Abstract
Angiogenesis is critical to tumor progression. The homeobox gene GAX inhibits angiogenesis in vascular endothelial cells (ECs). We have identified a microRNA (miR-130a) that regulates GAX expression and hypothesized that it plays a major role in modulating GAX activity in ECs. A 280-bp fragment from the GAX 3′-untranslated region (3′-UTR) containing 2 miR-130a targeting sites was observed to be required for the rapid down-regulation of GAX expression by serum and proangiogenic factors, whereas the activity of the GAX promoter did not vary with exposure to serum or proangiogenic factors. This same 280-bp sequence in the GAX 3′-UTR cloned into the psiCHECK2-Luciferase vector mediated serum-induced down-regulation of the reporter gene when placed 3′ of it. Finally, forced expression of miR-130a inhibits GAX expression through this specific GAX 3′-UTR sequence. A genome-wide search for other possible miR-130a binding sites revealed an miR-130a targeting site in the 3′-UTR of the antiangiogenic homeobox gene HOXA5, the expression and antiangiogenic activity of which are also inhibited by miR-130a. From these data, we conclude that miR-130a is a regulator of the angiogenic phenotype of vascular ECs largely through its ability to modulate the expression of GAX and HOXA5.
Introduction
Angiogenesis is critical to the growth, invasion, and metastasis of human tumors. Key to this process is the vascular endothelial cell (EC),1 which in health responds to a balance between proangiogenic and antiangiogenic factors secreted by various cells to maintain blood vessel homeostasis. This balance determines whether ECs become angiogenic in response to normal physiologic signals in processes as diverse as wound repair, the menstrual cycle, embryogenesis, and organogenesis.2,3 During carcinogenesis, tumors hijack angiogenesis by secreting proangiogenic factors to supply themselves with the oxygen and nutrients necessary for their continued growth, a transition known as the “angiogenic switch.”2,3 Because targeting angiogenesis has emerged as a promising avenue of treatment for malignancies,4 understanding the transcriptional regulation of the angiogenic phenotype in ECs has become increasingly important
Not surprisingly, given their diverse roles in development, homeobox genes are involved in the regulation of this transition between the resting and “activated,” or “angiogenic,” phenotype in ECs.5-19 Proangiogenic homeobox genes5-8,14,20 and antiangiogenic homeobox genes have been described,12,13,15-18 as have differences in homeobox gene expression and function in ECs from different vascular beds.17,19,21,22 The diverged homeodomain gene GAX (also known as MEOX2) is also expressed in ECs,12 and previous work in our laboratory has implicated GAX in inhibiting nuclear factor-κB (NF-κB) signaling as well as angiogenesis in ECs, both in vitro and in vivo.10,13,17 Most recently, we reported that GAX induces G0 cell- cycle arrest by activating the expression of p21WAF1/CIP110 through its binding to AT-rich sequences in the p21WAF1/CIP1 promoter and an enhancer site located approximately 13 kb upstream from the GAX start codon.10
Given its postulated role in regulating EC phenotype during angiogenesis, GAX represents a potentially important molecular target for the antiangiogenic therapy of cancer. Consequently, we wished to elucidate further how its expression is regulated in vascular ECs. Noting its long 3′-untranslated region (3′-UTR), we hypothesized that GAX expression is likely to be regulated, at least in part, by microRNAs. MicroRNAs are short, single-stranded RNAs transcribed from noncoding genes, which, after entry into the RNA interference pathway and maturation into approximately 22 base sequences, bind to identical or similar sequences in the 3′-UTRs of genes, resulting in inhibition of translation or cleavage of the mRNA target,23-26 including specific HOX genes.27 Although there is little known yet about the role of specific microRNAs in regulating angiogenesis, there is evidence implicating overall microRNA levels28,29 and at least one specific microRNA30 in regulating angiogenesis.
To test our hypothesis, we performed an in silico search for microRNA binding sites in the GAX 3′-UTR and identified consensus binding sites for multiple candidate microRNAs, of which only 1 (miR-130a) was expressed in proliferating ECs. Here, we report that miR-130a is largely responsible for the down-regulation of GAX expression attributable to mitogens and proangiogenic factors and antagonizes the antiangiogenic activity of GAX. Similar but less potent effects were observed for the antiangiogenic homeobox gene HOXA5,18 which contains one miR-130a consensus sequence. We thus propose that miR-130a activity is related, at least in part, to its ability to down-regulate the expression of 2 key antiangiogenic homeobox genes and that it is either a positive regulator of or permissive factor for the angiogenic phenotype in ECs. We further suggest that miR-130a may represent a promising target for the antiangiogenic therapy of cancer.
Methods
Cells and cell culture
Plasmid constructs
The construction of the Flag-tagged GAX expression vector (pcDNA3.1-GAX) has been described previously.10 The HOXA518 coding region containing the 3′-UTR and its miR-130a binding site was amplified by polymerase chain reaction (PCR) and inserted into the pcDNA3.1 expression vector with a Flag tag at the N-terminal end (pcDNA3.1-HOXA5). In addition, a fragment containing miR-130a was cloned by PCR (sense: 5′-GGA ATT GCA ATG CTG AGG AG-3′; antisense: 5′GCC GTT TTC TTT GAG GAC TG-3′) and inserted into pcDNA3.1 to produce pcDNA3.1–miR-130a.
To test their function, a 280-bp fragment containing the miR-130a targeting sites in the human GAX 3′-UTR31 was isolated from HUVEC total DNA by PCR and appended to the 3′ end of the GAX cDNA after its stop codon, and this fusion was inserted into pcDNA3.1 to produce pcDNA3.1-GAX-3′-UTR. This same 280-bp fragment of the GAX 3′-UTR containing the miR-130a target sequence was also cloned into the psiCHECK2 dual luciferase reporter plasmid (Promega, Madison, WI) at the 3′ end of the coding sequence of Renilla reniformis luciferase to produce psiCHECK2-GAX-3′-UTR. Finally, the GAX promoter (898 bp) was isolated from HUVECs by PCR and cloned upstream of luciferase in the pGL3 vector to produce pGAX-luciferase. All plasmid inserts were sequenced completely and microRNA and protein expression verified by Northern and Western blots, respectively.
Northern blots
Total RNA was isolated from cells using Trizol (Invitrogen, Carlsbad, CA), following a modification of the manufacturer protocol described previously,13,32,33 after which 40 μg of each sample was separated using 8 M urea/15% denaturing polyacrylamide gel electrophoresis, transferred to nylon membranes (Ambion, Foster City, CA), cross-linked with ultraviolet light, and baked in a vacuum at 80°C for 1 hour. Probes (Table) were end-labeled with γ-32P-ATP (300 Ci/mmol) using T4 polynucleotide kinase, and labeled probes were purified on a Sephadex G-25 column (GE Healthcare, Little Chalfont, United Kingdom). Blots were prehybridized in UltraHyb Oligo (Ambion) and then hybridized at 42°C. Membranes were washed twice with 2× standard saline citrate/0.5% sodium dodecyl sulfate (SDS) at 42°C for 30 minutes and exposed at −80°C to Kodak BioMax MR film (Eastman Kodak, Rochester, NY) using an intensifying screen. The sequence of the U6 probe was 5′-GCA GGG GCC ATG CTA ATC TTC TCT GTA T-3′.
Probe sequences for microRNAs in Gax 3′-UTR
| microRNA . | Sequence (5′ to 3′) . |
|---|---|
| miR-148b | ACA AAG TTC TGT GAT GCA CTG A |
| miR-148a | ACA AAG TTC TGT AGT GCA CTG A |
| miR-152 | CCC AAG TTC TGT CAT GCA CTG A |
| miR-301 | GCT TTG ACA ATA CTA TTG CAC TG |
| miR-130b | ATG CCC TTT CAT CAT TGC ACT G |
| miR-130a | ATG CCC TTT TAA CAT TGC ACT G |
| miR-30a-5p | CTT CCA GTC GAG GAT GTT TAC A |
| miR-30e-3p | GCT GTA AAC ATC CGA CTG AAA G |
| miR-206 | CCA CAC ACT TCC TTA CAT TCC A |
| miR-30c | GCT GAG AGT GTA GGA TGT TTA CA |
| miR-30b | AGC TGA GTG TAG GAT GTT TAC A |
| microRNA . | Sequence (5′ to 3′) . |
|---|---|
| miR-148b | ACA AAG TTC TGT GAT GCA CTG A |
| miR-148a | ACA AAG TTC TGT AGT GCA CTG A |
| miR-152 | CCC AAG TTC TGT CAT GCA CTG A |
| miR-301 | GCT TTG ACA ATA CTA TTG CAC TG |
| miR-130b | ATG CCC TTT CAT CAT TGC ACT G |
| miR-130a | ATG CCC TTT TAA CAT TGC ACT G |
| miR-30a-5p | CTT CCA GTC GAG GAT GTT TAC A |
| miR-30e-3p | GCT GTA AAC ATC CGA CTG AAA G |
| miR-206 | CCA CAC ACT TCC TTA CAT TCC A |
| miR-30c | GCT GAG AGT GTA GGA TGT TTA CA |
| miR-30b | AGC TGA GTG TAG GAT GTT TAC A |
Western blots
Protein was isolated from cells for Western blot as described previously10,17 and separated by electrophoresis in 10% SDS-polyacrylamide gels before transfer to polyvinylidene diflouride membranes. Membranes were blocked with phosphate-buffered saline (PBS) plus 5% nonfat dry milk and 0.1% Tween-20 before being incubated with primary antibody (mouse monoclonal anti-Flag, mouse monoclonal anti–α-tubulin [both from Sigma-Aldrich, St Louis, MO], or polyclonal rabbit anti-GAX),34 in blocking solution. Blots were washed with blocking solution and incubated with secondary antibody, either goat anti–mouse immunoglobulin G (IgG) or goat anti–rabbit IgG (Pierce Biotechnology, Rockford, IL) as appropriate and then washed again with blocking solution. Bands were visualized by chemiluminescence using the ECL-Plus reagent (GE Healthcare) and quantified using densitometry. Each band was normalized to α-tubulin as described previously.10
Transfections
Transfections were carried out using Trans-IT Jurkat Transfection Reagent (Mirus Bio, Madison, WI) according to a modification of the manufacturer instructions described previously.10,17 In general, a 1 μl:1 μg ratio of transfection reagent to DNA was used, and cells were exposed to reagent–DNA complexes for 1 to 3 hours, depending on the experiment, after which they were incubated 16 to 24 hours and then harvested for RNA or protein isolation. Transfection efficiency was measured using a pcDNA3.1 construct expressing green fluorescence protein (pcDNA3.1-GFP) and found to be between 30% and 40%.
In addition, a 2′-O-methyl-modified oligo-RNA inhibitor of miR-130a was designed according to principles described by Vermeulen et al35 with the following sequence: 5′-CUC UGA AAA GAG CUA UGC CCU UUU AAC AUU GCA CUG UCG AGA UUC GUC UC-3′. For experiments involving this miR-130a inhibitor, cells were transfected with 0 to 100 nM inhibitor and 0.125 μg/well of either the psiCHECK2-GAX-3′-UTR reporter plasmid or varying combinations of pcDNA3.1-GAX or pcDNA3.1-GAX-3′UTR and pcDNA3.1-miR130a using Mirus Jurkat reagent (1 μl:1 μg ratio) for 2 hours. Either empty pCHECK2 or pcDNA3.1 vector was used to equalize the total DNA amount as appropriate to the experiment. Cells were incubated in regular culture medium overnight, after which they were placed in medium containing 0.1% fetal bovine serum (FBS), 2%, or 10% FBS for 6 hours and then harvested for luciferase assay.
Dual luciferase reporter assays
Luciferase reporter assays were performed using the psiCHECK2-GAX-3′-UTR vector. Cells were grown to approximately 80% confluence in 6-well plates and cotransfected with psiCHECK2-GAX-3′-UTR or psiCHECK2 empty vector plus 0.5 μg pcDNA3.1-miR-130a as described above and previously.10 Cells were incubated with transfection reagent/DNA complex for 3 hours and then refreshed with fresh endothelial basal medium and supplements (Cambrex, East Rutherford, NJ) containing 2% FBS or 0.1% FBS overnight. In a separate set of experiments, cells were incubated for 12 hours after transfection with 10 ng/mL vascular endothelial growth factor (EGF), basic fibroblast growth factor (bFGF), and/or tumor necrosis factor-α (TNF-α). Firefly and Renilla luciferase activities were evaluated using the Dual-Luciferase Reporter Assay system (Promega), and Renilla luciferase activity was normalized to firefly luciferase activity. In the case of luciferase assays using pGAX-luciferase, luciferase activity was normalized to Renilla luciferase activity according to previously reported protocols.10
Quantitative reverse transcriptase real-time PCR
RNA (1.0 μg) used to synthesize cDNA using the iScrit cDNA Synthesis kit (Bio-Rad, Hercules, CA). cDNA was then subjected to quantitative real-time PCR using TaqMan probes to determine the GAX mRNA level as described previously17 using SYBR Green as the fluorophore. A Cepheid SmartCycler thermocycler, with associated SmartCycler v.2.0 software (Cepheid, Sunnyvale, CA), was used to analyze the data and determine the threshold count (Ct). PCR cycles started with an initial 1.5-minute denaturation step at 95°C, followed by 30 to 40 cycles of denaturation at 95°C for 10 seconds, annealing at 56°C for 20 seconds, and extension at 72°C for 30 seconds. Each sample was run in triplicate and Ct determined for the target gene. GAX levels were normalized to glyceraldehyde-3-phosphate dehydrogenase using the ΔΔCt method, as described previously.10,33,36-40
Migration and tube formation assays
Migration and tube formation assays were carried out as described previously.13,41 For migration assays, HUVECs were cotransfected with either pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX plus either pcDNA3.1-miR130a or pcDNA3.1 control empty vector. Eighteen hours later, 5 × 104 cells/well were plated onto gelatin-coated 8.0-μm pore size polycarbonate membrane in 24-well plates and allowed to attach for 1 hour, after which the medium in the upper chamber was replaced with low serum medium (LSM, which consisted of EGM-2 + 0.1% FBS lacking vascular EGF [VEGF], bFGF, and EGF), and the lower chamber with 10% FBS. After 5 hours, inserts were washed with PBS and the upper surfaces cleaned with a cotton swab to remove any cells that had not migrated. Cells were fixed with Diff-Quik Stain (Dade Behring, Deerfield, IL) and photographed for counting, 5 high-powered fields (hpf) per well. For tube formation assays, HUVECS were cotransfected as above. Eighteen hours later, 2 × 105 cells were plated on 6-well plates coated with Low Growth Factor Matrigel (BD Biosciences, San Jose, CA) and incubated overnight in the presence of serum and 10 ng/mL VEGF165 (R&D Systems, Minneapolis, MN). The number of tubes, defined as projections that connect 2 cell bodies per low-powered field (50×) for each well was determined for at least 5 fields by an observer blinded to experimental groups according to protocols used previously in our laboratory.13,17 This methodology is adequate to detect changes in tube formation of approximately 25% reliably. Finally, both migration and tube formation assays were carried out in the presence of 0 to 100 nM miR-130a inhibitor.
Immunofluorescence
HUVECs growing on cover slides were cotransfected with pcDNA3.1-GAX-3′-UTR, plus either pcDNA3.1-miR-130a or control empty vector (ratios described for each experiment), and then incubated in fresh culture medium overnight containing either varying concentration of FBS. Cells were fixed with 4% paraformaldehyde for 10 minutes at room temperature, followed by incubation in methanol for 20 minutes on ice. After 3 washes, the cells were permeabilized with 1% Triton X-100–PBS and further blocked with 5% goat serum. Immunostaining was performed using monoclonal antibody to Flag tag (Sigma-Aldrich) to identify exogenous protein using Alexa Fluor488–labeled goat anti-mouse IgG (Invitrogen) as the secondary antibody. Cell nuclei were stained with TO-PRO-3 Iodide (Invitrogen), and fluorescence was analyzed using a Nikon C1 Digital Eclipse (TE 2000-U) confocal microscope system and camera (Nikon Instruments, Melville, NY). Images were acquired under the same conditions for each experiment, and no digital manipulation of images was performed other than cropping.
Cell-cycle analysis
Flow cytometric and cell-cycle analyses were performed using HUVECs as described previously.42 In brief, HUVECs rendered quiescent by incubation in 0.1% FBS-containing medium for 24 hours were cotransfected with pcDNA3.1-GAX and pcDNA3.1-miR-130a and incubated for 24 hours in normal growth medium supplemented with 10 ng/mL VEGF. Cells were then harvested, fixed with −20°C cold absolute ethanol for 1 hour at 4°C, washed twice, and then incubated with 1 mL of 50 μg/mL propidium iodide (PI) staining solution before cell-cycle analysis on a Beckman Coulter Cytomics FC500 flow cytometer (Fullerton, CA).
Statistics and data analysis
Samples for quantitative reverse transcriptase real-time PCR (QRT-PCR) were always run in triplicate. All experiments were repeated at least a total of 3 times. Statistical significance was determined either by one-way analysis of variance (ANOVA) or the unpaired Students t test as appropriate, depending on the number of experimental groups analyzed.
Results
The GAX 3′-UTR contains consensus sequences for miR-130a
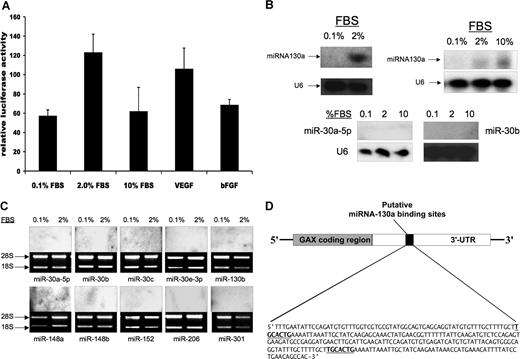
Given the length of the GAX 3′-UTR (1153 nucleotides),32 we considered it likely that sequences in the GAX 3′-UTR might represent a point of regulation of GAX expression by microRNAs. To estimate the contribution of transcriptional regulation through the GAX promoter to GAX expression, we transfected HUVECs with a pGAX-luciferase, a promoter construct containing the full-length GAX promoter linked to luciferase and then incubated the cells overnight in the presence of mitogens and proangiogenic factors. Neither serum nor various proangiogenic factors resulted in decreased luciferase activity (Figure 1A), implying that the rapid down-regulation of GAX expression is primarily post-transcriptional.
miR-130a, but not the other microRNAs with consensus binding sites in the 3′-UTR of the GAX cDNA, is expressed in HUVECs. (A) GAX promoter activity is not decreased by serum or proangiogenic factors. HUVECs were cotransfected with pGAX-Luciferase and pRL-SV (Renilla luciferase, Dual Luciferase Assay System) as described in “Transfections.” Luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Error bars represent SD. (B,C) Expression of 11 microRNAs in HUVECs in low serum and 2% serum. miR-130a is the only microRNA for which expression is detectable in HUVECs, and its expression is up-regulated by exposure to serum in a dose-dependent fashion. Northern blots were performed as described in “Northern blots” to detect these microRNAs. (D) Location of the 2 miR-130a consensus binding sequences in the GAX 3′-UTR.
miR-130a, but not the other microRNAs with consensus binding sites in the 3′-UTR of the GAX cDNA, is expressed in HUVECs. (A) GAX promoter activity is not decreased by serum or proangiogenic factors. HUVECs were cotransfected with pGAX-Luciferase and pRL-SV (Renilla luciferase, Dual Luciferase Assay System) as described in “Transfections.” Luciferase activity was normalized to Renilla luciferase activity to control for transfection efficiency. Error bars represent SD. (B,C) Expression of 11 microRNAs in HUVECs in low serum and 2% serum. miR-130a is the only microRNA for which expression is detectable in HUVECs, and its expression is up-regulated by exposure to serum in a dose-dependent fashion. Northern blots were performed as described in “Northern blots” to detect these microRNAs. (D) Location of the 2 miR-130a consensus binding sequences in the GAX 3′-UTR.
Next, we performed an in silico search for microRNA binding sites more than the full-length GAX cDNA sequence containing the 3′-UTR using the PicTar algorithm (http://pictar.bio.nyu.edu).43,44 Using this strategy, we identified 11 microRNA consensus sequences: miR-30a, miR-30b, miR-30c, miR-30e, miR-130a, miR-130b, miR-148a, miR-148b, miR-152, miR-206, and miR-301. Next, we subjected RNA from both rapidly growing and serum-starved HUVECs to Northern blot for these microRNAs (probe sequences in the Table), detecting expression of only one of these microRNAs: miR-130a (Figure 1B). Moreover, miR-130a expression was undetectable in quiescent HUVECs and strongly up-regulated after exposure to FBS (Figure 1B,C). We further noted that there is a 280-bp sequence in the GAX 3′-UTR beginning at nucleotide 1594 (417 bases beyond the GAX stop codon) that contains 2 consensus sequences for miR-130a (Figure 1D). Given the presence of 2 miR-130a consensus sequences in the GAX 3′-UTR and our observation that not only is miR-130a the only microRNA of these that is detectable in ECs by Northern blot but that it is up-regulated by exposure to mitogens, we initially conclude that it is likely that it is miR-130a that contributes to the rapid down-regulation of GAX expression that occurs in HUVECs in response to mitogenic stimuli.
miR-130a expression confers serum responsiveness to a reporter gene
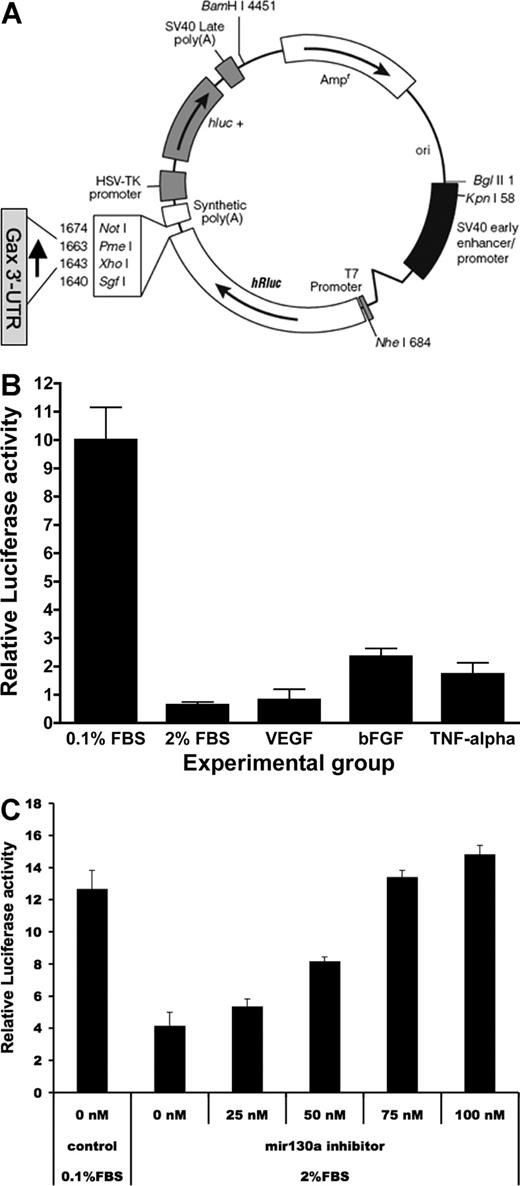
Next, to determine whether these 2 miR-130a sequences in the GAX 3′-UTR contribute to the rapid down-regulation of GAX expression, the 280-bp segment of the GAX 3′-UTR containing them was cloned into the psiCHECK2 at the 3′ end of the Renilla luciferase gene (Figure 2A). Quiescent HUVECs were transfected with either psiCHECK2 empty vector or psiCHECK2-GAX-3′-UTR, after which they were exposed to serum, proangiogenic factors (VEGF, bFGF, at 10 ng/mL each), or proinflammatory factors (10 ng/mL TNF-α) and incubated overnight. Cells were then harvested for dual luciferase assay as described in “Dual luciferase reporter assays,” with luciferase activities in cells transduced with psiCHECK2-GAX-3′-UTR normalized to those of corresponding HUVECs transduced with empty vector to correct for transfection-related nonspecific changes in luciferase activity. Exposure to FBS, VEGF, bFGF, and TNF-α all resulted in decreased luciferase activity in HUVECs transfected with psiCHECK2-GAX-3′-UTR (Figure 2B). Finally, we cotransfected HUVECs in serum-containing media with psiCHECK2-GAX-3′-UTR or psiCHECK2 empty vector and varying concentrations of miR-130a inhibitor, using HUVECs in LSM as a control and observed that inhibition of miR-130a blocked the decrease in luciferase activity because of exposure to serum (Figure 2C). From these data, we conclude that the 280-bp sequence in the GAX 3′-UTR containing the miR-130a consensus binding sites confers mitogen-responsive inhibition of expression to a reporter construct in HUVECs.
The GAX 3′-UTR confers responsiveness to serum and proangiogenic factors in ECs. (A) Construction of the psiCHECK2-GAX-3′-UTR. The 280-bp sequence in the GAX 3′-UTR containing 2 miR-130a consensus sequences was inserted into the vector at the 3′ end of the hRluc (Renilla luciferase) reporter gene. hRluc indicates Renilla reniformis luciferase. (B) Serum and proangiogenic factors suppress luciferase activity in HUVECs transfected with psiCHECK2-GAX-3′-UTR compared with psiCHECK2 empty vector. HUVECs were transduced with either psiCHECK2-GAX-3′-UTR or psiCHECK2 (control) and then incubated in 0.1% FBS, in 0.1% FBS supplemented with 10 ng/mL of either VEGF, bFGF, or TNF-α, or in 2% FBS. Renilla luciferase activities were measured as described in “Dual luciferase reporter assays.” In parallel, identical passaged HUVECs from the same split were transfected with psiCHECK2 empty vector and subjected to the same conditions. Results are expressed as a ratio of the signal from cells transfected with psiCHECK2-GAX-3′-UTR to the signal from cells transfected with psiCHECK2 empty vector. (C) Inhibition of miR-130a reverses the suppression of luciferase activity by serum in a dose-dependent fashion. HUVECs were treated as in B but were cotransfected with increasing concentrations of miR-130a inhibitor. A dose-dependent reversal of the down-regulation of reporter activity attributable to serum was observed. Error bars represent SEM.
The GAX 3′-UTR confers responsiveness to serum and proangiogenic factors in ECs. (A) Construction of the psiCHECK2-GAX-3′-UTR. The 280-bp sequence in the GAX 3′-UTR containing 2 miR-130a consensus sequences was inserted into the vector at the 3′ end of the hRluc (Renilla luciferase) reporter gene. hRluc indicates Renilla reniformis luciferase. (B) Serum and proangiogenic factors suppress luciferase activity in HUVECs transfected with psiCHECK2-GAX-3′-UTR compared with psiCHECK2 empty vector. HUVECs were transduced with either psiCHECK2-GAX-3′-UTR or psiCHECK2 (control) and then incubated in 0.1% FBS, in 0.1% FBS supplemented with 10 ng/mL of either VEGF, bFGF, or TNF-α, or in 2% FBS. Renilla luciferase activities were measured as described in “Dual luciferase reporter assays.” In parallel, identical passaged HUVECs from the same split were transfected with psiCHECK2 empty vector and subjected to the same conditions. Results are expressed as a ratio of the signal from cells transfected with psiCHECK2-GAX-3′-UTR to the signal from cells transfected with psiCHECK2 empty vector. (C) Inhibition of miR-130a reverses the suppression of luciferase activity by serum in a dose-dependent fashion. HUVECs were treated as in B but were cotransfected with increasing concentrations of miR-130a inhibitor. A dose-dependent reversal of the down-regulation of reporter activity attributable to serum was observed. Error bars represent SEM.
miR-130a binding sites in the GAX 3′-UTR mediate serum-induced down-regulation
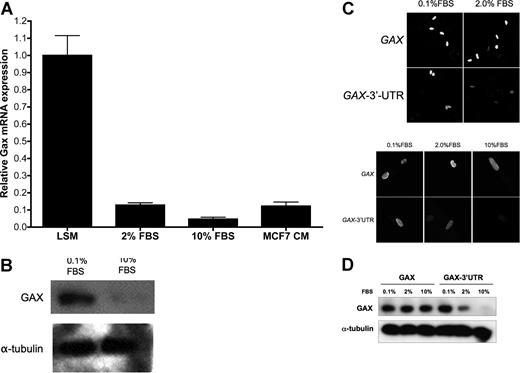
Quiescent HUVECs incubated for 16 to 18 hours in LSM (0.1% FBS) were exposed to either 0.1% (LSM), 2%, or 10% FBS, incubated overnight, and then harvested for total RNA and protein. GAX message was then measured by QRT-PCR, and it was observed that FBS down-regulated endogenous GAX expression in a dose-dependent fashion, as did conditioned media from the breast cancer cell line MCF7 (Figure 3A). A similar down-regulation of GAX protein by serum was observed using Western blot (Figure 3B).
The rapid down-regulation of GAX expression after exposure to mitogens requires the miR-130a sequences in the GAX 3′-UTR. (A) GAX mRNA expression is down-regulated by mitogens. Quiescent HUVECs incubated in LSM (0.1% FBS) were exposed to mitogens or conditioned medium from MCF7 cells, incubated overnight, and then harvested for isolation of total mRNA, which was subjected to real-time QRT-PCR. Mitogens strongly down-regulated GAX mRNA expression. (B) Mitogens down-regulate endogenous GAX protein expression. HUVECs were incubated in either 0.1% FBS or 10% FBS overnight and then harvested for total protein, which was subjected to Western blot with anti-Flag antibody. Bands were subjected to densitometry and normalized to α-tubulin levels miR-130a. (C,D) Down-regulation of GAX by mitogens depends on the presence of miR-130a sequences in its 3′-UTR. HUVECs were transfected with either pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX, incubated overnight in 0.1%, 2%, or 10% FBS, and then fixed for immunofluorescence (C) or harvested for Western blot (D) with anti-Flag antibodies. In panel C, the photomicrograph was taken at 400× magnification; fluorophores and confocal microscope are described in “Immunofluorescence.” Removing the 3′-UTR sequence almost completely abolishes the down-regulation of GAX attributable to serum stimulation. Error bars represent SEM.
The rapid down-regulation of GAX expression after exposure to mitogens requires the miR-130a sequences in the GAX 3′-UTR. (A) GAX mRNA expression is down-regulated by mitogens. Quiescent HUVECs incubated in LSM (0.1% FBS) were exposed to mitogens or conditioned medium from MCF7 cells, incubated overnight, and then harvested for isolation of total mRNA, which was subjected to real-time QRT-PCR. Mitogens strongly down-regulated GAX mRNA expression. (B) Mitogens down-regulate endogenous GAX protein expression. HUVECs were incubated in either 0.1% FBS or 10% FBS overnight and then harvested for total protein, which was subjected to Western blot with anti-Flag antibody. Bands were subjected to densitometry and normalized to α-tubulin levels miR-130a. (C,D) Down-regulation of GAX by mitogens depends on the presence of miR-130a sequences in its 3′-UTR. HUVECs were transfected with either pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX, incubated overnight in 0.1%, 2%, or 10% FBS, and then fixed for immunofluorescence (C) or harvested for Western blot (D) with anti-Flag antibodies. In panel C, the photomicrograph was taken at 400× magnification; fluorophores and confocal microscope are described in “Immunofluorescence.” Removing the 3′-UTR sequence almost completely abolishes the down-regulation of GAX attributable to serum stimulation. Error bars represent SEM.
Next, to determine whether the presence or absence of the GAX 3′-UTR miR-130a consensus binding sequences affects the down-regulation of GAX by serum, HUVECs rendered quiescent by incubation in 0.1% FBS overnight were transduced with either pcDNA3.1-GAX or pcDNA3.1-GAX-3′-UTR and then incubated overnight in varying serum concentrations before plasmid-dependent GAX expression was measured. To measure only the transduced GAX and verify that the transduced GAX was localizing appropriately to the nucleus, we first chose to study GAX expression in HUVECs by immunofluorescence using anti-Flag antibody. Strikingly, down-regulation of exogenous GAX expression in response to serum was greatly attenuated if only the coding sequence was used. However, adding back the consensus miR-130a sequences to the 3′ end of the GAX coding sequence restored this down-regulation because of serum (Figure 3C). Similar results were obtained in HUVECs treated identically and then subjected to Western blot with anti-Flag antibody (Figure 3D). From this, we conclude that the miR-130a consensus binding sequences in the GAX 3′-UTR mediate the down-regulation of GAX by mitogens and proangiogenic factors.
miR-130a down-regulates endogenous GAX expression in HUVECs
Next, we directly tested whether miR-130a regulates endogenous GAX expression. HUVECs were transfected with either pcDNA-3.1-miR-130a or empty vector and incubated overnight in 0.1% FBS to induce quiescence and maximal GAX expression, after which total RNA and protein were harvested for analysis. By both QRT-PCR (Figure 4A) and Western blot (Figure 4B), miR-130a strongly down-regulated endogenous GAX expression. Next, we cotransfected HUVECs with pcDNA-3.1-miR-130a and either pcDNA3.1-GAX or pcDNA3.1-GAX-3′-UTR, incubated the cells overnight, and then performed QRT-PCR to detect GAX message and Western blot with anti-Flag antibody to detect exogenous GAX protein. Consistent with our previous results showing that the GAX 3′-UTR miR-130a sequences conferred serum responsiveness to the psiCHECK2 reporter plasmid, miR-130a strongly down-regulated GAX expression in a manner dependent on the presence of its 3′-UTR miR-130a consensus sequences (Figure 4C,D). Finally, cotransfecting with a 2′-O-methyl-modified oligo-RNA miR-130a inhibitor more than reversed the down-regulation of endogenous GAX expression attributable to serum (Figure 4E). We conclude from these experiments that miR-130a regulates GAX expression through its consensus sequences in the 3′-UTR of the GAX gene.
miR-130a down-regulates GAX expression in HUVECs. (A) miR-130a down-regulates GAX mRNA expression. HUVECs were transfected with either empty vector (pcDNA3.1) or miR-130a expression vector (pcDNA3.1-miR-130a) and then incubated overnight in LSM ( = 0.1% FBS), after which total RNA was isolated for QRT-PCR. miR-130a expression markedly down-regulated the expression of endogenous GAX. (B) Representative Western blot of protein from the same experiment as in A. Band intensity was measured by densitometry and normalized to α-tubulin levels. Endogenous GAX protein is also down-regulated by miR-130a. * indicate P < .01. (C,D) Inhibition of GAX expression attributable to miR-130a depends on the presence of the miR-130a binding sites. HUVECs were cotransfected with either pcDNA3.1 or pcDNA3.1-miR-130a plus either pcDNA3.1 or pcDNA3.1-GAX-3′-UTR. Exogenous GAX protein from these plasmids was detected using anti-Flag antibody as described in “miR-130a binding sites in the GAX 3′-UTR mediate serum-induced down-regulation.” Down-regulation of GAX expression by miR-130a was only observed with the construct containing the 2 miR-130a consensus sequences in the 3′-UTR. (C = QRT-PCR; D = Western blot.) (E) Inhibition of miR-130a blocks the down-regulation of GAX attributable to mitogens. HUVECs were transfected with 0 to 100 nM mir-130a inhibitor and incubated in fresh culture medium overnight, after which they were placed in 0.1% FBS or 2% RBS medium for 6 hours. Total RNA was isolated for QRT-PCR as described in insert section. Error bars represent SEM.
miR-130a down-regulates GAX expression in HUVECs. (A) miR-130a down-regulates GAX mRNA expression. HUVECs were transfected with either empty vector (pcDNA3.1) or miR-130a expression vector (pcDNA3.1-miR-130a) and then incubated overnight in LSM ( = 0.1% FBS), after which total RNA was isolated for QRT-PCR. miR-130a expression markedly down-regulated the expression of endogenous GAX. (B) Representative Western blot of protein from the same experiment as in A. Band intensity was measured by densitometry and normalized to α-tubulin levels. Endogenous GAX protein is also down-regulated by miR-130a. * indicate P < .01. (C,D) Inhibition of GAX expression attributable to miR-130a depends on the presence of the miR-130a binding sites. HUVECs were cotransfected with either pcDNA3.1 or pcDNA3.1-miR-130a plus either pcDNA3.1 or pcDNA3.1-GAX-3′-UTR. Exogenous GAX protein from these plasmids was detected using anti-Flag antibody as described in “miR-130a binding sites in the GAX 3′-UTR mediate serum-induced down-regulation.” Down-regulation of GAX expression by miR-130a was only observed with the construct containing the 2 miR-130a consensus sequences in the 3′-UTR. (C = QRT-PCR; D = Western blot.) (E) Inhibition of miR-130a blocks the down-regulation of GAX attributable to mitogens. HUVECs were transfected with 0 to 100 nM mir-130a inhibitor and incubated in fresh culture medium overnight, after which they were placed in 0.1% FBS or 2% RBS medium for 6 hours. Total RNA was isolated for QRT-PCR as described in insert section. Error bars represent SEM.
miR-130a antagonizes the antiangiogenic activity of GAX
Next, we tested whether miR-130a expression plays a functional role in ECs. We therefore asked whether driving miR-130a expression can antagonize the known activities of GAX, specifically its ability to induce cell-cycle arrest10 and to inhibit EC tube formation on reconstituted basement membrane.10,13 Because EC proliferation in response to proangiogenic signals is a critical early step in angiogenesis, we first examined whether miR-130a can antagonize cell-cycle inhibition by GAX. Quiescent HUVECs were cotransfected with pcDNA-3.1-miR-130a plus either pcDNA3.1-GAX or pcDNA3.1-GAX-3′-UTR, exposed to 10% FBS for 24 hours, and then harvested for flow cytometry to determine cell-cycle distribution. miR-130a clearly antagonizes the ability of GAX to induce G0 cell-cycle arrest in a manner consistent with targeting the miR-130a consensus sequences in the GAX 3′-UTR (Figure 5A). Next, we measured whether miR-130a can antagonize the ability of GAX to inhibit HUVEC migration toward serum and tube formation on reconstituted basement membrane as described in Materials and methods. As with inhibition of cell-cycle progression, miR-130a also antagonized inhibition of migration (Figure 5C) and tube formation (Figure 5D) attributable to GAX. In both cases, the ability of miR-130a to antagonize GAX activity depended on the presence of the 2 miR-130a consensus sites in its 3′-UTR. Finally, we repeated the migration and tube formation experiments but added 100 nM miR-130a inhibitor during the transfection. miR-130a inhibitor reversed antagonism of GAX activity by miR-130a (Figure 5E,F).
miR-130a antagonizes the antiangiogenic activity of GAX. (A) miR-130a antagonizes G0/G1 cell-cycle arrest attributable to GAX depending on the presence of its consensus sequences in the GAX 3′-UTR. HUVECs rendered quiescent by serum starvation overnight were transfected with empty vector or pcDNA-3.1-miR-130a and either pcDNA3.1-GAX or pcDNA3.1-GAX-3′-UTR as for the tube formation experiment and exposed to 10% FBS for 24 hours, after which cells were harvested, stained with PI, and subjected to flow cytometry to determine cell-cycle distribution. By definition, the change in the G0/G1 fraction for the empty vector control is 0, and the G0/G1 fraction for the vector control was 0.38 in the experiment displayed. Statistical significance was determined by one-way ANOVA, and both the miR-130a and GAX-3′-UTR + miR-130a groups showed a statistically significant (P < .01) change from the others. (B) miR-130a antagonizes the antimigration activity of GAX. HUVECs were cotransfected with either pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX plus either pcDNA3.1-miR130a or pcDNA3.1 control empty vector and plated 18 hours later onto 8.0 μm pore size polycarbonate membranes in 24-well plates and allowed to attach, after which migration assays were carried out as described in “Migration and tube formation assays.” Cells were counted in 5 hpf per well (*P < .01 compared with empty vector control). (C) miR-130a antagonizes the antiangiogenic activity of GAX. HUVECs were cotransfected with either empty vector or pcDNA3.1-miR-130a and pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX and then incubated overnight, after which tube formation assays were carried out. The ratio of cotransfection was 2:1 miR-130a:GAX expression construct. Tube counts were determined as described in “Migration and tube formation assays.” (D) Quantification of tube number per low-powered field (tubes/LPF; *P < .01). (E and F) An inhibitor of miR-130a reverses its antagonism of GAX activity. Experiments described in panels C and D were repeated, but with the addition of 100 nM miR-130a inhibitor. * indicates P < .01. Error bars represent SEM.
miR-130a antagonizes the antiangiogenic activity of GAX. (A) miR-130a antagonizes G0/G1 cell-cycle arrest attributable to GAX depending on the presence of its consensus sequences in the GAX 3′-UTR. HUVECs rendered quiescent by serum starvation overnight were transfected with empty vector or pcDNA-3.1-miR-130a and either pcDNA3.1-GAX or pcDNA3.1-GAX-3′-UTR as for the tube formation experiment and exposed to 10% FBS for 24 hours, after which cells were harvested, stained with PI, and subjected to flow cytometry to determine cell-cycle distribution. By definition, the change in the G0/G1 fraction for the empty vector control is 0, and the G0/G1 fraction for the vector control was 0.38 in the experiment displayed. Statistical significance was determined by one-way ANOVA, and both the miR-130a and GAX-3′-UTR + miR-130a groups showed a statistically significant (P < .01) change from the others. (B) miR-130a antagonizes the antimigration activity of GAX. HUVECs were cotransfected with either pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX plus either pcDNA3.1-miR130a or pcDNA3.1 control empty vector and plated 18 hours later onto 8.0 μm pore size polycarbonate membranes in 24-well plates and allowed to attach, after which migration assays were carried out as described in “Migration and tube formation assays.” Cells were counted in 5 hpf per well (*P < .01 compared with empty vector control). (C) miR-130a antagonizes the antiangiogenic activity of GAX. HUVECs were cotransfected with either empty vector or pcDNA3.1-miR-130a and pcDNA3.1-GAX-3′-UTR or pcDNA3.1-GAX and then incubated overnight, after which tube formation assays were carried out. The ratio of cotransfection was 2:1 miR-130a:GAX expression construct. Tube counts were determined as described in “Migration and tube formation assays.” (D) Quantification of tube number per low-powered field (tubes/LPF; *P < .01). (E and F) An inhibitor of miR-130a reverses its antagonism of GAX activity. Experiments described in panels C and D were repeated, but with the addition of 100 nM miR-130a inhibitor. * indicates P < .01. Error bars represent SEM.
Antiangiogenic homeobox gene HOXA5 has an miR-130a binding site in its 3′-UTR and is also regulated by miR-130a
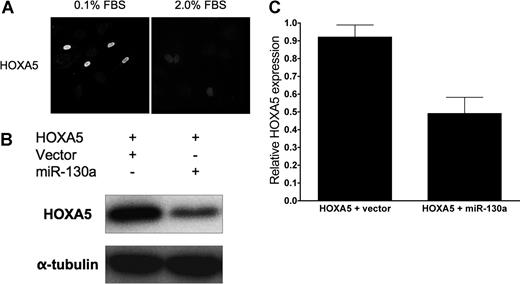
To elucidate further whether miR-130a regulates other homeobox genes involved in regulating EC phenotype during angiogenesis, we performed an in silico search for other possible downstream targets and found more than 400 candidates spanning nearly all gene functions, including transcription factors, genes involved in metabolism, and structural proteins. Of interest, we identified another antiangiogenic homeobox gene, HOXA518 , as a potential downstream target. Of note, HOXA5 up-regulates p53 expression in breast cancer cells45 and down-regulates the expression of several genes involved in angiogenesis, including VEGF receptor 2, ephrin A1, hypoxia inducible factor-1α, and cyclooxygenase-2.18 Examining the sequence of the HOXA5 cDNA for miR-130a binding sites, we found a consensus sequence 356 bp beyond the stop codon.46 Consequently, we wished to learn whether miR-130a also regulates HOXA5 expression in ECs. As in the case of GAX, we observed significant down-regulation of HOXA5 expression in response to incubation in medium supplemented with a relatively modestly increased serum concentration (Figure 6A). We also observed that miR-130a down-regulated HOXA5 expression but not as strongly as it did GAX (Figure 6B,C).
miR-130a down-regulates HOXA5 expression. (A) Mitogens down-regulate HOXA5 expression in HUVECs. HUVECs were transfected with full-length HOXA5 (pcDNA3.1-HOXA5), incubated overnight in either 0.1% FBS or 2.0% FBS, and then subjected to immunofluorescence with anti-Flag antibody. The photomicrograph was taken at 400× magnification; fluorophores and confocal microscope are described in “Immunofluorescence.” As with GAX and consistent with previously reported work,18 serum resulted in the down-regulation of HOXA5 expression.(B) miR-130a down-regulates HOXA5 expression. HUVECs were cotransfected with either pcDNA3.1 or pcDNA3.1-miR-130a plus pcDNA3.1-HOXA5, incubated overnight, and then harvested for Western blot with anti-Flag antibody. miR-130a strongly down-regulates HOXA5 expression. (C) Quantification. Western blots were subjected to densitometry and HOXA5 band densities normalized to those of the corresponding α-tubulin bands. Error bars represnt SEM.
miR-130a down-regulates HOXA5 expression. (A) Mitogens down-regulate HOXA5 expression in HUVECs. HUVECs were transfected with full-length HOXA5 (pcDNA3.1-HOXA5), incubated overnight in either 0.1% FBS or 2.0% FBS, and then subjected to immunofluorescence with anti-Flag antibody. The photomicrograph was taken at 400× magnification; fluorophores and confocal microscope are described in “Immunofluorescence.” As with GAX and consistent with previously reported work,18 serum resulted in the down-regulation of HOXA5 expression.(B) miR-130a down-regulates HOXA5 expression. HUVECs were cotransfected with either pcDNA3.1 or pcDNA3.1-miR-130a plus pcDNA3.1-HOXA5, incubated overnight, and then harvested for Western blot with anti-Flag antibody. miR-130a strongly down-regulates HOXA5 expression. (C) Quantification. Western blots were subjected to densitometry and HOXA5 band densities normalized to those of the corresponding α-tubulin bands. Error bars represnt SEM.
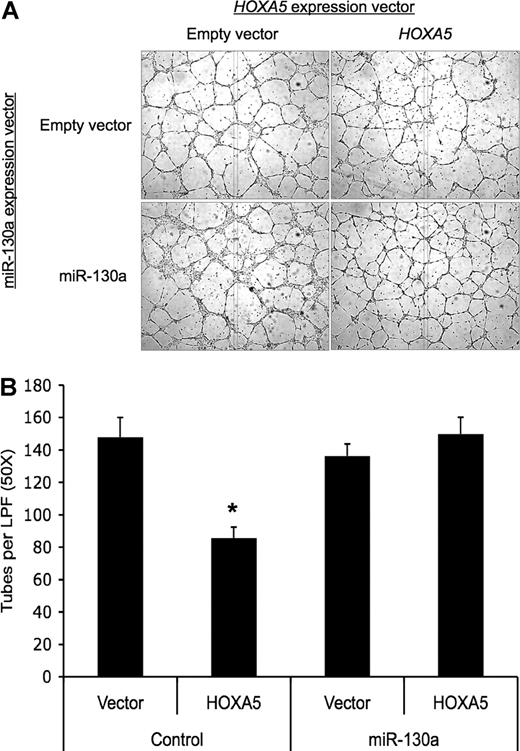
miR-130a antagonizes the antiangiogenic activity of HOXA5
Finally, we wished to determine whether miR-130a could antagonize the antiangiogenic activity of HOXA5, as it can for GAX. The antiangiogenic activity of HOXA5 was somewhat weaker than that of GAX in this assay (Figure 7). Nonetheless, miR-130a antagonized HOXA5 activity, although the antagonism seemed less pronounced than what was observed for GAX. This weaker effect of miR-130a on HOXA5 expression may be attributable to the presence of only 1 miR-130a consensus sequence in HOXA5 compared with the 2 in GAX.
miR-130a also antagonizes the antiangiogenic activity of HOXA5. (A) miR-130a antagonizes the antiangiogenic activity of HOXA5. HUVECs were cotransfected with either empty vector or pcDNA3.1-miR-130a and pcDNA3.1-HOXA5 and then incubated overnight before being subjected to tube formation assays. The ratio of cotransfection was 2:1 miR-130a:HOXA5 expression construct. Tube counts were determined as described in “Migration and tube formation assays.” (B) Quantification of tubes/LPF (*P < .01). Error bars represent SEM.
miR-130a also antagonizes the antiangiogenic activity of HOXA5. (A) miR-130a antagonizes the antiangiogenic activity of HOXA5. HUVECs were cotransfected with either empty vector or pcDNA3.1-miR-130a and pcDNA3.1-HOXA5 and then incubated overnight before being subjected to tube formation assays. The ratio of cotransfection was 2:1 miR-130a:HOXA5 expression construct. Tube counts were determined as described in “Migration and tube formation assays.” (B) Quantification of tubes/LPF (*P < .01). Error bars represent SEM.
Discussion
Cancer is an angiogenesis-dependent disease, which is why targeting tumor angiogenesis is of considerable interest as a therapeutic strategy. Key to this process is the cell that responds to these tumor-secreted factors, the vascular EC.47 When stimulated by angiogenic factors, an EC undergoes distinctive phenotypic changes, becoming capable of invading the basement membrane, migrating toward the angiogenic signal, proliferating, forming new tubes, and ultimately secreting factors to attract pericytes to form mature blood vessels.1
We have been interested in the role of homeobox genes in regulating this phenotypic conversion. Specifically, we have concentrated on the role of the homeobox gene GAX (MEOX2), which has many properties suggestive of a role as a major, if not master, regulator of EC phenotype in response to proantiogenic and antiangiogenic signals. GAX is expressed in both vascular smooth muscle cells and ECs, and, consistent with a role inhibiting cell growth and activation, GAX is expressed at its highest level in quiescent ECs and is rapidly down-regulated when ECs are exposed to mitogens, proangiogenic factors, or pro-inflammatory factors. Moreover, GAX expression induces G0 cell-cycle arrest through p53-independent up-regulation of p21WAF1/CIP1 expression10,42 and down-regulates the activity of NF-κB, resulting in the down-regulation of its downstream targets.17 In both in vitro and in vivo models in the peripheral vasculature, GAX expression inhibits angiogenesis.13,17 However, because GAX itself is antiangiogenic and a transcription factor, as a practical matter, targeting it therapeutically is likely to be difficult. If, on the other hand, it were possible to target factors that down-regulate GAX, such as microRNAs, this could represent a more practical strategy to keep the expression level of GAX high in the tumor vasculature by blocking its response to proangiogenic factors.
At present, little is known about the role that microRNAs probably play in regulating EC phenotype and function during angiogenesis. The importance of microRNAs in regulating cellular phenotype is becoming more and more clear in other cell types,24,48,49 and these small regulatory RNAs have been increasingly appreciated to have a major role in the pathogenesis of cancer.50-52 Evidence for the importance of microRNAs in the regulation of angiogenesis comes from observations that Dicer is required for embryonic angiogenesis and that knocking it out resulted in severely compromised embryos and yolk sacs, as well as decreased expression of several important positive regulators of angiogenesis.53 More recently, it has been observed that silencing Dicer29 or, to a lesser extent, Drosha,28 in adult ECs results in the down-regulation of positive regulators of the angiogenic phenotype and impaired tube formation on Matrigel. This observation suggests that microRNA expression in general is necessary for angiogenesis. Consistent with this observation, changes in microRNA expression profiles in response to angiogenesis have been reported,30 as has at least one individual microRNA (miR-221/222), the expression of which results in a profound effect on EC phenotype and angiogenesis.30
Our results suggest that at least one other microRNA (miR-130a) may be important in the regulation of EC conversion to the angiogenic phenotype. Very little is known about the function of this particular microRNA, but it does appear to be widely expressed in diverse cell types.54 During megakaryocyte development, it has been suggested that miR-130a targets MAFB, a transcription factor that is up-regulated during megakaryocytic differentiation and induces the GPIIb gene.55 In ECs, miR-130a expression is up-regulated by mitogens and proangiogenic factors (Figure 1A and data not shown) in a manner that is reciprocal to that of GAX, which is rapidly down-regulated by these factors.32 Moreover, miR-130a down-regulates GAX expression itself even in low serum, when GAX expression is normally at its highest (Figure 4), and functionally antagonizes the antiangiogenic activity of GAX in vitro (Figure 5). The ability of miR-130a to antagonize the antiangiogenic activity of GAX in these experiments is even more striking, given that our transfection protocol generally resulted in approximately 30% to 40% transfection efficiency (data not shown). Moreover, miR-130a is also able to down-regulate the expression of at least one other antiangiogenic homeobox gene, HOXA5, and antagonize its function in ECs as well. Although it does not appear to increase angiogenesis in vitro by itself, miR-130a does appear to down-regulate the expression of factors, such as GAX and HOXA5, that keep ECs in their resting state, and our results suggest it as an important regulator of EC phenotype. We thus speculate that miR-130a may play a permissive role during angiogenesis by down-regulating cellular “brakes” that keep ECs in a resting or nonangiogenic state, although we cannot rule out the possibility that miR-130a may indeed be proangiogenic and the limitations of our in vitro assays prevented us from detecting this activity.
We must also note that there are other microRNA consensus sites in the GAX 3′-UTR, including sites for miR-148/152, miR-1/206, and miR-409. However, most of these sites are also within the 280-bp segment of the GAX 3′-UTR, and we were unable to detect the expression of any of these other than miR-130a in our Northern blot screen (Figure 1A). Although we cannot entirely rule out the possibility that these micro-RNAs are also involved in the down-regulation of GAX expression in response to serum or proangiogenic factors, given our results, this seems an unlikely possibility. With that caveat in mind, it is tempting to envision a model in which different microRNAs or sets of microRNAs modulate GAX expression in response to different mitogens, proangiogenic factors, or proinflammatory factors in different vascular cell types in which GAX is expressed. Given the manner in which miRNA-130a down-regulates both GAX and HOXA5, as well as functionally antagonizing their activities with respect to angiogenesis, we consider it likely that miR-130a is an important, if not the most important, regulator of GAX expression, and, to a lesser extent, HOXA5 expression. Such a result is not without precedent, as several homeobox genes have now been shown to be regulated by microRNAs. For example, miR-181 has been shown to target HOXA11 during myoblast differentiation,56 and miR-196 directs the cleavage of HOXB827 .
In conclusion, our results suggest that microRNA sequences, specifically miR-130a, are important regulatory sequences in the GAX 3′-UTR that control the down-regulation of GAX and HOXA5 expression in response to mitogens, proangiogenic factors, and proinflammatory factors. Given that GAX appears to function to keep ECs in the resting, quiescent state, and that its down-regulation occurs rapidly on stimulation with mitogens, suggesting that its down-regulation is a necessary precondition for ECs to re-enter the cell cycle and become angiogenic, it is possible that miR-130a might represent a promising molecular target for the antiangiogenic therapy of cancer.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Prof Arnold Rabson (University of Medicine and Dentistry of New Jersey-Robert Wood Johnson Medical School and The Cancer Institute of New Jersey) for his helpful input and critical reading of this manuscript.
The work described in this article was supported by grants from the National Cancer Institute (1 R01 CA111344) and the United States Department of Defense (DAMD17-03-1-0292).
Authorship
Contribution: D.H.G. and Y.C. both contributed to designing the experiments, and Y.C. performed the experiments. D.H.G. and Y.C. both analyzed the data and made the figures. D.H.G. wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: David H. Gorski, MD, PhD, Assistant Professor of Surgery, Division of Surgical Oncology, UMDNJ-Robert Wood Johnson Medical School, The Cancer Institute of New Jersey, 195 Little Albany St, New Brunswick, NJ 08901; e-mail: gorskidh@umdnj.edu.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal