Abstract
We previously showed that side population (SP) cells, characterized by specific Hoechst dye efflux pattern in flow cytometric analysis, were present in teleost kidney hematopoietic tissue, and that kidney SP cells were enriched in hematopoietic stem cells (HSCs). ABCG2/Abcg2 is an ATP-binding cassette (ABC) transporter that is known to be associated with Hoechst dye efflux activity of mammalian HSCs. In the present study, we examined the expression and function of Abcg2 in kidney SP cells from zebrafish (Danio rerio). Although the zebrafish genome has 4 paralogous copies of ABCG2 (zAbcg2a, b, c, and d), zAbcg2a and zAbcg2c mRNA was expressed in kidney SP cells. Transfection of COS-7 cells with zAbcg2a and zAbcg2c showed that zAbcg2a was directly associated with the SP phenotype. These results indicate that zAbcg2a mRNA is a useful marker for zebrafish HSCs. In situ hybridization in kidney tissue showed that zAbcg2a-positive cells were sporadically localized on the surface of renal tubules, and tightly adhered to renal tubule epithelial cells. This result suggests that teleost HSCs adhere to the surface of renal tubules, and that renal tubule epithelial cells are a key component of HSC niche in teleosts.
Introduction
Side population (SP) cells are identified from bone marrow based on their capacity to efflux of the fluorescent dye Hoechst 33342 (Hoechst), and are enriched in hematopoietic stem cells (HSCs).1 Since the Hoechst dye efflux activity of HSCs is widely conserved in mammals, SP cell purification has been extensively used to enrich mammalian HSCs.2-4 Recently, we showed that the hematopoietic activity of teleost SP cells isolated from kidney, the main hematopoietic organ in teleosts,5,6 using the transplantation model system of clonal ginbuna (Carassius auratus langsdorfii, S3n strain) and ginbuna goldfish (Carassius auratus) hybrids (S4n strain). The SP cell population was clearly identified from ginbuna kidney. When the kidney SP cells from S3n were injected into S4n, all types of donor-derived blood cells (erythrocytes, neutrophils, basophils, monocytes, thrombocytes, and T and B lymphocytes) were detected in the recipient blood over a period of 9 months.7 These findings indicate that SP cell purification can be used to enrich HSCs from not only mammals but also teleosts
Zebrafish (Danio rerio) has recently emerged as a powerful genetic model in which forward genetic screens can uncover novel genes involved in development, hematopoiesis, and human disease.8 Although many homologues of human genes, including hematopoietic genes, have been identified,9,10 stem-cell markers have not been shown in zebrafish. Therefore, little is known about the localization of HSCs and about stem-cell niche, a microenvironment supporting the maintenance and self-renewal of stem cells, in teleost kidney hematopoietic tissue. In murine bone marrow, ABCG2/Abcg2, an ATP-binding cassette (ABC) transporter associated with Hoechst dye efflux activity of HSCs,11-15 is highly expressed in the most primitive HSCs and is sharply down-regulated with differentiation.11 In addition, the expression of Abcg2 gene in stem cells is highly conserved from various sources such as mouse skin, skeletal muscle, mammary grand, and testis, as well as human bone marrow.11,16-18 Abcg2 is thus used as a general marker of stem cells.
In the present study, we wished to determine whether Abcg2 is also marker of teleost HSCs. In zebrafish, 4 paralogous copies of ABCG2 gene (zAbcg2a, b, c, and d) have been already identified in its genome.19 We examined the relationship between Hoechst dye efflux activity and zAbcg2 expression in zebrafish kidney SP cells. We also examined the localization of zAbcg2-positive HSCs in kidney hematopoietic tissue by in situ hybridization.
Methods
Fish maintenance
Wild-type zebrafish (Danio rerio) were obtained from a local pet supplier, maintained in aerated tanks at 28°C and fed commercial pellet food twice daily.
Alignment and phylogenetic analysis
Four types of zebrafish Abcg2 (zAbcg2) sequences were obtained from the National Center for Biotechnology Information (NCBI) database.20 The deduced amino acid sequences of zAbcg2 and human ABCG2 were aligned using ClustalW (European Bioinformatics Institute, Cambridge, United Kingdom). A phylogenetic tree of ABCG subfamily proteins was constructed using the neighbor-joining method and displayed by Tree View (Win16, v.1.40, http://taxomony.xoalogy.gla.ac.uk/rod/rod.html). The GenBank accession numbers of ABCG subfamily genes are as follows: zAbcg2a, NM 001039066; zAbcg2b, NM 001039066; zAbcg2c, NM 001039639; zAbcg2d, NM 001042772; human (Homo sapiens) ABCG2, XM 526633; mouse (Mus musculus) Abcg2, BC053730; dog (Canis familiaris) Abcg2, DQ222459; cow (Bos taurus) Abcg2, AJ871176; chicken (Gallus gallus) Abcg2, XM 421638; Xenopus (Xenopus tropicalis) Abcg2, NM 001045762; pufferfish (Tetraodon nigroviridis) Abcg2, CAAE01014665; human ABCG1, NM 004915; mouse Abcg1, NM 009593; dog Abcg1, XM 544902; human ABCG4, NM 022169; mouse Abcg4, AF425077; dog Abcg4, XM 848138; zebrafish Abcg4, XM 682593; mouse Abcg5, NM 031884; human ABCG8, BC113657; mouse Abcg8, AY196216; and zebrafish Abcg8, XM 679554.
Cell preparation
Hematopoietic cells were obtained from zebrafish body kidney as previously described.5 Briefly, zebrafish were anesthetized with 0.01% benzocaine (Sigma, St Louis, MO), and were killed by decapitation before kidney collection. Body kidney was dissected, and hematopoietic cells were obtained by macerating the kidney on the stainless steel mesh with forceps in 5 mL ice-cold Hanks balanced salt solution (HBSS) containing 2% fetal bovine serum (FBS). Cells from body kidney were pelleted by centrifugation. After discarding the supernatant, the pellet was gently mixed with 1 mL distilled water by pipetting to lyse the erythrocytes by osmotic shock. Subsequently, 1 mL 2 × HBSS was added and the cells were washed twice with HBSS by centrifugation. Cells were resuspended at a density of 106 cells/mL in HBSS.
Hoechst staining and flow cytometry
Kidney hematopoietic cells from 6 to 20 zebrafish were separately stained with Hoechst 33342 (Hoechst; Molecular Probes, Eugene, OR) according to the procedure for ginbuna kidney hematopoietic cells with some modification.7 After preliminary staining with 1, 3, 5, 7.5, and 10 μg/mL Hoechst for 60, 90, and 120 minutes at 25°C, a staining procedure with 3 μg/mL Hoechst for 90 minutes was used in the majority of experiments. For inhibition experiments, hematopoietic cells were stained with Hoechst in the presence of 250 μM verapamil (Sigma), an inhibitor of ABC transporters. After staining, cells were washed by centrifugation, adjusted to 107 cells/mL in HBSS, and kept on ice until use. Just before flow cytometric (FCM) analysis, propidium iodide (Molecular Probes) solution was added at a final concentration of 2 μg/mL to identify nonviable cells. FCM analysis and sorting were performed on a dual laser (488 nm and UV) flow cytometer (EPICS ALTRA; Beckman Coulter, Fullerton, CA) as previously described.7 Cells were sorted into 100% FBS, and were used for morphologic analysis. FlowJo software (version 4.3.2; TreeStar, Ashland, OR) was used for the analysis of FCM data.
RT-PCR analysis
For expression analysis of zAbcg2 genes in various tissues, head kidney, body kidney, spleen, liver, intestine, and gill of 3 zebrafish were pooled by tissue type, and total RNA was extracted using a RNeasy Mini Kit (Qiagen, Valencia, CA). Total RNA (500 ng) from various tissues was reverse transcribed into cDNA using TaqMan Reverse Transcription Reagents with Random Hexamer (Applied Biosystems, Foster City, CA). Expression analysis by reverse-transcription–polymerase chain reaction (RT-PCR) with the specific primer sets was performed using BIOTAQ (Bioline, London, United Kingdom). The primer sequences are in Table 1. PCR condition of each gene was as follows: one cycle of 94°C for 2 minutes, 23 to 35 cycles of 94°C for 15 seconds, 58°C for 15 seconds, and 72°C for 60 seconds, and finally 72°C for 2 minutes.
Primer nucleotide sequences
| Primer name . | Forward primers (5′ → 3′) . | Reverse primers (5′ → 3′) . |
|---|---|---|
| zAbcg2a | ATCGCAGAGAGGAAACTGTTTGTACATG | TCATGTGAACTTCTTAATGAAGCGCAG |
| zAbcg2b | CATTAATGAGCGAGCAATTTTTGTGCATG | CCTGCGTAACTGTATGTATGCAAATGTC |
| zAbcg2c | AGCGAGAGAGAGCTCTTCATCCAT | GACACAACTGCACATACGCCAGAA |
| zAbcg2d | GCATACAAAACAGGCGAGTTGATCAC | CTACATGAGGACGTGTGCAGCAG |
| EF1α | ATGTGGCTGGAGACAGCAAGAAC | CTTTGTGACCTTGCCAGCACCA |
| β-actin* | CCTGACTGACTACCTCATGAAGATC | ATCCACATCTGCTGGAAGGTGGA |
| Primer name . | Forward primers (5′ → 3′) . | Reverse primers (5′ → 3′) . |
|---|---|---|
| zAbcg2a | ATCGCAGAGAGGAAACTGTTTGTACATG | TCATGTGAACTTCTTAATGAAGCGCAG |
| zAbcg2b | CATTAATGAGCGAGCAATTTTTGTGCATG | CCTGCGTAACTGTATGTATGCAAATGTC |
| zAbcg2c | AGCGAGAGAGAGCTCTTCATCCAT | GACACAACTGCACATACGCCAGAA |
| zAbcg2d | GCATACAAAACAGGCGAGTTGATCAC | CTACATGAGGACGTGTGCAGCAG |
| EF1α | ATGTGGCTGGAGACAGCAAGAAC | CTTTGTGACCTTGCCAGCACCA |
| β-actin* | CCTGACTGACTACCTCATGAAGATC | ATCCACATCTGCTGGAAGGTGGA |
The sequences from vervet monkey (Cercopithecus aethiops) (Accession number: AB004047).
For expression analysis in sorted cells, sorted cells were pooled by cell type in lysis buffer, and total RNA were extracted using Absolutely RNA Nanoprep Kit (Stratagene, La Jolla, CA). Total RNA from 5 × 103 cells of each subset was amplified using MessageAmp II aRNA Amplification Kit (Applied Biosystems). Reverse transcription and RT-PCR were performed with amplified RNA as described in the previous paragraph.
Transient expression
The HindIII and BamHI linker-linked insert cDNA fragments of zAbcg2a or zAbcg2c were digested with HindIII and BamHI, and ligated into pcDNA3 vectors (Invitrogen, Carlsbad, CA). The pcDNA3 vector containing ginbuna CD8α (gCD8α) was kindly supplied by Dr T. Somamoto (Kyushu University, Fukuoka, Japan) and used as a control. COS-7 cells were transiently transfected with 6 μg of the above constructs in 25-cm2 plastic tissue culture flasks using FuGENE HD Transfection Reagent (Roche Molecular Biochemicals, Mannheim, Germany). Transfected COS-7 cells were grown for 2 days and then harvested with PBS containing 0.01% trypsin and 0.02% EDTA. These cells were washed by centrifugation, resuspended in HBSS, and stained with 10 μg/mL Hoechst for 90 minutes at 37°C. FCM analysis, inhibition experiment, and RT-PCR analysis in these COS-7 cells were performed as described above.
In situ hybridization
The zAbcg2a PCR product that amplified with cDNA from zebrafish intestine using the same primer of expression analysis was ligated into the pCR II TOPO vector (Invitrogen). These plasmids were linearized with NotI or BamHI for the antisense or sense probes, respectively. RNA labeled with digoxigenin (DIG) was transcribed from these linearized plasmids as recommended by the manufacturer (Dig RNA Labeling Kit; Roche Molecular Biochemicals). The body kidney was dissected from zebrafish and fixed in 4% paraformaldehyde (PFA) for 24 hours at 4°C. After being washed in PBS-T, the tissue was stored in methanol at −20°C until use. The tissue was then dehydrated sequentially in ethanol and xylene and embedded in paraffin. Sections of 3 μm in thickness were prepared and mounted onto MAS-coated slides (Matsunami, Osaka, Japan). Tissue sections were deparaffinized, rehydrated, and washed twice in PBS for 5 minutes. Hybridization was performed using DIG-labeled oligonucleotide antisense or sense probes in hybridization solution (5 × SSC, 50% deionized formamide, 0.25 mg/mL Baker yeast tRNA, 0.5 mg/mL fish sperm DNA, 1 × Denhardt solution) for 16 hours at 42°C in a humidified chamber. Following hybridization, the sections were washed twice in 2× SSC for 15 minutes at 42°C, twice in 1 × SSC for 15 minutes at 42°C, and twice in PBS for 5 minutes at room temperature. After incubation of the sections with blocking solution (1% sheep serum, 0.1% Triton X-100) for 30 minutes, they were incubated overnight at 4°C with antidigoxigenin-AP conjugated antibody (Roche Molecular Biochemicals) at a dilution (in blocking solution) of 1:500. The color reaction was developed over 6 to 12 hours in the dark at room temperature using nitroblue tetrazolium chloride and 5-bromo-4-chloro-3-indolyl-phosphate (NBT/BCIP). Visible light imaging was performed on Olympus BX-51 microscope using 40× oil objective lens and Olympus DP20 digital camera and software (Olympus, Melville, NY).
Results
Characterization of 4 types of zebrafish Abcg2 (zAgbg2)
Four types of zebrafish Abcg2 (zAbcg2a, b, c, and d) sequences were obtained from the NCBI database. The deduced amino acid sequences of zAbcg2a, b, c, d, and human ABCG2 were aligned using ClustalW software (Figure 1). High amino acid identities were observed in ATP-binding cassette region, which contains 3 motifs, Walker A, Walker B, and ABC signature. Comparison of the deduced amino acid sequences showed that, of the 4 zAbcg2 types, zAbcg2a showed the highest amino acid identity with other vertebrate Abcg2 (Table 2). To further examine the relationship between zAbcg2 and other vertebrate Abcg2, a phylogenetic tree was constructed using neighbor-joining method (Figure 2). Although each of the zAbcg2 clustered with other vertebrate Abcg2, zAbcg2a and zAbcg2d were more closely related to other vertebrate Abcg2 than zAbcg2b and zAbcg2c. An RT-PCR analysis in zebrafish tissues showed that zAbcg2a and zAbcg2b were strongly expressed in intestine, zAbcg2c was strongly expressed in all tissue examined except liver, and zAbcg2d was not detected in any of the tissues examined (Figure 3).
Multiple alignment of the deduced amino acid sequences of zebrafish Abcg2 (zAbcg2) and human ABCG2 (hABCG2). The multiple alignment was produced using ClustalW. * indicates positions that have a single, fully conserved residue; colons and periods, positions that have strong and weak similarity, respectively; dashes, gaps; the solid line under the alignments, the ATP-binding cassette region; and shaded boxes, the 3 motifs, Walker A, Walker B, and ABC signature.
Multiple alignment of the deduced amino acid sequences of zebrafish Abcg2 (zAbcg2) and human ABCG2 (hABCG2). The multiple alignment was produced using ClustalW. * indicates positions that have a single, fully conserved residue; colons and periods, positions that have strong and weak similarity, respectively; dashes, gaps; the solid line under the alignments, the ATP-binding cassette region; and shaded boxes, the 3 motifs, Walker A, Walker B, and ABC signature.
Amino acid sequence identity of zebrafish Abcg2 (zAbcg2)
| Identity (%) . | ||||
|---|---|---|---|---|
| Species | zAbcg2a | zAbcg2b | zAbcg2c | zAbcg2d |
| Human | 60.4 | 44.1 | 45.1 | 48.1 |
| Mouse | 58.1 | 42.7 | 43.9 | 50 |
| Chicken | 57.9 | 41.2 | 50 | 45.6 |
| Xenopus | 60.8 | 43.8 | 42.6 | 47.9 |
| Pufferfish | 66.4 | 42.8 | 44.2 | 55.8 |
| Identity (%) . | ||||
|---|---|---|---|---|
| Species | zAbcg2a | zAbcg2b | zAbcg2c | zAbcg2d |
| Human | 60.4 | 44.1 | 45.1 | 48.1 |
| Mouse | 58.1 | 42.7 | 43.9 | 50 |
| Chicken | 57.9 | 41.2 | 50 | 45.6 |
| Xenopus | 60.8 | 43.8 | 42.6 | 47.9 |
| Pufferfish | 66.4 | 42.8 | 44.2 | 55.8 |
Human, Homo sapiens; Mouse, Mus musculus; Chicken, Gallus gallus; Xenopus, Xenopus tropicalis; Pufferfish, Tetraodon nigroviridis.
Phylogenetic analysis of ABCG subfamily proteins. The phylogram was constructed by the neighbor-joining method, based on the amino acid alignment (ClustalW) of full-length Abcg2 proteins from human (Hosa-ABCG2), mouse (Mumu-Abcg2), dog (Cafa-Abcg2), cow (Bota-Abcg2), chicken (Gaga-Abcg2), pufferfish (Teni-Abcg2), and zebrafish (zAbcg2a, b, c, and d). For comparison, the Abcg1 proteins from human, mouse, and dog; Abcg4 proteins from human, mouse, dog, and zebrafish; Abcg5 proteins from mouse; and Abcg8 proteins from human, mouse, and zebrafish are also shown. The Abcg5 protein from mouse was used as an out-group. The numbers at the relevant branches refer to bootstrap values of 1000 replications.
Phylogenetic analysis of ABCG subfamily proteins. The phylogram was constructed by the neighbor-joining method, based on the amino acid alignment (ClustalW) of full-length Abcg2 proteins from human (Hosa-ABCG2), mouse (Mumu-Abcg2), dog (Cafa-Abcg2), cow (Bota-Abcg2), chicken (Gaga-Abcg2), pufferfish (Teni-Abcg2), and zebrafish (zAbcg2a, b, c, and d). For comparison, the Abcg1 proteins from human, mouse, and dog; Abcg4 proteins from human, mouse, dog, and zebrafish; Abcg5 proteins from mouse; and Abcg8 proteins from human, mouse, and zebrafish are also shown. The Abcg5 protein from mouse was used as an out-group. The numbers at the relevant branches refer to bootstrap values of 1000 replications.
Expression analysis of zAbcg2a, b, c, and d in zebrafish tissues. RT-PCR was performed with total RNA from various tissues (ie, head kidney, body kidney, spleen, liver, intestine, and gill) using specific primers for zAbcg2a, b, c, d, and zebrafish EF1α. EF1α was used as an internal control. NC indicates negative control.
Expression analysis of zAbcg2a, b, c, and d in zebrafish tissues. RT-PCR was performed with total RNA from various tissues (ie, head kidney, body kidney, spleen, liver, intestine, and gill) using specific primers for zAbcg2a, b, c, d, and zebrafish EF1α. EF1α was used as an internal control. NC indicates negative control.
Isolation of zebrafish kidney SP cells
To isolate SP cells from zebrafish kidney, hematopoietic cells obtained from the body kidney were stained with Hoechst and analyzed by FCM. Typical results of FCM analysis are shown in Figure 4. When the hematopoietic cells were displayed in a Hoechst red versus Hoechst blue pseudocolor plot, SP cells, which show a high level of dye efflux activity, were readily identified as a small streak of cells with low Hoechst red/blue fluorescence (Figure 4A). The percentage of SP cells in zebrafish body kidney was 0.056% ± 0.008% (n = 6).
Flow cytometric analysis of zebrafish kidney hematopoietic cells. Hematopoietic cells from zebrafish body kidney were stained with Hoechst 33342 (Hoechst) and analyzed by flow cytometry. (A) Hoechst fluorescence (Hoechst blue vs Hoechst red) of kidney hematopoietic cells is shown. Gated region indicates SP population. (B) Hoechst fluorescence of kidney hematopoietic cells that were stained with Hoechst in the presence of 250 μM verapamil is shown. (C) Scatter profile of SP cells is shown. Lymphoid indicates the FSlow, SSlow fraction; myeloid, the FShigh fraction. (D) Morphologic analyses of lymphoid-SP (Lym-SP) (i) and myeloid-SP (Mye-SP) (ii) are shown (May-Grünwald Giemsa, scale bar = 5 μm).
Flow cytometric analysis of zebrafish kidney hematopoietic cells. Hematopoietic cells from zebrafish body kidney were stained with Hoechst 33342 (Hoechst) and analyzed by flow cytometry. (A) Hoechst fluorescence (Hoechst blue vs Hoechst red) of kidney hematopoietic cells is shown. Gated region indicates SP population. (B) Hoechst fluorescence of kidney hematopoietic cells that were stained with Hoechst in the presence of 250 μM verapamil is shown. (C) Scatter profile of SP cells is shown. Lymphoid indicates the FSlow, SSlow fraction; myeloid, the FShigh fraction. (D) Morphologic analyses of lymphoid-SP (Lym-SP) (i) and myeloid-SP (Mye-SP) (ii) are shown (May-Grünwald Giemsa, scale bar = 5 μm).
To investigate whether low Hoechst fluorescence of SP cells is mediated by ABC transporters, hematopoietic cells were stained with Hoechst in the presence of verapamil, an inhibitor of ABC transporters. As shown in Figure 4B, the percentage of SP cells was significantly decreased by verapamil treatment (0.008% ± 0.004%, n = 6, P < .01), indicating that the low Hoechst fluorescence of SP cells reflects the activity of ABC transporters.
To further characterize the properties of SP cells, we examined the scatter profile and morphology of SP cells. As shown in Figure 4C, 56% of SP cells fell into the lymphoid fraction (FSlow, SSlow), while the rest was in the myeloid fraction (FShigh). These lymphoid SP (Lym-SP) and myeloid SP (Mye-SP) cells were sorted and morphologically examined (Figure 4D). Almost all the Lym-SP cells had a thin-layered cytoplasm and round nucleus (Figure 4Di), which are typical features of HSCs.21,22 In contrast, Mye-SP cells were heterogeneous, consisting of both mature and immature myeloid cells (Figure 4Dii).
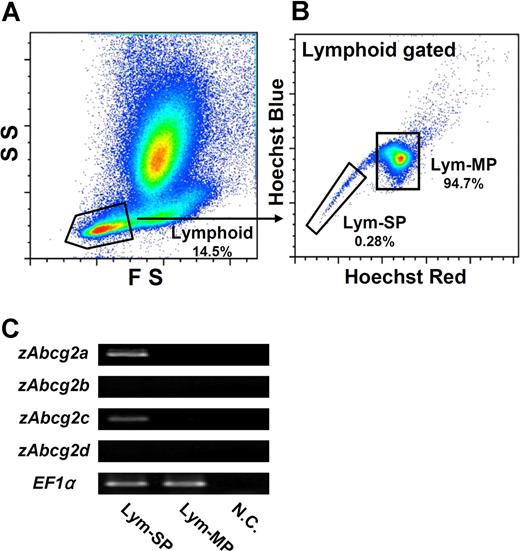
Expression analysis of zAbcg2 in Lym-SP
Because Lym-SP cells showed the typical features of HSCs, we next examined which zAbcg2 genes are expressed in Lym-SP cells. To compare the expression levels of zAbcg2 genes between SP and MP (main population, non-SP) cells, the lymphoid cells, which are in a FSlow, SSlow subset (Figure 5A), were subdivided into 2 populations, Lym-SP and Lym-MP, by Hoechst fluorescence intensity (Figure 5B). These cells were sorted and examined by RT-PCR (Figure 5C). The expressions of zAbcg2a and zAbcg2c in the Lym-SP cells were higher than those in Lym-MP cells. In contrast, zAbcg2b and zAbcg2d were not expressed in either Lym-SP or Lym-MP cells. These results suggest that zAbcg2a and/or zAbcg2c are associated with the Hoechst dye efflux activity of Lym-SP cells.
Expression analysis of zAbcg2 genes in Lym-SP and Lym-MP cells. (A) Scatter profile of kidney hematopoietic cells is shown. Gated region indicates lymphoid cells (FSlow, SSlow). (B) Hoechst fluorescence of lymphoid cells is shown. The lymphoid cells were subdivided into 2 populations, Lym-SP and Lym-MP. (C) The results of RT-PCR analysis of zAbcg2 genes in Lym-SP and Lym-MP are shown. EF1α was used as an internal control. NC indicates negative control.
Expression analysis of zAbcg2 genes in Lym-SP and Lym-MP cells. (A) Scatter profile of kidney hematopoietic cells is shown. Gated region indicates lymphoid cells (FSlow, SSlow). (B) Hoechst fluorescence of lymphoid cells is shown. The lymphoid cells were subdivided into 2 populations, Lym-SP and Lym-MP. (C) The results of RT-PCR analysis of zAbcg2 genes in Lym-SP and Lym-MP are shown. EF1α was used as an internal control. NC indicates negative control.
Transfection of COS-7 cell lines with zAbcg2
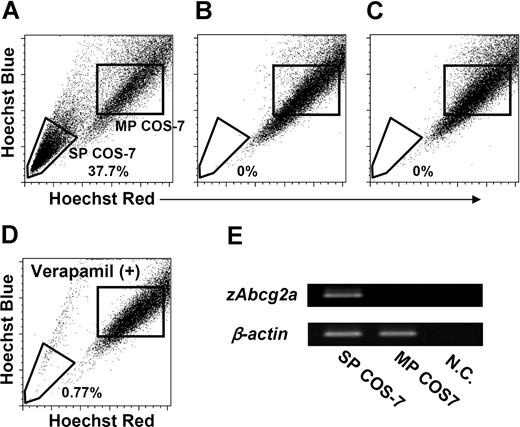
To confirm that zAbcg2a and zAbcg2c have the capacity to expel Hoechst dye, COS-7 cells were transiently transfected with the pcDNA3 vectors containing zAbcg2a, zAbcg2c, or ginbuna CD8α (gCD8α) as a control. These transfected cells, zAbcg2a/COS-7, zAbcg2c/COS-7, and gCD8α/COS-7, were used for the expression analysis of zAbcg2a, zAbcg2c, and gCD8α mRNA, respectively (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Thereafter, the COS-7 cells that expressed zAbcg2a, zAbcg2c, or gCD8α mRNA were stained with Hoechst and analyzed by FCM (Figure 6A-C). A large proportion (37.7%) of zAbcg2a/COS-7 cells showed low Hoechst fluorescence (Figure 6A). This population is referred to as SP COS-7 cells. In contrast, SP COS-7 cells were not detected in zAbcg2c/COS-7 or gCD8α/COS-7 (Figure 6B,C).
Transfection analysis of zAbcg2. COS-7 cells were transiently transfected with the pcDNA3 vectors containing zAbcg2a, zAbcg2c, or ginbuna CD8α (gCD8α) as a control, and these cells, zAbcg2a/COS-7, zAbcg2c/COS-7, and gCD8α/COS-7, were stained with Hoechst 33342 (Hoechst) and analyzed by flow cytometry. The zAbcg2a/COS-7 (A), zAbcg2c/COS-7 (B), and gCD8α/COS-7 (C) cells are displayed in Hoechst red versus Hoechst blue plot. (D) Hoechst fluorescence of zAbcg2a/COS-7 stained with Hoechst in the presence of 250 μM verapamil is shown. (E) The results of RT-PCR analysis of zAbcg2a in SP COS-7 and MP COS-7 cells from zAbcg2a/COS-7 cells are shown. The β-actin from vervet monkey (Cercopithecus aethiops) was used as an internal control. NC indicates negative control.
Transfection analysis of zAbcg2. COS-7 cells were transiently transfected with the pcDNA3 vectors containing zAbcg2a, zAbcg2c, or ginbuna CD8α (gCD8α) as a control, and these cells, zAbcg2a/COS-7, zAbcg2c/COS-7, and gCD8α/COS-7, were stained with Hoechst 33342 (Hoechst) and analyzed by flow cytometry. The zAbcg2a/COS-7 (A), zAbcg2c/COS-7 (B), and gCD8α/COS-7 (C) cells are displayed in Hoechst red versus Hoechst blue plot. (D) Hoechst fluorescence of zAbcg2a/COS-7 stained with Hoechst in the presence of 250 μM verapamil is shown. (E) The results of RT-PCR analysis of zAbcg2a in SP COS-7 and MP COS-7 cells from zAbcg2a/COS-7 cells are shown. The β-actin from vervet monkey (Cercopithecus aethiops) was used as an internal control. NC indicates negative control.
We investigated whether the SP COS-7 population in zAbcg2a/COS-7 is eliminated by verapamil treatment as has been demonstrated for zebrafish kidney SP cells. As shown in Figure 6D, the percentage of SP COS-7 in zAbcg2a/COS-7 was decreased by verapamil treatment (0.8%). We further examined the expression level of zAbcg2a mRNA in SP COS-7 and main population (MP) COS-7 cells. As shown in Figure 6E, relatively high level of zAbcg2a mRNA was detected in SP COS-7 cells compared with MP COS-7 cells. These results indicate that zAbcg2a has the capacity to efflux the Hoechst dye, and is directly associated with the SP phenotype in zebrafish.
Localization of zAbcg2a-positive cells in body kidney
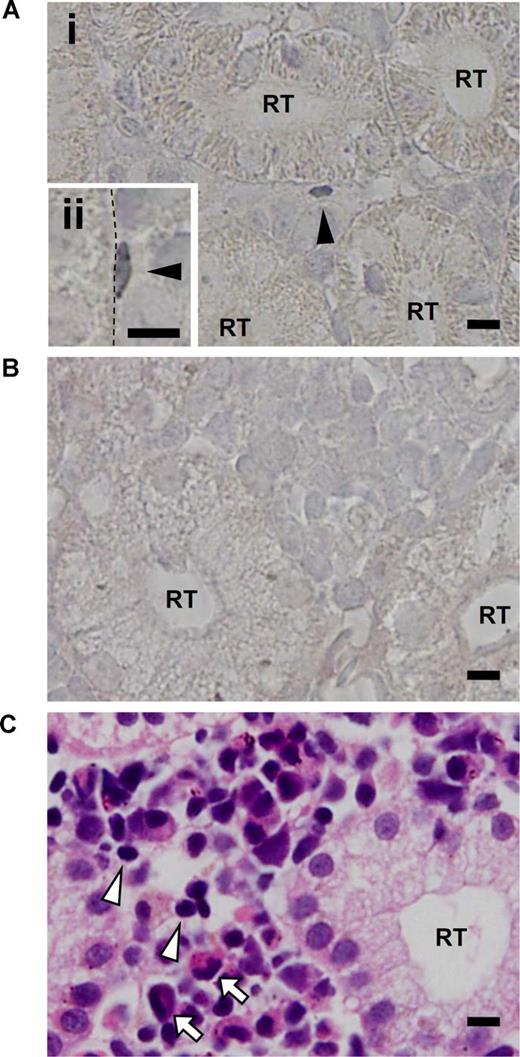
Based on the correlation between the SP phenotype and zAbcg2a expression, we used the zAbcg2a mRNA as a marker for SP cells, and examined the expression pattern of zAbcg2a in kidney by in situ hybridization. In zebrafish kidney, zAbcg2a mRNA expression was detected by antisense RNA probe in kidney interrenal cells, which contain hematopoietic cells, and was not detected in renal tubules and glomeruli, while zAbcg2a expression was not detected by the sense probe (Figure 7A,B). We found that 68.1% of zAbcg2a-positive cells were sporadically localized on the surface of renal tubules (Figure 7Ai arrowhead). Based on their size, zAbcg2a-positive cells present on the renal tubule surface were more similar to lymphoid cells (Figure 7C arrowheads) than to myeloid cells (Figure 7C arrows). In addition, some of the zAbcg2a-positive cells displayed a flattened morphology and tightly adhered to the renal tubule epithelial cells (Figure 7Aii arrowhead). These results indicate that zAbcg2a-expressing Lym-SP cells adhere to the renal tubule surface, and suggest that renal tubule epithelial cells play a role in supporting HSCs as a stem cell niche in zebrafish kidney.
Results of in situ hybridization of zAbcg2a in zebrafish kidney. (A) Tissue section in situ hybridizations with antisense RNA probe for zAbcg2a on body kidney are shown. Arrowheads indicate zAbcg2a-positive cells. Dashed line indicates the surface of renal tubule. (B) Tissue section in situ hybridization with sense RNA probe for zAbcg2a on body kidney is shown. (C) Body kidney section stained with hematoxylin eosin (H-E) is shown. Arrowheads and arrows indicate lymphoid cells and myeloid cells, respectively. All scale bars indicate 5 μm. RT indicates renal tubule.
Results of in situ hybridization of zAbcg2a in zebrafish kidney. (A) Tissue section in situ hybridizations with antisense RNA probe for zAbcg2a on body kidney are shown. Arrowheads indicate zAbcg2a-positive cells. Dashed line indicates the surface of renal tubule. (B) Tissue section in situ hybridization with sense RNA probe for zAbcg2a on body kidney is shown. (C) Body kidney section stained with hematoxylin eosin (H-E) is shown. Arrowheads and arrows indicate lymphoid cells and myeloid cells, respectively. All scale bars indicate 5 μm. RT indicates renal tubule.
Discussion
In the present study, we identified a zebrafish HSC marker, zAbcg2a, and used it to examine the localization of HSCs in kidney hematopoietic tissue. Our data showed that zAbcg2a-positive HSCs were localized on the surface of renal tubules and tightly adhered to the renal tubule epithelial cells. These results suggest that the renal tubule epithelial cells have an important role in supporting HSCs as a stem cell niche. This is the first report of the HSC niche in nonmammalian vertebrate hematopoietic tissue.
The ABCG2 gene was originally identified as a drug-resistance gene in human tumor cell lines.23,24 This gene is also expressed in a variety of normal tissues such as placenta, brain, and intestine.23,25 Although the normal physiologic role is unclear, ABCG2 is associated with resistance to mitoxantrone, a chemotherapeutic drug that intercalates into DNA and induces mutagenesis, in tumor cell lines23,24,26,27 and HSCs.28 Cellular detoxification is an important physiologic role of ABCG2,29,30 suggesting that ABCG2 protects the HSC genome from exposure to environmental toxins. On the other hand, overexpression of human ABCG2 in murine bone marrow cells significantly blocked hematopoietic development,11 leading to speculation that ABCG2 also plays a role in early stem-cell self-renewal by blocking differentiation. In this report, we showed that the same transporter was expressed in teleost HSCs and similarly conferred Hoechst dye efflux activity. The conserved expressions of ABCG2 among vertebrate HSCs may reflect an important role in self-protecting and/or self-renewal of HSCs.
In various fish species, some genes appear to be present in multiple copies, possibly as a result of gene duplication.31-33 These duplicated genes generally retain their original functions, whereas some copies may acquire new functions or lose their functions.19,34 Since the 4 copies of Abcg2 genes are present in the zebrafish genome, we examined which gene is similar to the human ABCG2 gene with regard to amino acid sequences and expression patterns. Although zAbcg2a showed the highest amino acid identity with human ABCG2, zAbcg2a formed a cluster with zAbcg2d in phylogenetic analysis, and these 2 genes were more closely related to human ABCG2 than zAbcg2b and zAbcg2c. We determined the genomic location for the zAbcg2 genes to examine whether a synteny occurred between zAbcg2 and human ABCG2 genomic loci. The zAbcg2d has flanking genes that are clear orthologs of human genes mapping close to the human ABCG2 genes (Figure S2). These results suggest that zAbcg2d is the true ortholog of human ABCG2, and that zAbcg2a has been duplicated from zAbcg2d after the divergence from the tetrapod lineage. However, the expression analysis of zAbcg2 mRNA in zebrafish tissues showed that zAbcg2a was strongly expressed in intestine as has been described for human ABCG2,23,25 but zAbcg2d was not expressed in any tissues examined. These observations suggest that zAbcg2a, rather than zAbcg2d, functions as an ABCG2.
SP cells in murine bone marrow are highly homogeneous with respect to cell-surface markers found on HSCs and are enriched in long-term repopulating cells.1,2 Furthermore, SP cells are present in the bone marrow of all mammalian species examined.2-4 Although the expression of stem-cell markers has not been shown in ginbuna kidney SP cells, the evidence that ginbuna kidney SP cells have the ability to perform long-term and multilineage repopulation7 led us to use the kidney SP cells for the purification of HSCs from other fish species. The cyprinid fish have 2 main hematopoietic organs, head (pronephric) kidney and body (mesonephric, trunk) kidney.5,35 Our previous study7 showed that SP cells were abundantly present in body kidneys compared with head kidneys in ginbuna and common carp (Cyprinus carpio). Therefore, in this report, we focused on the body kidney to identify zebrafish SP cells. Furthermore, we used only female zebrafish to isolate SP cells, because, in many cases, hematopoietic cells isolated from male zebrafish body kidney were contaminated with the testicular cells that were clearly identified as a Hoechst very low population, which is observed only in male body kidney and testis, but not in female body kidney (Figure S3). We found that Lym-SP cells in zebrafish kidney had a very similar appearance to ginbuna kidney SP cells and murine bone marrow SP cells, which are uniform round cells of small size with a very thin rim of cytoplasm and round nucleus.7,21,22 These Lym-SP cells were eliminated by verapamil treatment (data not shown), indicating that the low Hoechst fluorescence of these cells was mediated by ABC transporters. Thus, Lym-SP cells showed the typical features of HSCs, and we used Lym-SP as a HSC population in zebrafish.
Zhou et al reported that although several ABC transporters (Abcb1, Abcc1, Abcc3, Abcc4, and Abcg2) are expressed in murine bone marrow SP cells, only Abcg2 was directly associated with Hoechst dye efflux activity of SP cells.11 Based on this observation, we examined the correlation between Hoechst dye efflux activity and zAbcg2 expression in Lym-SP cells. Our results showed that although zAbcg2a and zAbcg2c were expressed in Lym-SP, only zAbcg2a had the capacity to efflux Hoechst dye. These results indicate that the Hoechst dye efflux activity of zebrafish kidney SP cells is attributed to the expression of zAbcg2a, and that zAbcg2a mRNA is a useful marker for zebrafish HSCs. However, it is unclear why zAbcg2c was not able to expel Hoechst dye. In phylogenetic analysis, zAbcg2a and zAbcg2c formed distinct clades. In addition, the expression patterns of zAbcg2a and zAbcg2c in zebrafish tissues were different, suggesting that zAbcg2c has a different function from zAbcg2a.
The regulation of self-renewal and differentiation of HSCs requires a specific microenvironment of surrounding cells known as a stem-cell niche.36,37 Recently, 2 groups reported that a subpopulation of osteoblasts was a definitive regulatory component of the HSC niche in murine bone marrow.38,39 Arai et al showed that Tie2+ HSCs that were quiescent and had antiapoptotic properties were equivalent to SP cells and adherent to osteoblasts at the surface of trabecular bone in mouse.40 These observations suggest that HSCs in a niche show the SP phenotype, and that these cells can be detected by the expression of Abcg2. In zebrafish kidney, approximately 70% of zAbcg2a-positive cells, which were probably equivalent to Lym-SP cells, were localized on the surface of renal tubules, and interestingly, some of these zAbcg2a-positive cells displayed a flattened morphology and tightly adhered to the renal tubule epithelial cells. The tight adherence of these cells is in agreement with a previous report on the adherence of Tie2+ HSCs in murine bone marrow.40 Based on these observations, we propose the hypothesis that zAbcg2a-positive HSCs adhere to the surface of renal tubules, and renal tubule epithelial cells may be a key component of the HSC niche in teleost kidney. There is additional evidence in support of this hypothesis. The ginbuna kidney SP cells labeled with carboxyfluorescein diacetate succinimidyl ester (CFSE) were injected into syngenic recipients. One day after transplantation, CFSE-labeled SP cells were detected in recipient body kidneys. We found that 46% of these cells detected in recipient body kidneys were localized on the surface of renal tubules (Figure S4), suggesting that HSCs home to the stem-cell niches. Furthermore, our previous report7 showed that lower percentages of SP cells were observed in the head kidneys of ginbuna and carp, which have fewer renal tubules and glomeruli than the body kidneys.41,42 Thus, there is a close relation between HSCs and renal tubules in teleosts. However, the molecular mechanisms underlying the interaction between HSCs and renal tubules remain elusive.
The zebrafish has proven utility as a genetically tractable vertebrate organism, and many transgenic and mutant fish have been used to study hematopoiesis.43-45 Although the key regulatory genes responsible for generating hematopoietic cells or lineage-committed cells have been analyzed in zebrafish,46 genetic analyses of primitive HSCs have not been well studied. The zebrafish kidney SP cells are particularly suitable for the study of HSC biology, and can be used to identify genes that regulate self-renewal or differentiation of HSCs as well as to study the interactions between HSCs and stem-cell niches.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr T. Somamoto (Kyushu University, Japan) for providing the pcDNA3 vector containing gCD8α.
National Institutes of Health
Authorship
Contribution: I.K. designed and performed research and wrote the paper; K.S. performed research; T.M. and T.N. designed research; and K.A. and F.T. contributed analytical tools.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Tadaaki Moritomo, Department of Veterinary Medicine, Nihon University, Kameino 1866, Fujisawa, Kanagawa, Japan; e-mail: moritomo@brs.nihon-u.ac.jp.