).
Authorship
Contribution: V.G. coordinated the study and data analyses and wrote the manuscript; P.B. performed data and statistical analyses and contributed to write the manuscript; M.I.D.P. provided clinical data of patients and contributed to data analysis; R.B., M.D.B., and M.D. performed the IGHV gene mutation and contributed to data analyses; A.Z. performed part of immunophenotypical studies, contributed to data analysis, and wrote the manuscript; L.M. and F.B provided clinical data of patients; F.M.R and F.L. performed part of immunophenotypical studies and contributed to data analysis; S.A. organized the clinical activities and data collection; and G.D.P. coordinated the study and data analyses and contributed to write the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Valter Gattei, Clinical and Experimental Onco-Hematology Unit, Centro di Riferimento Oncologico, IRCCS, Via Franco Gallini 2, Aviano (PN), Italy; e-mail: vgattei@cro.it.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal