Abstract
It has been shown that the expression of osteoprotegerin (OPG) is up-regulated in tumor-associated endothelial cells as well as in the sera of patients affected by both solid tumors and hematologic malignancies. We now report that sera of p53−/− mice contain higher levels of OPG with respect to p53+/+ mice and that endothelial cells, in which p53 was knocked down by siRNA, release increased levels of OPG with respect to mock-transfected cells. Conversely, activation of the p53 pathway by the MDM2 small molecule antagonist Nutlin-3 significantly attenuated both spontaneous and tumor necrosis factor-α (TNF-α)–induced OPG mRNA and protein release in endothelial cell cultures. OPG promoter functional assays and chromatin immunoprecipitation experiments revealed inhibitory effects of Nutlin-3 on the TNF-α-induced NF-κB DNA binding activity to the OPG promoter. Because OPG inhibits the pro-tumoricidal activity of TNF-related apoptosis-inducing ligand, our findings suggest that, besides its well-documented functions within the malignant cancer cells, the ability of p53 to down-modulate OPG production by endothelial cells may be an additional important mechanism whereby it exerts non–cell-autonomous tumor suppression function.
Introduction
Osteoprotegerin (OPG) is a soluble member of the tumor necrosis factor (TNF) receptor superfamily, whose best characterized activity is the inhibition of receptor activator of NF-κB ligand (RANKL)-stimulated formation of osteoclasts.1 OPG also interacts with TNF-related apoptosis-inducing ligand (TRAIL),2 a death-inducing ligand whose extracellular domain shares a 35% homology with RANKL. Mounting evidence indicates that the ability of OPG to inhibit TRAIL cytotoxicity might represent an important mechanism in promoting the survival of prostate cancer, breast cancer, colon cancer, and multiple myeloma cells at least in vitro.3-5 More importantly, for the purpose of this study, it has also been shown that OPG is produced in vitro by vascular endothelial cells, is overexpressed by tumor-associated endothelial cells, and is able to promote the survival/proliferation of endothelial cells in a paracrine or autocrine manner.6-10 Moreover, elevated levels of serum OPG have been detected in both solid tumors and hematologic malignancies.11 Although the relative contribution of both normal and tumor-associated endothelial cells to serum OPG remains to be established,2 both the stromal and the tumoral vasculature is engaged in extensive interactions with the cancer cells, probably contributing to the growth and invasiveness of the tumor.12,13
The p53 protein is a sequence-specific transcription factor that functions as a major tumor suppressor in mammals.13,14 In response to various types of oncogenic stresses, p53 is activated to promote cell-cycle exit, apoptosis, or replicative senescence, thereby preventing the propagation of incipient cancer cells. Consequently, p53 is very often disabled within cancer cells, either by direct mutational inactivation of the TP53 gene or by alterations in other genes of which the products impinge on p53. Whereas the effect of p53 has been widely investigated in a cell-autonomous context, much less attention has been given to its non–cell-autonomous functions, although it has been recently demonstrated that the stromal compartment of the tumors is able to modulate the latency of tumorigenesis in a p53-dependent manner.12,13 On these bases, the aim of this study was to investigate the effect of p53 knock-down and/or induction on the expression and release of OPG in human endothelial cells.
Methods
Cell cultures and treatments
Human umbilical vein endothelial cells (HUVECs) were purchased from BioWhittaker (Walkersville, MD) and grown on 0.2% gelatin-coated tissue culture plates in M199 endothelial growth medium supplemented with 20% fetal bovine serum, 10 μg/mL heparin, and 50 μg/mL ECGF (endothelial cell growth factor; all from BioWhittaker). In all experiments, cells were used between the 3rd and 5th passages in vitro, as previously described.15 For endothelial cell treatments, the following reagents have been used: Nutlin-3 (Cayman Chemical, Ann Arbor, MI), TNF-α (R&D Systems, Minneapolis, MN), and Aphidicolin (Alexis Biochemicals, Lausen, Switzerland).
Recombinant TRAIL was prepared as previously described,16 and recombinant OPG was purchased from R&D Systems.
Enzyme-linked immunosorbent assays
OPG levels were measured in mouse serum samples as well as in endothelial cell culture supernatants using sandwich-type enzyme-linked immunosorbent assays (ELISA) kits. In particular, mouse OPG serum levels were measured in sera from sex-matched p53+/+ (n = 10) and p53−/− (n = 10) C57black mice (8-week-old) using a mouse OPG ELISA kit purchased from R&D Systems, whereas OPG levels in culture supernatants were measured using a human OPG ELISA kit purchased from Alexis Biochemicals. Release of GDF15/MIC-1 in supernatants of HUVEC cultures was measured using a commercially available ELISA kit purchased from R&D Systems. All the assays were used according to the manufacturer's instructions. Measurements were done in duplicates, and the results were read at an optical density of 450 nm using an Anthos 2010 ELISA reader (Anthos Labtec Instruments, Salzburg, Austria).
Flow cytometric analyses
Cell-cycle profile was analyzed by incubating cell samples with 50 μM bromodeoxyuridine (BrdU) (Sigma-Aldrich, St Louis, MO) at 37°C for 1 hour. Antibody anti-BrdU (BD Pharmingen, San Jose, CA) was bound to BrdU incorporated into neosynthesized DNA, and the complex was detected by FITC-conjugated secondary antibody. Then, cells were stained with propidium iodide (PI; 50 μg/mL) and analyzed by flow cytometry. To avoid nonspecific fluorescence from dead cells, live cells were gated tightly using forward and side scatter. Apoptosis was quantitatively evaluated by double staining with annexin-V and PI, followed by flow cytometry analysis. For this purpose, cells were stained with FITC-conjugated annexin-V (Alexis Biochemicals) and PI, according to the manufacturer's instructions, and analyzed as previously detailed.17,18
Transfection experiments
Confluent HUVECs were detached and resuspended in the specified electroporation buffer to a final concentration of 7 × 105 cells/mL 0.2 μg plasmid DNA (EGFP-construct or luciferase-reporter plasmids) or 1 μg siRNA cocktails were mixed with 0.1 mL of cell suspension, transferred to electroporation cuvettes and nucleofected with the human HUVEC nucleofector kit (Amaxa, Cologne, Germany) using the program A-034 of the nucleofector device (Amaxa NucleofectorII apparatus). After electroporation, cells were immediately transferred to complete M199 medium and cultured in 24-well plates or T25 flasks at 37°C until analysis. Transfection efficiency was monitored in each experiment by scoring the percentage of fluorescent EGFP-positive cells. The cells were visualized by a fluorescence microscope (Eclipse TE200 inverted; Nikon, Tokyo, Japan; 10×/0.25 NA Ph1 DL objective) and images were acquired with a CoolSNAP camera (Photometrics, Tucson, AZ) using CoolSNAP version 1.1 software (Photometrics).
siRNAs were designed and manufactured by Ambion (Woodward Austin, TX) according to the current guidelines for effective knock-down by this method. The following siRNA cocktails were used: target 1 for p53, 5′-GGGAGUUGUCAAGUCUUGCtt-3′ (sense) and 5′-GCAAGACUUGACAACUCCCtc-3′ (antisense); target 2 for p53, 5′-GGGUUAGUUUACAAUCAGCtt-3′ (sense) and 5′-GCUGAUUGUAAACUAACCCtt-3′ (antisense). Ambion's Silencer Negative Control siRNAs (“scrambled siRNAs”) were used to demonstrate that the transfection did not induce nonspecific effects on gene expression. A cocktail of 3 different negative control siRNAs, each composed of a 19-bp scrambled sequence with 3′ dT overhangs, was used. The sequences have no significant homology to any known gene sequence from human, and they have been previously tested for the lack of nonspecific effects on gene expression (Ambion).
To generate the OPG-promoter reporter plasmid (pOPG-Luc), a 1172 bp fragment of the OPG gene 5 flanking region was amplified from HUVEC genomic DNA by polymerase chain reaction (PCR) using the following primers 5-AGATCTCTGGAGACATATAACTTGAACACT-TGGCCC-3 and 5-GAATTCTGTGGTCCCCGGAAACCTCAGG-3 (restriction sites are italicized). This fragment was cloned into BglII/EcoRI sites in the pMetLucReporter Vector (Clontech, Mountain View, CA). HUVECs were grown to 80% confluence and then transfected with 2 μg of either the pOPG-Luc or with the control vector (pMetLuc-Reporter, Clontech). After seeding, cells were allowed to recover before treatment with Nutlin-3 or TNF-α in the absence or presence of Nutlin-3 for 18 hours. The secreted luciferase was measured with the ready-to-glow secreted luciferase reporter system (Clontech) according to the manufacturer's instructions at 405 nm in a luminometer. In each experiment, a β-galactosidase-control plasmid under the control of a constitutive promoter (SV40), was cotransfected to normalize transfection efficiency (β-gal Reporter System, Clontech).
Real-time reverse transcription-PCR analysis
To validate efficiency and specificity of the siRNA-mediated gene knockdown, TP53 gene expression was quantitatively measured in HUVEC cultures at 48 hours after transfection. Moreover, expression levels of selected genes (P21 and OPG) were evaluated in HUVEC cultures treated with Nutlin-3 alone or TNF-α in the absence or presence of Nutlin-3 for 24 hours. For these purposes, RNA was isolated from HUVEC cultures using the Qiagen RNeasy mini kit (Qiagen, Hilden, Germany) according to the supplier's instructions, and the quality of the total RNA preparation was verified by agarose gel. Then, amplification for target gene expression was performed with a 7500 Real Time PCR System (Applied Biosystems, Foster City, CA) using SYBR Green based-technology and using the SuperArray Bioscience's RT2 Real-Time Gene Expression Assays, which include specific validated primer sets and PCR master mixes (SuperArray Bioscience, Frederick, MD). All samples were run in triplicate, and gene expression levels in treated cultures were compared with the matched control (untreated) samples.
Western blot analysis
HUVECs were lysed with a buffer containing 1% Triton X-100, Pefablock (1 mM), aprotinin (10 mg/mL), pepstatin (1 mg/mL), leupeptin (10 mg/mL), NaF (10 mM), and Na3VO4 (1 mM), as previously described.19 After protein determination performed with the Bradford assay (Bio-Rad, Richmond, CA), equal amounts of proteins for each sample were migrated in 10% SDS-PAGE, blotted onto nitrocellulose filters, and incubated with the following antibodies: anti-p53, anti-p21 (both from Santa Cruz Biotechnology, Santa Cruz, CA), and antitubulin (Sigma-Aldrich) as loading control. After incubation with peroxidase-conjugated antirabbit IgG, specific reactions were revealed with the ECL detection reagent (Amersham, Arlington Heights, IL). Densitometry values were estimated by the ImageQuant TL software (Amersham). Multiple film exposures were used to verify the linearity of the samples analyzed and to avoid saturation of the film.
Assay for NF-κB DNA binding and chromatin immunoprecipitation (ChIP) analysis
NF-κB induction by TNF-α in the absence or presence of Nutlin-3 was measured using the Trans-AM NF-κB p65 kit (Active Motif, Rixensart, Belgium), which measures the level of active form of NF-κB contained in cell extracts, able to specifically bind to an oligonucleotide containing the NF-κB consensus site (5′-GGGACTTTCC-3′), attached to a 96-well plate. Assays were performed in duplicates, according to the manufacturer's instructions. NF-κB DNA binding activity was determined as absorbance values measured using an Anthos 2010 ELISA reader (Anthos Labtec Instruments). Increase in fluorescence was linear over extract concentration.
Chromatin immunoprecipitation was performed using the ChIP-IT kit from Active Motif as described by the manufacturer. After the treatment of HUVEC with Nutlin-3 and TNF-α in the presence or absence of Nutlin-3, extracts were normalized according to their DNA concentration, and separate aliquots from each chromatin preparation were incubated overnight at 4°C with 3 μg of anti-p65 antibody, or with nonimmune rabbit IgG antibody. An aliquot was also retained and used as a loading control for the PCR (input control). PCR was performed with total DNA and immunoprecipitated DNA using OPG promoter-specific primers (5′-TGGAGGAGACACAAGCACAG-3 and 5′-GTGCAGAAAGCTCCAGGATT-3′) flanking the consensus κB sites (5-GGGRNNYYCC-3). The length of the amplified product was 271 bp. The PCR products were subjected to electrophoresis on 2% agarose gel.
Statistical analysis
The median, minimum, and maximum values were calculated for each group of data. For each set of experiments, values are reported as mean plus or minus SD. For selected experiments, results are reported as box plots showing the median, minimum, and maximum values and 25th to 75th percentiles. Data were analyzed by ANOVA and with the Mann-Whitney rank-sum test. Comparison of group means was performed by Bonferroni method. Statistical significance was defined as P less than .05.
Results
p53 knock-down increases the OPG release both in vivo and in vitro in endothelial cells
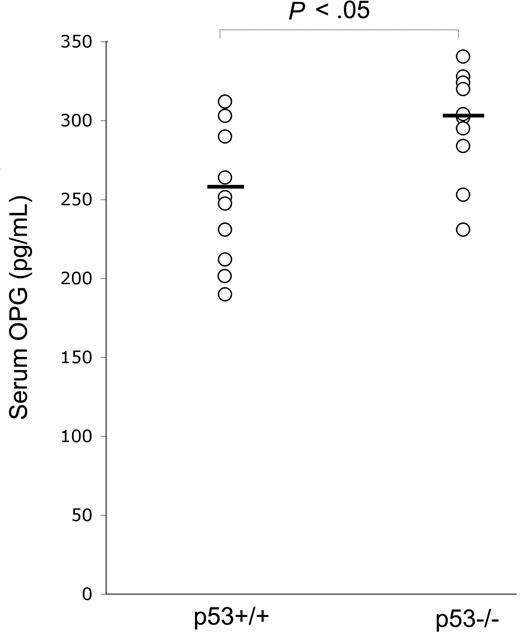
In view of the potentially important role of OPG in promoting tumor cell survival, in the first group of experiments, we have compared the serum levels of OPG between p53−/− and p53+/+ wild-type C57black mice. As shown in Figure 1, p53−/− mice, which are known to be more prone to develop a variety of cancers and in particular hematologic malignancies,20 showed significantly (P < .05) higher levels of serum OPG with respect to normal littermates. To exclude the possibility that OPG elevation might be the result of the presence of tumors in p53−/− mice, the determination of serum OPG was performed in 2-month-old mice, much earlier than the appearance of tumors, which typically develop 6 to 7 months after birth in p53−/− mice.20 Although we cannot exclude the presence of subclinical inflammation in p53−/− mice, the weight and the general conditions of these animals were indistinguishable with respect to p53 wild-type littermates.
p53 knock-out up-regulates serum OPG levels. Serum OPG was measured in 8-week-old p53−/− (n = 10) and p53+/+ (n = 10) mice. OPG level of each mouse is plotted in a scatter diagram; horizontal bars are medians.
p53 knock-out up-regulates serum OPG levels. Serum OPG was measured in 8-week-old p53−/− (n = 10) and p53+/+ (n = 10) mice. OPG level of each mouse is plotted in a scatter diagram; horizontal bars are medians.
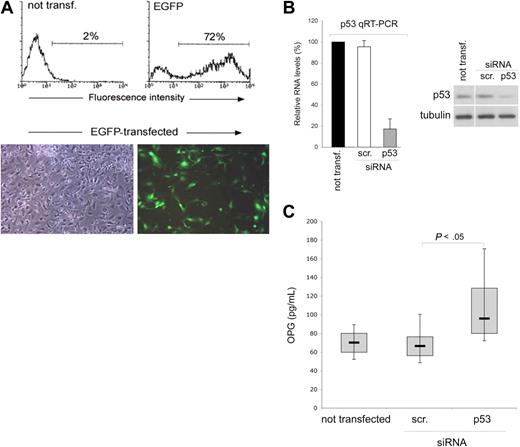
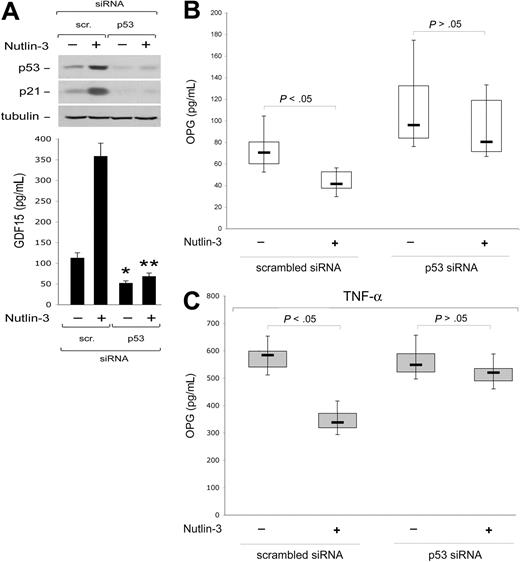
Serum OPG can be released by a variety of cell types, including vascular endothelial cells.2 Therefore, we next assessed the effect of p53 on OPG expression and release in endothelial cell cultures, using a gene knock-down approach to specifically attenuate p53 expression. Of note, the transfection procedure was very efficient in endothelial cells, with a range of transfected cells ranging between 65% and 80%, as evaluated by flow cytometry and fluorescence microscopy analyses of EGFP-transfected HUVEC (Figure 2A). With these optimal experimental conditions, HUVECs were transfected with siRNA specific for TP53 or with control scrambled siRNA and the efficiency and specificity of the p53 knock-down was documented at both the mRNA level as well as at protein level, at 48 hours after transfection (Figure 2B). Of note, knock-down of p53 resulted in significantly (P < .05) higher release of secreted OPG in the medium with respect to scrambled-transfected endothelial cells (Figure 2C).
p53 knock-down up-regulates OPG release in endothelial cultures. HUVECs were transfected with either an EGFP construct or with the indicated siRNA (scr., control scrambled). (A) Efficiency of transfection in each experiment was monitored by flow cytometric and microscopy analyses of EGFP-transfected HUVEC. Representative flow cytometric profiles and microscopy fields (original magnification ×10) of HUVEC cultures transfected with the EGFP-plasmid are shown. (B) Efficiency and specificity of the p53 knock-down were documented by real-time reverse transcription-PCR analyses (qRT-PCR) and Western blot. Results, from amplifications done in triplicate, are expressed as relative RNA levels calculated, after normalization for GAPDH, with respect to control not transfected cultures, which were set to 100. One of 3 Western blot experiments with similar results is shown. Error bars are SD. (C) OPG released in culture supernatants was measured in HUVEC cultures, either not transfected or transfected with the indicated siRNA. Results were obtained from 6 independent experiments, each performed in duplicate. Horizontal bars represent median, upper, and lower edges of box (75th and 25th percentiles); lines extending from box, 10th and 90th percentiles.
p53 knock-down up-regulates OPG release in endothelial cultures. HUVECs were transfected with either an EGFP construct or with the indicated siRNA (scr., control scrambled). (A) Efficiency of transfection in each experiment was monitored by flow cytometric and microscopy analyses of EGFP-transfected HUVEC. Representative flow cytometric profiles and microscopy fields (original magnification ×10) of HUVEC cultures transfected with the EGFP-plasmid are shown. (B) Efficiency and specificity of the p53 knock-down were documented by real-time reverse transcription-PCR analyses (qRT-PCR) and Western blot. Results, from amplifications done in triplicate, are expressed as relative RNA levels calculated, after normalization for GAPDH, with respect to control not transfected cultures, which were set to 100. One of 3 Western blot experiments with similar results is shown. Error bars are SD. (C) OPG released in culture supernatants was measured in HUVEC cultures, either not transfected or transfected with the indicated siRNA. Results were obtained from 6 independent experiments, each performed in duplicate. Horizontal bars represent median, upper, and lower edges of box (75th and 25th percentiles); lines extending from box, 10th and 90th percentiles.
Nutlin-3 down-regulates the release of OPG independently of its effects on cell-cycle progression
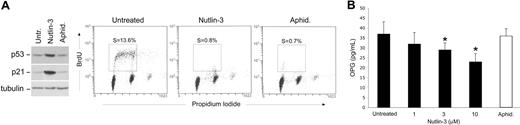
In the absence of cellular stress, the activation of p53 is repressed by murine double minute 2 (MDM2) gene. Recently, potent and selective small molecule inhibitors of p53-MDM2 interaction, the Nutlins, have been discovered.21 These compounds bind MDM2 in the p53 binding pocket with high selectivity and can release p53 from negative control, leading to effective stabilization of p53 and activation of the p53 pathway.21 Therefore, to further evaluate the involvement of p53 in modulating the release of OPG, endothelial cells were exposed to Nutlin-3. A rapid activation of the p53 pathway was induced after exposure to Nutlin-3, with a marked effect obtained with 10 μM of Nutlin-3, as documented in Western blot analysis by p53 protein accumulation and up-regulation of the cyclin-dependent kinase inhibitor p21 (Figure 3A), a relevant p53 target gene also in endothelial cells.22 Of note, Nutlin-3 treatment induced a dose-dependent decrease of the OPG release in HUVEC culture supernatants (Figure 3B). It has been clearly shown that p53 initiates several programs that ultimately arrest proliferation and prevent the generation of genetically altered cells. The spectrum of p53-based cell fate decisions ranges from a transient cell-cycle arrest enabling damage repair to an irreversible block of proliferation through senescence or apoptosis.14 Thus, to exclude the possibility that the decreased OPG release by endothelial cells treated with Nutlin-3 merely reflects a cytostatic effect, we have also tested the release of OPG in endothelial cells treated with Aphidicolin, a well-known molecule able to arrest the cell-cycle progression.23 Despite the comparable ability of Aphidicolin and Nutlin-3 to drastically decrease the percentage of endothelial cells entering the S-phase of the cell cycle (Figure 3A), Aphidicolin did not significantly affect the release of OPG by endothelial cells (Figure 3B). In addition, no significant increase of apoptosis over the background levels was observed in response to Nutlin-3. Taken together, these data exclude the possibility that the Nutlin-3-mediated down-regulation of OPG might merely reflect the cytostatic/cytotoxic activity of Nutlin-3 in endothelial cells.
Nutlin-3 down-modulates OPG release in endothelial cells. HUVECs were left untreated or treated with Nutlin-3 (10 μM) or Aphidicolin (1 μg/mL) for 24 hours. (A) Comparison between the effects of Nultin-3 and Aphidicolin on the p53 pathway and cell cycle. Levels of p53 and p21 proteins were assessed by Western blot analysis in cell lysates. Tubulin staining is shown as loading control. In parallel, cell cycle was analyzed by BrdU labeling and PI staining. In each panel, the rectangle represents the cells in S phase of the cell cycle, which have incorporated BrdU. Representative examples of 3 to 6 independent experiments of Western blot results and cell-cycle profiles analyzed by flow cytometry. (B) OPG levels in culture supernatants were assessed by ELISA. Data are mean plus or minus SD of results from 4 independent experiments, each performed in duplicate (*P < .05, compared with untreated).
Nutlin-3 down-modulates OPG release in endothelial cells. HUVECs were left untreated or treated with Nutlin-3 (10 μM) or Aphidicolin (1 μg/mL) for 24 hours. (A) Comparison between the effects of Nultin-3 and Aphidicolin on the p53 pathway and cell cycle. Levels of p53 and p21 proteins were assessed by Western blot analysis in cell lysates. Tubulin staining is shown as loading control. In parallel, cell cycle was analyzed by BrdU labeling and PI staining. In each panel, the rectangle represents the cells in S phase of the cell cycle, which have incorporated BrdU. Representative examples of 3 to 6 independent experiments of Western blot results and cell-cycle profiles analyzed by flow cytometry. (B) OPG levels in culture supernatants were assessed by ELISA. Data are mean plus or minus SD of results from 4 independent experiments, each performed in duplicate (*P < .05, compared with untreated).
Nutlin-3 efficiently counteracts the release of OPG in response to inflammatory cytokines
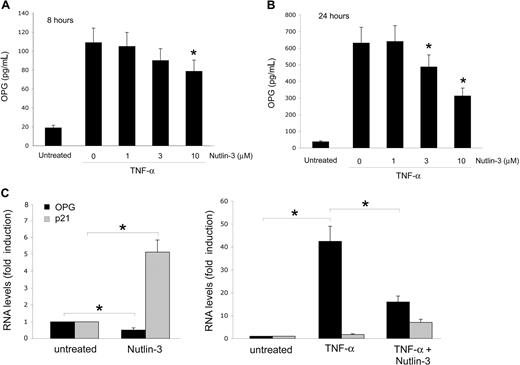
It has been clearly shown that chronic inflammation represents an important risk factor for promoting neoangiogenesis and cancer development,24 and that OPG can directly contribute to inflammation by promoting the adhesion of leukocytes to endothelial cells.25,26 Therefore, the next experiments were performed to investigate whether Nutlin-3 was able to interfere also with the release of OPG induced in response to the exposure of endothelial cells to inflammatory cytokines. As expected,26 the inflammatory cytokine TNF-α (10 ng/mL) up-regulated the release of OPG in HUVEC culture medium already after 8 hours (Figure 4A), and this effect further significantly increased after 24 hours (Figure 4B). The concomitant treatment with Nutlin-3 dose- and time-dependently down-regulated OPG release in the culture supernatants (Figure 4A,B), clearly indicating that p53 activation induced by Nutlin-3 was able to suppress the production and release of OPG also in response to inflammatory cytokines. It is noteworthy that, at the end of the experiments (24 hours of treatments), no significant differences were observed in the number of viable endothelial cells among the cultures treated with TNF-α (10 ng/mL) in the absence or presence of 10 μM Nutlin-3 (data not shown).
Nutlin-3 counteracts the induction of OPG by TNF-α. HUVECs were left untreated or treated with TNF-α in the presence or absence of the indicated concentrations of Nutlin-3. OPG levels in culture supernatants were assessed by ELISA after 8 (A) and 24 (B) hours of treatment. Data are expressed as mean plus or minus SD of results from 4 independent experiments, each performed in duplicate (*P < .05, compared with TNF-α alone). (C) The relative levels of OPG mRNA, and for comparison, also of p21 mRNA were determined by real-time reverse transcription-PCR. Results from amplifications done in triplicate are expressed as relative RNA levels calculated, after normalization for GAPDH, with respect to control untreated cultures, which were set to 1 (*P < .05).
Nutlin-3 counteracts the induction of OPG by TNF-α. HUVECs were left untreated or treated with TNF-α in the presence or absence of the indicated concentrations of Nutlin-3. OPG levels in culture supernatants were assessed by ELISA after 8 (A) and 24 (B) hours of treatment. Data are expressed as mean plus or minus SD of results from 4 independent experiments, each performed in duplicate (*P < .05, compared with TNF-α alone). (C) The relative levels of OPG mRNA, and for comparison, also of p21 mRNA were determined by real-time reverse transcription-PCR. Results from amplifications done in triplicate are expressed as relative RNA levels calculated, after normalization for GAPDH, with respect to control untreated cultures, which were set to 1 (*P < .05).
To elucidate whether Nutlin-3 down-regulated OPG at the transcriptional level, the steady-state OPG mRNA levels were next evaluated by quantitative RT-PCR. As shown in Figure 4C, Nutlin-3 significantly (P < .05) down-regulated the mRNA levels of OPG in endothelial cells, counteracting (P < .05) also the marked OPG mRNA up-regulation induced by TNF-α.
Nutlin-3 down-regulates OPG in a p53-dependent manner
Next, to establish whether Nutlin-3 down-regulated OPG expression/release through p53, endothelial cell cultures transfected with siRNA specific for TP53 were treated with Nutlin-3. As documented in Figure 5A, p53 knock-down specifically abrogated the ability of Nutlin-3 to increase both p53 and p21 protein levels in cell lysates as well as the release in culture supernatants of GDF15, a relevant p53 target gene that has been shown to accurately correlate with the degree of activation of wild-type p53.27 On the other hand, TP53 siRNA, but not scrambled siRNA, efficiently (P < .05) counteracted the inhibitory activity of Nutlin-3 on the spontaneous (Figure 5B) and TNF-α-induced (Figure 5C) OPG release, strongly suggesting that the inhibitory activity of Nutlin-3 on OPG release was mediated by p53.
Nutlin-3 down-modulates OPG release in a p53-dependent manner. HUVECs were transfected with control scrambled siRNA or p53 siRNA before treatment with Nultin-3 (10 μM) or TNF-α in the absence and presence of Nultin-3, as indicated. (A) Levels of p53 and p21 proteins were assessed by Western blot analysis of cell lysates; tubulin staining is shown as loading control. Representative examples of Western blot results of 3 to 5 independent experiments are shown. In parallel, activation of GDF15 p53 target gene was assessed by ELISA on supernatants from cell cultures (40 000 cells/mL). Data are mean plus or minus SD of the results from 3 to 5 independent experiments each performed in duplicate (*P < .05, compared with scrambled-transfected and left untreated; **P < .05, compared with scrambled-transfected and treated with Nutlin-3). OPG release in supernatants of the transfected cultures was assessed by ELISA. Results were obtained from 3 (C) or 7 (B) independent experiments, each performed in duplicate. Horizontal bars represent median, upper, and lower edges of box (75th and 25th percentiles); lines extending from box, 10th and 90th percentiles.
Nutlin-3 down-modulates OPG release in a p53-dependent manner. HUVECs were transfected with control scrambled siRNA or p53 siRNA before treatment with Nultin-3 (10 μM) or TNF-α in the absence and presence of Nultin-3, as indicated. (A) Levels of p53 and p21 proteins were assessed by Western blot analysis of cell lysates; tubulin staining is shown as loading control. Representative examples of Western blot results of 3 to 5 independent experiments are shown. In parallel, activation of GDF15 p53 target gene was assessed by ELISA on supernatants from cell cultures (40 000 cells/mL). Data are mean plus or minus SD of the results from 3 to 5 independent experiments each performed in duplicate (*P < .05, compared with scrambled-transfected and left untreated; **P < .05, compared with scrambled-transfected and treated with Nutlin-3). OPG release in supernatants of the transfected cultures was assessed by ELISA. Results were obtained from 3 (C) or 7 (B) independent experiments, each performed in duplicate. Horizontal bars represent median, upper, and lower edges of box (75th and 25th percentiles); lines extending from box, 10th and 90th percentiles.
Nutlin-3 inhibits TNF-α–induced NF-κB recruitment to the OPG gene promoter
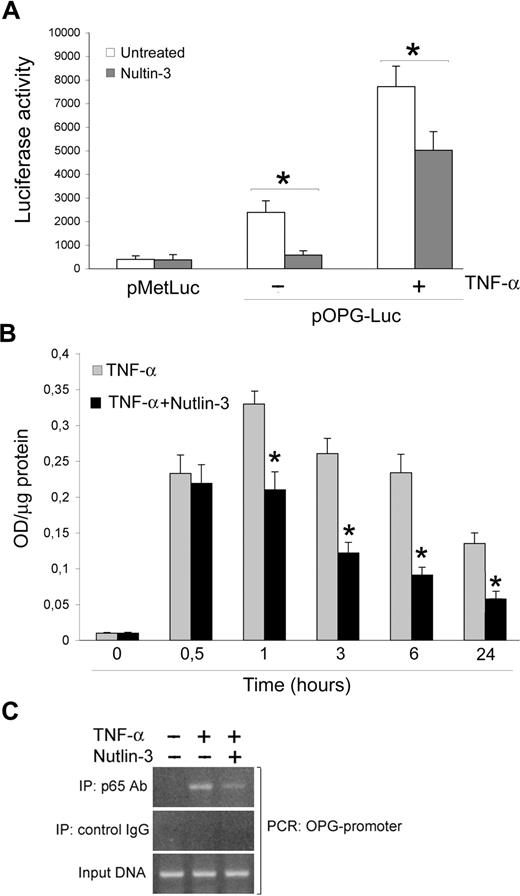
Taking into account that OPG is an NF-κB-dependent gene and that a molecular cross-talk between the p53 and NF-κB pathways has been established,28,29 the next experiments were carried out to evaluate whether Nutlin-3 down-regulated OPG expression/release by modulating the NF-κB pathway. This was initially investigated by assessing the modulation of the human OPG promoter, containing consensus NF-κB sites, in functional assays (Figure 6A). For this purpose, a promoter-reporter construct containing the OPG promoter upstream of a luciferase reporter gene (pOPG-Luc) was generated as described under “Transfection experiments.” HUVECs were transfected with this construct, or with the empty control vector, and then were treated with or without Nutlin-3 and TNF-α in the absence or presence of Nutlin-3 for 18 hours before assessing reporter gene activity. As shown in Figure 6A, pOPG-Luc was significantly expressed in HUVEC and was induced by TNF-α stimulation. Of note, Nutlin-3 treatment significantly (P < .05) reduced pOPG-Luc expression in both unstimulated and TNF-α stimulated cultures.
Nutlin-3 inhibits TNF-α–induced NF-κB recruitment to the OPG promoter. (A) HUVECs were transfected with pOPG-Luc or empty pMetLuc-Reporter vector. After recovery, cells were stimulated as indicated, and after 18 hours luciferase activity was assessed. Results, expressed as RLU, represent mean plus or minus SD of 3 independent experiments (*P < .05). (B) HUVECs were either left untreated or stimulated with TNF-α in the absence and presence of Nutlin-3. NF-κB–p65 DNA binding activity was determined at the indicated time points as absorbance values per microgram of cell lysate protein. Results are mean plus or minus SD of 3 independent experiments performed in duplicates (*P < .05, compared with TNF-α). (C) Cells were stimulated with TNF-α in the absence and presence of Nutlin-3. ChIP analysis was performed using anti-p65 or control IgG antibodies for immunoprecipitation followed by OPG promoter-specific PCR with primers flanking the consensus κB sites. The PCR products were subjected to electrophoresis on 2% agarose gel. These results are representative of 3 independent experiments.
Nutlin-3 inhibits TNF-α–induced NF-κB recruitment to the OPG promoter. (A) HUVECs were transfected with pOPG-Luc or empty pMetLuc-Reporter vector. After recovery, cells were stimulated as indicated, and after 18 hours luciferase activity was assessed. Results, expressed as RLU, represent mean plus or minus SD of 3 independent experiments (*P < .05). (B) HUVECs were either left untreated or stimulated with TNF-α in the absence and presence of Nutlin-3. NF-κB–p65 DNA binding activity was determined at the indicated time points as absorbance values per microgram of cell lysate protein. Results are mean plus or minus SD of 3 independent experiments performed in duplicates (*P < .05, compared with TNF-α). (C) Cells were stimulated with TNF-α in the absence and presence of Nutlin-3. ChIP analysis was performed using anti-p65 or control IgG antibodies for immunoprecipitation followed by OPG promoter-specific PCR with primers flanking the consensus κB sites. The PCR products were subjected to electrophoresis on 2% agarose gel. These results are representative of 3 independent experiments.
In parallel, we assessed the ability of NF-κB to bind to an oligonucleotide containing the κB consensus site (TransAM). For this purpose, DNA binding activity of p65/RelA was checked at various time points in TNF-α-treated cultures supplemented or not with Nutlin-3 (Figure 6B). A strong induction of NF-κB-DNA binding over untreated cultures was observed starting after 30 minutes, peaking at 1 hour of TNF-α treatment and showing a slow decline thereafter. The simultaneous addition of Nutlin-3 significantly (P < .05) reduced the TNF-α-induced p65/RelA DNA binding activity from 1 hour onward (Figure 6B).
In an additional set of experiments, a ChIP approach was used to examine whether the association of p65 with the OPG promoter induced by TNF-α-mediated NF-κB activation in human endothelial cells was affected by Nutlin-3. HUVECs were stimulated with TNF-α in the absence or presence of Nutlin-3 for 6 hours before immunoprecipitation of nuclear lysates with anti-p65 antibody. Analysis of immunoprecipitated DNA by PCR indicated that the treatment of endothelial cells with Nutlin-3 inhibited TNF-α-induced p65 recruitment to the OPG promoter (Figure 6C). The specificity of OPG promoter enrichment was confirmed by the absence of GAPDH promoter in p65 immunoprecipitates (data not shown).
Discussion
Several studies2,6-10 have demonstrated that recombinant OPG supports the survival of both normal and tumor-associated endothelial cells. In particular, OPG has been shown to mediate the integrin-dependent survival of serum-deprived endothelial cells6 as well as to prevent anoikis of epithelial cells.30 Engagement of integrins on the endothelial cell surface triggers an NF-κB–dependent generation of OPG that is essential for conveying the antiapoptotic activity of NF-κB in both endothelial and epithelial cells.6,30 Consistent with a central role of the NF-κB pathway in mediating the up-regulation of OPG, it has been demonstrated that OPG expression and release by endothelial cells are markedly up-regulated in response to inflammatory cytokines.31 Because these previous data support a potential biologic role for OPG in the development and/or maintenance of normal as well as of tumor-associated vasculature, our present demonstration, that activation of the p53 pathway by Nutlin-3 attenuates the expression and release of OPG by endothelial cells, is particularly noteworthy. The ability of Nutlin-3 to down-regulate the expression of gene products relevant for endothelial cell biology is not unprecedented. Indeed, 2 recent studies have demonstrated that Nutlin-3 is able to decrease the release of stromal derived factor-1/CXCL12 by normal fibroblasts32 as well as of vascular endothelial growth factor by normal endothelial cells.33 The relevance of these previous studies together with our present findings is underlined by the fact that normal and leukemic hematopoietic stem and progenitor cells are found adjacent to the vascular niches, represented by bone marrow endothelial cells,34 and that recent data support the role of the vascular niches also in initiating metastasis.35
It should also be emphasized that OPG significantly inhibited the proapoptotic activity of recombinant TRAIL in both multiple myeloma and prostate cancer cell lines, acting in an autocrine/paracrine manner.36,37 Thus, an inappropriate elevation of OPG at the bone marrow microenvironment might contribute to the escape of malignant multiple myeloma cells or metastatic prostate cancer cells from the killing by TRAIL. Consistent with this hypothesis, it has been recently demonstrated that the affinity of OPG for TRAIL is similar to that for RANKL.38 It is also interesting that not only endothelial cells but also bone marrow stromal cells increased the release of OPG in response to TNF-α (data not shown), suggesting that a chronic inflammation state may favor leukemogenesis by inducing the release of protective concentrations of OPG able to neutralize the proapoptotic activity of TRAIL. An additional mechanism, which may link OPG to hematologic malignancies, is its ability to favor the adhesion of both normal and leukemic cells to endothelial cells.25,26
Although the molecular mechanism involved in the down-regulation of OPG expression by Nutlin-3 requires further investigation, we could demonstrate that it was p53-dependent and that Nutlin-3 down-regulated the TNF-α–induced NF-κB activation in endothelial cells. In this respect, while our study was submitted for publication, it has been demonstrated that Nutlin-3 inhibits the NF-κB pathway also in a lung cancer cell model.29 In this respect, several studies have previously suggested that wild-type p53 acted as a repressor of NF-κB both in vitro and in vivo28,39 whereas mutated p53 potentiated the NF-κB transcriptional activity.40 How exactly p53 affects NF-κB remains an open question, although in line with previous studies,28,41 we have demonstrated that p53 interferes with the transcriptional activity of p65/RelA. It should also be underlined that the ability of Nutlin-3 to down-regulate OPG release was not confined to endothelial cells but was also observed when Nutlin-3 was added to bone marrow stromal cells (data not shown).
Thus, besides exerting a direct cytotoxic effect in various types of hematologic malignancies, including AML, multiple myeloma, B-cell chronic lymphocytic leukemia, and lymphomas, as well as in solid tumors,42-48 Nutlin-3 may also act indirectly against hematologic malignancies by attenuating the release of OPG, by endothelial and/or bone marrow stromal cells, in the bone marrow microenvironment.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the Italian Association for Cancer Research, the Italian Ministry of University and Research (PRIN and FIRB RBIN045LT8), and the Carife Foundation.
National Institutes of Health
Authorship
Contribution: G.Z., P.S., and F.C. designed and performed research, analyzed data, and wrote the paper; E.R., C.C., and M.G.d.I. performed research; A.R. and G.D.S. contributed experimental tools.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Paola Secchiero, Department of Morphology and Embryology, Human Anatomy Section, University of Ferrara, Via Fossato di Mortara 66, 44100 Ferrara, Italy; e-mail: secchier@mail.umbi.umd.edu.
References
Author notes
P.S. and F.C. contributed equally to this study.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal