Abstract
Early tumor detection and intervention are important determinants of survival in patients with cancer. We have recently reported that the “platelet angiogenesis proteome” may be used to detect microscopic tumors in mice. We now present evidence that changes in platelet-associated platelet factor-4 (PF-4) detect malignant growth across a spectrum of human cancers in mice. A deregulated expression of an 8206-Da protein was observed by surfaceenhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-ToF MS) proteomic comparison of platelets from normal and tumor-bearing mice. The differentially expressed protein was identified as PF-4 by tandem mass spectrometry and ProteinChip immunoassay using anti–PF-4 antibody. The platelet-associated PF-4 appeared to be up-regulated in early growth of human liposarcoma, mammary adenocarcinoma, and osteosarcoma. A 120-day follow-up study of liposarcoma revealed a sustained 2-fold or higher increase of platelet-associated PF-4 at 19, 30, and 120 days. In contrast, only an insignificant change of PF-4 was observed in the plasma of mice bearing the different human tumor xenografts, and throughout the 120 days of the liposarcoma study. We conclude that platelet-associated PF-4, but not its plasma counterpart, may represent a potential biomarker of early tumor presence.
Introduction
The identification of biomarkers of early tumor recurrence, growth, and therapeutic response has been of great interest in oncology. Considerable effort is currently focused on methods for early tumor detection, including those involving detection of specific proteins or proteomic profiles in the serum,1-4 DNA in stool samples,5-8 and gene expression profiles in lesional biopsies.9-12 The realization that angiogenesis is a critical part of tumor progression of solid13 and liquid14,15 tumors led, over the past few decades, to numerous attempts to correlate plasma and serum levels of angiogenic proteins with disease progression.16,17 While helpful in the identification of patients with disseminated disease, the reliability of serum and plasma levels of VEGF, bFGF, or other angiogenesis regulatory proteins in early-stage tumors remains uncertain.18,19
Our previous report that platelets may serve as a reservoir of biomarkers20 introduced the finding that the platelet protein content may reflect the presence of a tumor. Further proteomic analysis of platelets from tumor-bearing and non–tumor-bearing mice revealed that the majority of differentially expressed proteins were angiogenesis regulators, rather than the more abundant proteins such as albumin and fibrinogen. The levels of albumin and fibrinogen contained in the platelets were equal in tumor-bearing and non–tumor-bearing mice. This finding suggests a very selective sequestration of angiogenesis regulating proteins by platelets. We also showed that the enhanced sequestration of angiogenesis regulators in the platelet may enable us to detect tumors as small as 1 mm3 in mice.
Platelets may sequester these proteins and protect them from plasma proteolytic enzymes. As a result of this sequestration, platelet-associated proteins, and PF-4 in particular, may be more reliable in detecting early cancer growth than their respective plasma or serum counterparts.
Platelet factor-4 (PF-4) is a tetrameric, lysine-rich member of the CXC chemokine family produced almost exclusively by megakaryocytes. Under physiological conditions, only a small amount of platelet factor-4 is taken up by circulating platelets, therefore the bulk of the PF-4 protein originates in megakaryocytes. PF-4 was originally cloned from a human erythroleukemia cell line,21 and its genetic mapping and polymorphisms were discovered soon thereafter.22,23 PF-4 is stored within the α-granules of platelets and secreted at high concentrations in the vicinity of injured blood vessels following platelet activation.24 Platelet factor-4 was discovered to inhibit angiogenesis in 1982.25 By 1990, it was shown to inhibit tumors in mice.26 In 1995, platelet factor-4 was reported to bind preferentially to vascular endothelium in vivo27 and to bind selectively to regions of active angiogenesis in vivo.28 By 1998, PF-4 was revealed to be a marker of new vessel formation in xenografts of human breast cancer.29
In the absence of any known receptor for PF-4, its antiangiogenic effect30,31 is presumed to be due to its ability to bind stimulatory chemokines such as IL-832,33 and to compete with other growth factors for heparin binding.34,35 The heterodimer of IL-8 and PF-4 enhances the antiproliferative activity of PF-4 and attenuates the stimulatory effects of IL-8.32 PF-4 also modulates the effect of proangiogenic growth factors. It binds with high affinity to vascular endothelial growth factor (VEGF165), preventing the interaction of VEGF165 with its receptor (VEGFR-2) thus inhibiting angiogenesis.34,36 A PF-4 derivative generated by peptide bond cleavage between Thr16 and Ser17 exhibits a 30- to 50-fold greater growth inhibitory activity on endothelial cells than PF-4 itself.37 The antitumor effect of PF-4 is also revealed by a decrease in the number and size of lung metastases of B16F10 melanoma38 and a decrease in growth of HCT-116 human colon carcinoma.39 PF-4 modifies the mitogenic effect of bFGF on fibroblasts,40 inhibits the proliferation of activated human T cells41 and tumor-infiltrating lymphocytes, and inhibits cytokine release by tumor stroma.42
Here we present new data demonstrating that changes in the platelet concentration of PF-4 may be used as a novel biomarker to detect human tumor xenografts that range from microscopic to macroscopic size. These tumors in mice include human liposarcoma, mammary adenocarcinoma, and osteosarcoma.
Methods
Human-tumor xenografts
All of the cancer cell lines used exhibit either nonangiogenic (microscopic, dormant tumors) or angiogenic (rapidly growing tumors) phenotypes in immunodeficient mice. The nonangiogenic and angiogenic cell lines have been previously described by Folkman and colleagues (Almog et al43 ; Achilles et al44 ; and Naumov et al45 ). For each of the 3 parent cell lines (liposarcoma [SW872], osteosarcoma [KHOS-24OS], and mammary adenocarcinoma [MDA-MB-436]) 2 phenotypes exist, a nonangiogenic tumor and an angiogenic one.
All cell lines were cultured in DMEM containing 10% heat-inactivated fetal bovine serum (HyClone, Logan, UT), 1% antibiotics (penicillin, streptomycin), and 0.29 mg/mL l-glutamine in a humidified 5% CO2 incubator at 37°C. For injections into mice, 80% to 90% confluent tumor cells were rinsed in phosphate-buffered saline (PBS; Sigma-Aldrich, St Louis, MO), briefly trypsinized and suspended in serum-free DMEM. Five million viable cells from each of the tumor cell lines were suspended in 200 μL serum-free media and implanted subcutaneously (SW872 and KHOS-24OS cell lines) into the flanks of 6- to 8-week-old male severe combined immunodeficient (SCID) mice. For the human breast adenocarcinoma (MDA-MB-436) cell line, 1 million viable cells were suspended in 50 μL serum-free media and implanted in the mammary fat pad through a 0.75- to 1-cm incision. The corresponding sham operation was a 0.75- to 1.0-cm incision. The mice were terminally bled under isoflurane anesthesia at 30 days after implantation to collect the platelets. The mice were obtained from the Massachusetts General Hospital (MGH; Boston, MA). Animals and tumors were monitored daily as per institutional guidelines. Data were analyzed using Student t test to determine the P values for differences between means; the graphs in Figures 4 and 5 are therefore expressed as means plus or minus SEM.
Platelet and plasma processing for SELDI-ToF mass spectrometry
Blood samples were processed according to standard methods for platelet collection. Briefly, mice were anaesthetized using 2% isofluorane/1 L O2 flow system. Whole blood (1 mL) was collected by terminal cardiac bleed into 105 mM sodium citrate (pH 5) anticoagulant at a ratio of 1:9 (vol/vol) buffer to blood. The first centrifugation step at 180g for 20 minutes at room temperature allowed for the collection of platelet-rich plasma (PRP). A second centrifugation at 900g for 30 minutes at room temperature separated the platelets and the upper phase, platelet-poor plasma (PPP). This resulted in 2 separate phases for processing and analysis by surface-enhanced laser desorption/ionization time-of-flight mass spectrometry (SELDI-ToF MS) technology (Ciphergen Biosystems, Fremont, CA), platelet pellets, and PPP. The pellets and 20 μL PPP from each mouse were processed in 25 μL and 40 μL, respectively, U9 buffer (2% CHAPS [3-[(3-cholamidopropyl) dimethylammonio]-1-propansulfonate], 50 mM Tris-HCl, pH 9; Ciphergen Biosystems) for 1 hour at room temperature. Platelet lysates were then centrifuged at 10 000g for 1 minute at 4°C. Both platelet extracts and PPP were fractionated by anion-exchange chromatography modified after the expression difference mapping (EDM) serum fractionation protocol (Ciphergen Biosystems). The fractionation was performed in a 96-well format filter plate on a Biomek 2000 Laboratory Work Station (Beckman Coulter, Fullerton, CA) equipped with a Micromix 5 shaker (Siemens Medical Solutions Diagnostics, Deerfield, IL). An aliquot of 20 μL platelet and 60 μL denatured plasma diluted with 100 μL 50 mM Tris-HCl (pH 9) was transferred to a filter-bottom 96-well microplate prefilled with Q Ceramic HyperD F sorbent beads (Pall, East Hills, NY) rehydrated and pre-equilibrated with 50 mM Tris-HCl (pH 9). All liquids were removed from the filtration plate using a multiscreen vacuum manifold (Millipore, Bedford, MA) into respective wells of 96-well microtiter plates with the capture of the initial flow-through as fraction 1. This step was repeated for subsequent incubations with 2 × 100 μL of the following buffers: pH 7.5 (1 M urea, 0.1% CHAPS, 50 mM NaCl, 2.5% acetonitrile, 50 mM Tris-HCl [50 mM HEPES]); pH 5 (1 M urea, 0.1% CHAPS, 50 mM NaCl, 2.5% acetonitrile 50 mM NaAcetate); pH 4 (1 M urea, 0.1% CHAPS, 50 mM NaCl, 2.5% acetonitrile 50 mM NaAcetate); pH 3 (1 M urea, 0.1% CHAPS, 500 mM NaCl, 2.5% acetonitrile 50 mM NaCitrate), which yielded the respective fractions 2, 3, 4, and 5. A final organic wash with 33% isopropanol/16.7% acetonitrile/8% formic acid represents fraction 6.
Expression difference mapping (EDM) on ProteinChip arrays was carried out using weak cationic exchange chromatography protein arrays (WCX2 and CM10 ProteinChip arrays; Ciphergen Biosystems) by loading sample fractions onto a 96-well bioprocessor, and equilibrating with 50 mM sodium acetate 0.1% octyl glucoside (Sigma), pH 5. A further dilution of 40 μL anion exchange chromatography fraction into 100 μL of the same buffer on each array spot was incubated for 1 hour. Array spots were washed for 3 minutes with 100 μL 50 mM sodium acetate 0.1% octyl glucoside (pH 5). After rinsing with water, 1 μL sinapinic acid matrix solution was added twice to each array spot. For protein profiling, all fractions were diluted 1:2.5 in their respective buffers used to pre-equilibrate ProteinChip arrays. This step was followed by readings using the Protein Biology System II (PBSII) and Protein Ciphergen System 4000 (PCS4000) SELDI-ToF mass spectrometer (Ciphergen Biosystems) and processed with the ProteinChip Software Biomarker Edition, Version 3.2.0 (Ciphergen Biosystems). After baseline subtraction, spectra were normalized by a total ion current method. Peak detection was performed using Biomarker Wizard software (Ciphergen Biosystems) using a signal-to-noise ratio of 3.
For immunocapture experiments, anti–PF-4 antibody (rabbit, affinity-purified polyclonal antibody; R&D, Minneapolis, MN) was immobilized on preactivated ProteinChip array (RS100; Ciphergen Biosystems). After blocking and washing of excess antibody, platelet extract diluted in BSA Triton X100 PBS was incubated with the immobilized antibody. After washing with PBS containing urea and CHAPS, the captured proteins were detected by SELDI. To confirm a full capture of the protein, the mobile phase was also incubated on a preactivated spot for a second time to verify immunodepletion. A final confirmation of the protein identity was obtained using murine PF-4 enzyme-linked immunosorbent assay (ELISA; data included as Figure S2).
Protein identification
Candidate protein biomarker was purified by affinity chromatography on beads with immobilized IgG spin columns and by reverse-phase chromatography. The purity of each step was monitored using normal phase (NP) ProteinChip arrays. The enriched fractions were reduced by 5 mM DTT in Tris-HCL buffer (pH 9) and alkylated with 25 mM iodoacetamide. The alkylated preparation was finally purified using 16% tricine sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE). The gel was stained by Colloidal Blue Staining Kit (Invitrogen, Carlsbad, CA). Selected protein bands were excised, washed with 200 μL 50% methanol/10% acetic acid for 30 minutes, dehydrated with 100 μL acetonitrile (ACN) for 15 minutes, and extracted with 70 μL 50% formic acid, 25% ACN, 15% isopropanol, and 10% water for 2 hours at room temperature with vigorous shaking. The candidate biomarkers in extracts were again verified by analysis of 2 μL on a normal phase ProteinChip array (NP20). The remaining extract was digested with 20 μL of 10 ng/μL modified trypsin (Roche Applied Science, Indianapolis, IN) in 50 mM ammonium bicarbonate (pH 8) for 3 hours at 37°C. Single MS and MS/MS spectra were acquired on a QSTAR mass spectrometer (Applied Biosystems, Foster City, CA) equipped with a Ciphergen PCI-1000 ProteinChip Interface. A 1-μL aliquot of each protease digest was analyzed on an NP20 ProteinChip array in the presence of CHCA matrix (Ciphergen Biosystems). Spectra were collected from m/z values of 900 to 3000 in single MS mode. After reviewing the spectra, specific ions were selected and subjected to collision-induced dissociation (CID). The CID data were submitted to the database-mining tool Mascot (Matrix Science, Boston, MA) for identification (Table 1; Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Amino acid sequences of identified peptides
| m/z . | Amino acid sequence . | Mowse score* . | Significan homology score† . | Identity or extensive homology score‡ . |
|---|---|---|---|---|
| Peptide I: 1350.75 | HCAVPQLIATLK + CAM§ | 60 | >17 | >22 |
| Peptide II: 1677.90 | HCAVPQLIATLKNGR + CAM | 14 | >13 | >15 |
| m/z . | Amino acid sequence . | Mowse score* . | Significan homology score† . | Identity or extensive homology score‡ . |
|---|---|---|---|---|
| Peptide I: 1350.75 | HCAVPQLIATLK + CAM§ | 60 | >17 | >22 |
| Peptide II: 1677.90 | HCAVPQLIATLKNGR + CAM | 14 | >13 | >15 |
Ion score is −10*Log(P), where P is probability that the match is a random event.
Individual ion scores higher than a number displayed in the column indicate significant homology.
Individual ion scores higher than a number displayed in the column indicate identity or extensive homology (P < .05).
Carbamidomethyl cysteine.
Results
Identification of the differentially expressed PF-4 in tumor-bearing mice
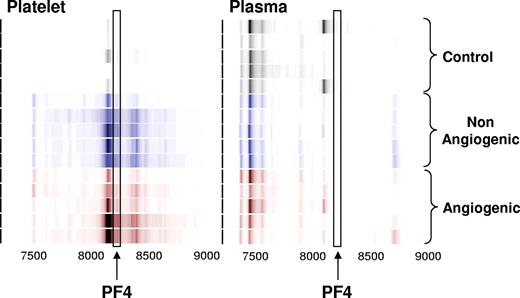
Platelet lysates from healthy non–tumor-bearing mice and those bearing nonangiogenic or angiogenic xenografts of human liposarcoma, mammary adenocarcinoma, and osteosarcoma for a minimum of 30 days were subjected to a standard biomarker discovery protocol. Among the several unknown differentially expressed proteins, elevation was observed in the platelet content of a polypeptide with an apparent molecular weight of 8206 Da (Figures 1,2). This protein was later found to be consistently elevated across 3 repeated experiments, and across several tumor types.
Identification of the platelet- and plasma-derived candidate proteins. Platelets were harvested from mice bearing nonangiogenic or angiogenic xenografts of human liposarcoma at 30 days after tumor implantation and compared with those of their littermate controls using a standard SELDI-ToF biomarker discovery method. A spectral readout from SELDI-ToF MS is presented here in gel view format, and groups are color-coded for clarity. Gray represents protein content of platelets from control animals; blue, from mice bearing the nonangiogenic dormant clone; and red, from mice bearing the angiogenic clone. A differentially expressed protein was observed at 8206 Da. The candidate peptide (↑) was later analyzed and identified as PF-4. Each horizontal strip represents an individual mouse sample (n = 5), and the color intensity corresponds to the height of the protein peak. The experiment was reproduced on 2 independent occasions for a total of 15 mice per group.
Identification of the platelet- and plasma-derived candidate proteins. Platelets were harvested from mice bearing nonangiogenic or angiogenic xenografts of human liposarcoma at 30 days after tumor implantation and compared with those of their littermate controls using a standard SELDI-ToF biomarker discovery method. A spectral readout from SELDI-ToF MS is presented here in gel view format, and groups are color-coded for clarity. Gray represents protein content of platelets from control animals; blue, from mice bearing the nonangiogenic dormant clone; and red, from mice bearing the angiogenic clone. A differentially expressed protein was observed at 8206 Da. The candidate peptide (↑) was later analyzed and identified as PF-4. Each horizontal strip represents an individual mouse sample (n = 5), and the color intensity corresponds to the height of the protein peak. The experiment was reproduced on 2 independent occasions for a total of 15 mice per group.
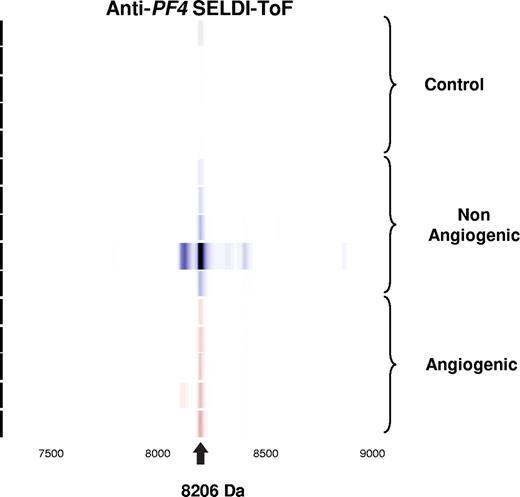
Validation of candidate biomarker by immunoprecipitation. Spectral readout in gel view format obtained from arrays prepared with an anti–PF-4 antibody prior to incubation with platelet extracts from mice within the indicated groups. The labeled arrow indicates both the presence and theoretic mass of PF-4.
Validation of candidate biomarker by immunoprecipitation. Spectral readout in gel view format obtained from arrays prepared with an anti–PF-4 antibody prior to incubation with platelet extracts from mice within the indicated groups. The labeled arrow indicates both the presence and theoretic mass of PF-4.
A careful analysis was therefore performed to identify the protein of interest. The 8206-Da protein was purified using chromatography and SDS-PAGE. Gel-purified protein was digested with trypsin, and unique tryptic fragments were analyzed by tandem MS (data in Figure S1). The ion with m/z of 1350.75 was identified as Cys-carbamidomethylated peptide HCAVPQLIATLK with Mowse score46 of 60 (ion score > 17 indicated significant homology; > 22 indicated identity or extensive homology). The ion with m/z of 1677.90 was identified as Cys-carbamidomethylated peptide HCAVPQLIATLKNGR with Mowse score of 14 (ion score > 13 indicated significant homology; > 15 indicated identity or extensive homology). Both peptides corresponded to unique tryptic fragments of mouse PF-4 (SwissProt accession no. Q9Z126) previously identified.47-49 Theoretic molecular weight of mouse PF-4 is 8210.71 Da, however considering 2 Cys-Cys bridges in the polypeptide molecule, the expected MW is 8206.71 Da. The latter value is very close to the observed experimental molecular weight of the candidate biomarker.
Confirmation of identity of platelet-derived PF-4 by ProteinChip immunoassay using anti–PF-4 antibody
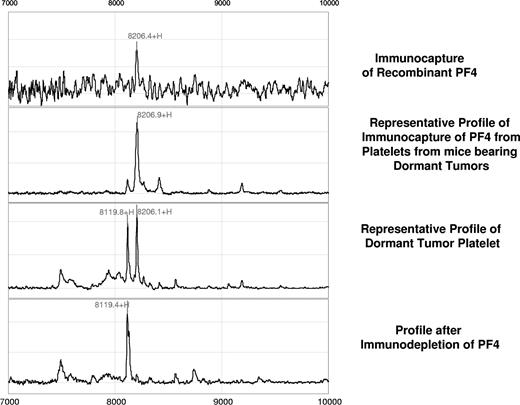
Further validation of this candidate biomarker was obtained by immunoprecipitation using rabbit anti–PF-4 antibody. Figure 2 represents the spectral readout obtained from arrays coated with an immobilized anti–PF-4 antibody prior to incubation with the platelet extracts from mice. The presence of a thick protein peak at 8206 Da (arrow) validates both the presence and theoretic mass of PF-4 (Figure 2). The identity of the differentially expressed protein was further confirmed by immunocapture/immunodepletion of the protein. The protein captured using the PF4-specific antibody has a peak identical to that of the recombinant protein (upper 2 panels of Figure 3), and the peak is absent in the mobile phase of the spotted lysate (Figure 3).
Confirmation of the PF-4 identity by immunocapture and immunodepletion. For immunocapture experiments, anti–PF-4 antibody was immobilized on a preactivated ProteinChip array, followed by incubation with platelet extracts derived from mice bearing the dormant clone of liposarcoma. Comparison of a profile generated by the recombinant PF-4 (first panel) with that generated by platelet lysates of dormant liposarcoma-bearing mice reveals an identical molecular weight and isoelectric point of the protein in question (second panel). Immunodepletion of the PF-4 protein is confirmed by the absence of its respective peak from the mobile phase (fourth panel).
Confirmation of the PF-4 identity by immunocapture and immunodepletion. For immunocapture experiments, anti–PF-4 antibody was immobilized on a preactivated ProteinChip array, followed by incubation with platelet extracts derived from mice bearing the dormant clone of liposarcoma. Comparison of a profile generated by the recombinant PF-4 (first panel) with that generated by platelet lysates of dormant liposarcoma-bearing mice reveals an identical molecular weight and isoelectric point of the protein in question (second panel). Immunodepletion of the PF-4 protein is confirmed by the absence of its respective peak from the mobile phase (fourth panel).
Validation of PF-4 as a surrogate marker of tumor presence
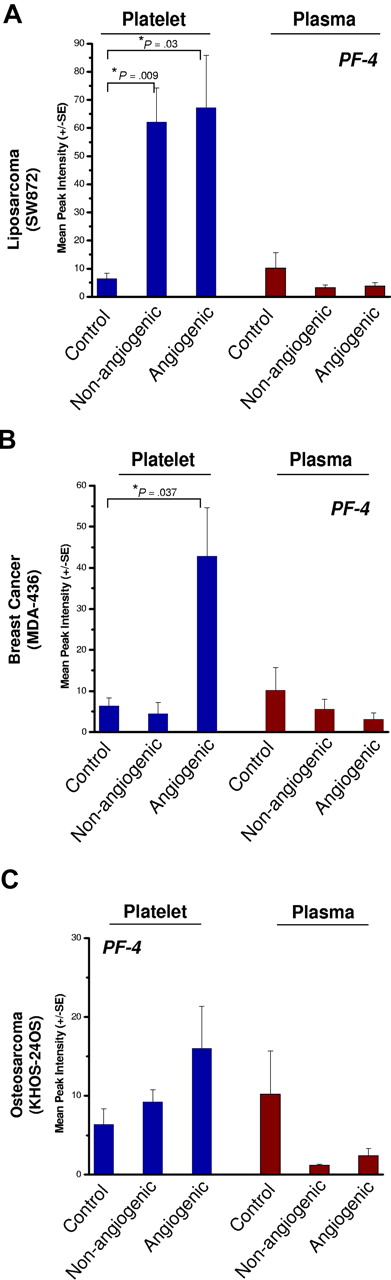
Platelets of nonangiogenic or angiogenic human liposarcoma xenografts, SW872, exhibited a 7-fold elevation of platelet-derived PF-4 compared with non–tumor-bearing controls at 30 days after implantation (Figure 4A) without a corresponding increase of PF-4 in the plasma. In this model, platelets of mice bearing the nonangiogenic xenografts (tumors less than 1 mm) contained PF-4 levels comparable with its angiogenic counterpart. Platelets of mice bearing the angiogenic mammary adenocarcinoma, MDA-MB-436 (Figure 4B), or the angiogenic osteosarcoma, KHOS-24OS (Figure 4C), revealed similar trends at 4- and 2-fold up-regulation, respectively. The nonangiogenic xenografts did not show an elevation of PF-4 in platelets. The platelet-associated PF-4 content was consistently found to be higher than that of the corresponding plasma, even though the degree of elevation varied based on tumor type (Figure 4A-C). Angiogenic tumors were associated with the greatest differences between platelet-derived PF-4 versus that of plasma, even though platelets of mice bearing the nonangiogenic tumors of liposarcoma also showed PF-4 elevation. Liposarcoma xenografts exhibited the greatest platelet content of PF-4, while the increases in PF-4 for mammary adenocarcinoma and osteosarcoma were not as large (Figure 4A-C).
PF-4 in platelets of mice bearing human-tumor xenografts. Platelets of mice bearing xenografts of SW872 liposarcoma (A), MDA-MB-436 mammary adenocarcinoma (B), or KHOS-24 osteosarcoma (C) were analyzed using SELDI-ToF. The blue bars represent whole platelet extracts and brown bars represent plasma. The platelets of non–tumor-bearing control mice served as a reference for the endogenous levels of platelet- and plasma-derived PF-4. The control group is shared by all 3 experiments. Platelets of nonangiogenic or angiogenic human liposarcoma xenografts, SW872, exhibited a 7-fold elevation of platelet-derived PF-4 compared with non–tumor-bearing controls at 30 days after implantation (A) without a corresponding increase of PF-4 in the plasma. Platelets of mice bearing the angiogenic mammary adenocarcinoma, MDA-MB-436 (B), also had significant elevation of platelet but not plasma PF-4. In the case of angiogenic osteosarcoma, KHOS-24OS (C), a similar trend at 4- and 2-fold up-regulation can be observed, but the value did not reach significance. Each bar represents the mean peak intensities corresponding to the level of the protein (± SEM) of 5 to 10 mice per experiment. Student t test was used to compare means of the groups. Each experiment was repeated twice.
PF-4 in platelets of mice bearing human-tumor xenografts. Platelets of mice bearing xenografts of SW872 liposarcoma (A), MDA-MB-436 mammary adenocarcinoma (B), or KHOS-24 osteosarcoma (C) were analyzed using SELDI-ToF. The blue bars represent whole platelet extracts and brown bars represent plasma. The platelets of non–tumor-bearing control mice served as a reference for the endogenous levels of platelet- and plasma-derived PF-4. The control group is shared by all 3 experiments. Platelets of nonangiogenic or angiogenic human liposarcoma xenografts, SW872, exhibited a 7-fold elevation of platelet-derived PF-4 compared with non–tumor-bearing controls at 30 days after implantation (A) without a corresponding increase of PF-4 in the plasma. Platelets of mice bearing the angiogenic mammary adenocarcinoma, MDA-MB-436 (B), also had significant elevation of platelet but not plasma PF-4. In the case of angiogenic osteosarcoma, KHOS-24OS (C), a similar trend at 4- and 2-fold up-regulation can be observed, but the value did not reach significance. Each bar represents the mean peak intensities corresponding to the level of the protein (± SEM) of 5 to 10 mice per experiment. Student t test was used to compare means of the groups. Each experiment was repeated twice.
Platelet PF-4 in early tumor detection
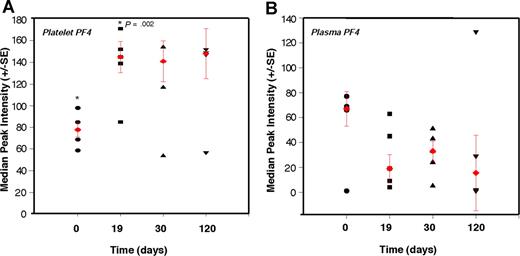
To test whether platelet content of angiogenesis regulators can be used in detection of early tumor growth, we explored the ability of PF-4 to predictably detect nonangiogenic (dormant) microscopic tumors in mice over an extended period of time. We conducted a time-course analysis of platelet-associated PF-4 in mice bearing a subcutaneous xenograft of the nonangiogenic (dormant) clone of human liposarcoma (SW872). The malignant progression of liposarcoma has been previously described,43 and it is known that the nonangiogenic (dormant) clone undergoes a spontaneous switch to the angiogenic phenotype, begins to grow, and becomes detectable by gross examination at a median of approximately 133 days after implantation. We show that PF-4 remains significantly elevated throughout a period of 120 days of observation of the nonangiogenic (dormant) state. Even without a palpable tumor, at 19 days, the median level of PF-4 in platelets is 1.7-fold higher than baseline without a corresponding increase in plasma level of the protein (Figure 5A,B). The plasma and platelet levels of PF-4 were similar at the time of implantation. However, while platelet PF-4 rose in the first 2 weeks of tumor growth, and remained elevated for the duration of the 120 days of the experiment, plasma PF-4 continued to decline (Figure 5B). The size of the tumor did not exceed 1 mm for the duration of the experiment.
Elevation of platelet-derived PF-4 correlates with the presence of microscopic tumors. Platelets and plasma from mice bearing a nonangiogenic subclone of the human liposarcoma (SW872) were analyzed at the indicated times using SELDI-ToF. The relative levels of PF-4 protein in platelets of non–tumor-bearing mice at time 0 (●; ie, before the implantation of the tumors) were compared with platelet-associated PF-4 on day 19 (■), day 30 (▲), and day 120 (▼). At 19 days, without a palpable tumor, the median level of PF-4 in platelets is 1.7-fold higher than baseline without a corresponding increase in plasma level of the protein. The red symbols within the cluster analysis represent the median peak intensity of 5 to 6 mice plus or minus SEM.
Elevation of platelet-derived PF-4 correlates with the presence of microscopic tumors. Platelets and plasma from mice bearing a nonangiogenic subclone of the human liposarcoma (SW872) were analyzed at the indicated times using SELDI-ToF. The relative levels of PF-4 protein in platelets of non–tumor-bearing mice at time 0 (●; ie, before the implantation of the tumors) were compared with platelet-associated PF-4 on day 19 (■), day 30 (▲), and day 120 (▼). At 19 days, without a palpable tumor, the median level of PF-4 in platelets is 1.7-fold higher than baseline without a corresponding increase in plasma level of the protein. The red symbols within the cluster analysis represent the median peak intensity of 5 to 6 mice plus or minus SEM.
Discussion
Numerous angiogenesis regulatory proteins are present in platelets.50 While the relative concentrations of these proteins remain stable under physiologic conditions, their levels change significantly in the presence of a tumor. There are other reports documenting angiogenesis regulatory factors in platelets of cancer patients,51-53 and continuing controversy persists as to whether serum or plasma levels of angiogenesis factors are more accurate for the measurement of angiogenesis-related diseases.54 In a study of paired serum and plasma samples,55,56 VEGF levels correlated with platelet count in 116 patients with colorectal cancer, but not in controls. Support can be found for both serum or plasma measurements.55,57,58
The diagnostic use of angiogenic proteins such as VEGF, bFGF, or PF-4 in early disease has been hindered in part by the minute levels of the proteins and their short half-lives in the circulation. The finding of platelet sequestration of angiogenesis regulatory proteins suggests a new modality for early detection of human cancer. We propose that angiogenesis regulators are not released from platelets into the circulation. Instead, these proteins are exchanged locally at sites of platelet adhesion and aggregation, where they remain bound to glycosaminoglycans such as heparan sulfate in tissues. As such, the levels of these proteins in plasma or serum increase significantly only in the presence of a large tumor load that generates sufficient angiogenesis regulatory proteins to saturate the mass of circulating platelets. We provide evidence that at least one of these platelet proteins, PF-4, can reliably predict the presence of a microscopic, nonangiogenic (dormant) human tumor in mice and circulates predominantly in platelets early in the disease process. Its relative absence in plasma may explain why the proteomic search for plasma and serum markers of patients with various cancers59-61 did not identify this marker. We emphasize that while only PF-4 is being presented here, other angiogenesis regulatory proteins sequestered in platelets may have the same diagnostic capacity and remain to be identified.
We introduce PF-4 as one of the platelet-associated angiogenesis regulators that may serve as an early tumor biomarker. We show that platelet-associated PF-4 can be detected as early as 19 days after implantation, and that a steady elevation of the protein can be observed throughout 120 days (Figure 5). While this paper does not provide sufficient data to support a functional role of PF-4 in tumor angiogenesis, there is sufficient published evidence that PF-4 is an angiogenesis suppressor and a tumor growth suppressor.25,26,38,39,62-64 PF-4 may modulate tumor growth by modifying VEGF effects35 or by binding and neutralizing heparin65 and related sulfated glycosaminoglycans42 required for the binding of proangiogenic factors.30,40,66 The binding and neutralization of heparin down-regulates angiogenesis mainly by preventing the binding of other angiogenesis regulators to heparan sulfate in tissues and by interfering with VEGF and bFGF signaling pathways.67 The high levels of VEGF and bFGF secreted by the nonangiogenic clone of SW872 liposarcoma43 may be counterbalanced by PF-4 leading to tumor quiescence and dormancy. The increase in platelet-associated PF-4 in dormant (nonangiogenic) tumors may therefore be reflective of the functional inhibition of angiogenesis in liposarcoma, which secretes large amounts of VEGF and bFGF.43 A feedback loop may exist in animals bearing tumors, such that increased VEGF and bFGF induce megakaryocyte synthesis of PF-4. This is supported by the finding that tumors that do not secrete large amounts of VEGF and bFGF, such as the nonangiogenic clones of MDA-MB-436 mammary adenocarcinoma and the KHOS-24OS osteosarcoma,45 do not manifest a marked elevation of PF4 (Figure 4). These tumors may use other means of tumor growth suppression. PF-4 appears to be a marker of angiogenesis and was present in the platelets of all of the tested angiogenic tumor models.
The changes in platelet-associated PF4 may have the potential to convey valuable clinical information about the angiogenic potential of the tumor, and a serial measurement of platelet PF-4 levels may provide us with the ability to detect tumor progression in an otherwise healthy subject.
Numerous reports have suggested an active role of platelets in cancer progression68,69 and in tumor growth and metastasis.70-72 Most investigators assume that platelets act as a reservoir of angiogenic proteins that are released into the sera.53,73 However, we show here that platelets actively sequester select proteins in tumor-bearing animals, and that this process is distinct from the nonspecific uptake of proteins such as albumin. One of the main reasons previous proteomic analysis on platelets74-78 may not have detected the differential expression of angiogenesis-related proteins was because these studies analyzed normal platelets and not platelets of cancer patients. We report for the first time, to our knowledge, the changes in the “platelet angiogenesis proteome” in response to the presence of a tumor in experimental animals.
Platelet-associated PF-4 may be a potential tumor biomarker. Platelet-associated PF-4 should be explored in other mouse models of cancer and in high-risk patient populations predisposed to early tumor progression due to mutations in APCC, p53, PTEN, or BRCA1. If validated, it may improve our ability to intervene very early in recurrent cancer, keep cancers in a dormant stage, and possibly convert cancer into a chronic, more manageable disease.79,80 As biologic modifiers, including angiogenesis inhibitors, which are relatively less toxic, become available for cancer therapy, early treatment may be much more possible than it has been in the past. A long-term goal would be to “treat the biomarker” until it returns to normal, before the onset of symptoms of recurrent tumor and before the tumor can be anatomically located.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors acknowledge the generosity of Ciphergen Biosystems, for conducting the mass spectrometry assays identification of proteins free of charge, and for sharing much of the available scientific expertise. The authors are grateful for the helpful comments and suggestions provided by Dr Sean Downing.
This work was supported by grants from the Breast Cancer Research Foundation (J.F.), the Department of Defense (DOD grant no. W81XWH-04-1-0316; J.F.), and NASA (grant no. NNH04ZUU002N).
Authorship
Contribution: D.C. wrote the initial draft of the paper and (along with T-T.Y.) executed, analyzed, and evaluated the mass spectrometry data; T-T.Y. performed, analyzed, and evaluated the mass spectrometry data and provided invaluable expertise with the SELDI ToF MS technology; N.B. executed in vitro data and assisted with animal experiments; V.N.P. performed the purification and identification of candidate biomarkers; J.P. and A.A.-S. have been instrumental in the development of a murine PF4 ELISA and quantified the PF-4 protein; G.N.N. provided the breast and osteosarcoma models; E.B. assisted with the animal experiments; N.A. designed and executed the liposarcoma experiment and advised on the dormancy models; J.E.I. contributed to the evaluation of the data and the paper revisions; J.F. provided mentorship for the team and expertise in preparation of the paper; G.L.K. designed and performed the research, provided guidance for the group, analyzed the data, and revised the original paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Giannoula Klement, Children's Hospital Boston, Karp Family Research Laboratories, Rm 11.211, One Blackfan Circle, Boston, MA 02115; e-mail: giannoula.klement@childrens.harvard.edu.
References
Author notes
D.C. and T.-T.Y. contributed equally to this work.