Abstract
Combined factor V and factor VIII deficiency (F5F8D) is a rare, autosomal recessive coagulation disorder. F5F8D is genetically linked to mutations in the transmembrane lectin ERGIC-53 and its soluble interaction partner MCFD2. The ERGIC-53/MCFD2 protein complex functions as transport receptor of coagulation factors V and VIII by mediating their export from the endoplasmic reticulum (ER). Here, we studied a F5F8D patient who was found to be a compound heterozygote for 2 novel mutations in MCFD2: a large deletion of 8.4 kb eliminating the 5′UTR of the gene and a nonsense mutation resulting in the deletion of only 3 amino acids (ΔSLQ) from the C-terminus of MCFD2. Biochemical and structural analysis of the ΔSLQ mutant demonstrated impaired binding to ERGIC-53 due to modification of the 3-dimensional structure of MCFD2. Our results highlight the importance of the ERGIC-53/MCFD2 protein interaction for the efficient secretion of coagulation factors V and VIII.
Introduction
Combined deficiency of coagulation factors V and VIII (F5F8D, Online Mendelian inheritance in Man no. 227300) is a rare autosomal recessive bleeding disorder first described by Oeri et al in 1954.1 Individuals affected with this disorder present with a moderate bleeding tendency and plasma levels of factors V and VIII in the range of 5% to 30% of normal. Extensive genetic analysis of F5F8D patients linked the disorder to causative mutations in the 2 genes LMAN1 and MCFD2.2-7 LMAN1 encodes the transmembrane mannose-lectin ERGIC-53 (ER-Golgi intermediate compartment protein of 53 kDa),8 whereas MCFD2 is a soluble luminal protein with 2 calmodulin-like EF hand motifs.7 MCFD2 interacts with ERGIC-53 in a calcium dependent manner and the complex recycles between the endoplasmic reticulum (ER) and the ER-Golgi intermediate compartment (ERGIC).9 The ERGIC-53/MCFD2 complex is believed to capture factor V and factor VIII in the ER and to package the 2 coagulation factors into transport vesicles that mediate transport to the ERGIC. Indeed, chemical crosslinking identified a triple complex of ERGIC-53, MCFD2 and factor VIII.10 In the absence of the ERGIC-53/MCFD2 complex, the secretion of factor V and factor VIII is inefficient resulting in low plasma levels and bleeding
Study design
The study was approved by the Ethical Committee of the University Hospitals, Geneva, Switzerland.
Patient
The patient, a 10-year-old girl with factor V levels of 10% to 13% and factor VIII levels of 5% to 19%, is a single child of nonconsanguineous parents of South American origin. The patient suffers from easy bruising, epistaxis, and gingival bleeding. Her mother did not report any bleeding problem, nor did any other member of the maternal family. Her father had several episodes of epistaxis during childhood but apparently no major bleeding manifestation.
Mutation screening
Blood samples were obtained from the affected child and her mother after informed consent was obtained in accordance with the Declaration of Helsinki. The father was unavailable for analysis. Primers used for polymerase chain reaction (PCR) amplification and sequencing of LMAN1 have been previously described.6 The MCFD2 gene was analyzed by PCR amplification of the 4 exons and intron-exon junctions followed by sequencing. Primer sequences are available on request.
Southern blot analysis
Southern blot analysis was performed according to standard procedures. Genomic DNA (4 μg) from the patient, her mother, and 2 controls was digested with Sac I or HindIII. A 510-bp probe containing MCFD2 exon 1 and part of intron 1 was obtained by PCR using oligonucleotide primers: MCFD2×1F' 5′GGGGCGAAGCCGAGGAAGA3′ and MCFD2×1R: 5′CAGAGAGGGAATACAACAGG3′ and cloned using pDRIVE (Qiagen, Basel, Switzerland). The deleted allele was confirmed and sequenced by PCR using 2 forward primers: MCFD2delF2: 5′ATTGCTTGAGCCCAGAGCTCGAG3′ and MCFD2delF3: 5′CCAATGAATAGTAAGGTACTC3′ and the reverse primer MCFD2×1R.
ERGIC-53 interaction study
Cloning of pcDNA3[HA-MCFD2 WT] was described previously.9 pcDNA3[HA-MCFD2 ΔSLQ] was generated by changing Ser144 into a stop codon using PCR-mutagenesis. Constructs were transfected into HeLa cells (CCL-2, American Type Culture Collection, Manassas, VA) using FuGENE6 (Roche Applied Science, Rotkreuz, Switzerland). Forty-eight hours after transfection, cells were harvested, lysed, and subjected to immunoprecipitation using mouse monoclonal antibody G1/93 against human ERGIC-53 (ALX-804-602; Alexis, Lausen, Switzerland), covalently coupled to protein A Sepharose.9,11 Immunoblotting was performed with G1/93 and antihemagglutinin (HA; Covance, Princeton, NJ).
Circular dichroism spectroscopy
DNA fragments encoding MCFD2 WT and ΔSLQ were cloned into the pET-16b plasmid (N-terminal polyhistidine tag) and expressed in the Escherichia coli BL21(DE3) codonplus strain induced with 0.5 mM isopropyl β-D-thiogalactopyranoside. Polyhistidine-tagged proteins were purified from lysates with Ni2+-nitriltriacetic acid columns (GE Healthcare, Uppsala, Sweden), cleaved by incubation with factor Xa (Novagen, Madison, WI) and further purified using a Benzamidine Sepharose column (Amersham Biosciences, Uppsala, Sweden). Circular dichroism (CD) measurements were conducted on a Jasco J-725 spectropolarimeter (Jasco, Tokyo, Japan) at room temperature using protein samples of 0.15 mg/mL in 150 mM NaCl, 5 mM CaCl2, and 10 mM Tris-HCl, at pH 7.5.
Results and discussion
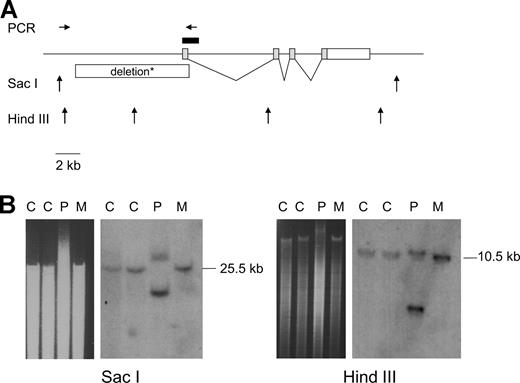
To identify the cause of F5F8D in a young patient, we screened the LMAN1 and MCFD2 genes for mutations. None were identified in LMAN1. Instead, the patient was found to be heterozygous for a novel nonsense mutation in MCFD2 exon 4 (c.431C > G; p.Ser144X), transmitted by the mother. Because no other mutation was identified by sequencing all MCFD2 exons (including the noncoding exon 1) and intron-exon junctions, we performed Southern blot analysis of genomic DNA digested with Sac I using a probe containing MCFD2 exon 1 and part of intron 1 (Figure 1A). For the mother and 2 controls, an expected band of 25.5 kb was detected (Figure 1B left). In contrast, for the patient, a smaller band of approximately 17 kb was detected in addition to the normal band (Figure 1B left, the patient's DNA sample (P) migrates more slowly than the samples from the controls (C) and from the patient's mother (M) so that the patient's normal band appears to be of greater size). Southern blot analysis with HindIII was also performed using the same probe. Again, a smaller band was detected for the patient, compatible with a deletion of approximately 8.5 kb from the 5′ of MCFD2 (Figure 1B right panel, one HindIII site is included in the deletion). This was confirmed by PCR amplification of the deleted allele followed by sequencing. The deletion eliminates 8.4 kb of DNA 5′ of MCFD2 including exon 1 which contains nearly the entire 5′UTR and is unlikely to result from a single recombination event because 2 small sequences of 55 bp (in the 5′ upstream region) and 96 bp (in intron 1) are retained in the deleted allele in inverted orientation. There is no doubt that this is a null mutation because it knocks out the MCFD2 promoter sequence. Notably, this is the first large deletion identified in a patient with F5F8D.
Identification of an 8.4-kb deletion upstream of MCFD2. (A) Schematic representation of the genomic region containing MCFD2 on human chromosome 2 (negative strand). Sac I and HindIII restriction sites are indicated by ↑, PCR oligonucleotide primers (not to scale) used to amplify and sequence the deleted allele are indicated by →←. The probe used for Southern blotting is shown as a black box. *The deletion eliminates 8.4 kb of DNA including exon 1 but retains 2 small sequences of 55 bp (in the 5′ genomic region, approximately 3.4 kb upstream of exon 1) and 96 bp (starting 9 bp inside intron 1) in inverted orientation. (B) Southern blot analysis with Sac I and HindIII. An image of the gel electrophoresis is shown to the left of each Southern blot, the patient's DNA sample (P) migrates more slowly than the samples from the controls (C) and from the patient's mother (M) so that the patient's normal band in both blots appears to be of greater size.
Identification of an 8.4-kb deletion upstream of MCFD2. (A) Schematic representation of the genomic region containing MCFD2 on human chromosome 2 (negative strand). Sac I and HindIII restriction sites are indicated by ↑, PCR oligonucleotide primers (not to scale) used to amplify and sequence the deleted allele are indicated by →←. The probe used for Southern blotting is shown as a black box. *The deletion eliminates 8.4 kb of DNA including exon 1 but retains 2 small sequences of 55 bp (in the 5′ genomic region, approximately 3.4 kb upstream of exon 1) and 96 bp (starting 9 bp inside intron 1) in inverted orientation. (B) Southern blot analysis with Sac I and HindIII. An image of the gel electrophoresis is shown to the left of each Southern blot, the patient's DNA sample (P) migrates more slowly than the samples from the controls (C) and from the patient's mother (M) so that the patient's normal band in both blots appears to be of greater size.
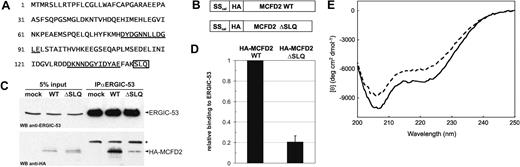
The remaining MCFD2 allele contains the novel nonsense mutation p.Ser144X. The effect of this mutation on the MCFD2 protein is comparatively mild because only the last 3 C-terminal amino acids are truncated. To determine the underlying molecular mechanism responsible for F5F8D, we analyzed if MCFD2 ΔSLQ can still bind ERGIC-53. To this end, HA-tagged versions of wildtype (WT) and ΔSLQ MCFD2 (Figure 2B) were transiently expressed in HeLa cells and coimmunoprecipitated with ERGIC-53. Although equally expressed as MCFD2 WT, MCFD2 ΔSLQ coprecipitated only minimally with ERGIC-53 (Figure 2C). Quantification revealed that the ΔSLQ mutation in MCFD2 reduced binding to ERGIC-53 by 80% compared with MCFD2 WT (Figure 2D). To examine if impaired binding to ERGIC-53 may be due to a structural defect, recombinant MCFD2 WT and ΔSLQ were expressed in E coli, purified, and analyzed by circular dichroism (CD). The CD spectrum of MCFD2 WT in the presence of Ca2+ is typical for an α-helical conformation with minima at around 207 and 222 nm. However, the CD spectrum of the MCFD2 ΔSLQ mutant showed a significantly lower α-helical content (Figure 2E). We conclude that the C-terminal ΔSLQ deletion perturbs the 3-dimensional structure of MCFD2 and as a consequence impairs binding to ERGIC-53 and causes F5F8D.
MCFD2 ΔSLQ displays impaired binding to ERGIC-53 due to a structural defect. (A) Amino acid sequence of MCFD2. EF-hand motifs are underlined and the ΔSLQ mutation is boxed. (B) Schematic representation of HA-tagged constructs. The endogenous signal sequence of MCFD2 was replaced by the signal sequence of calreticulin (SScal). (C) Cell lysates (5% input) and anti-ERGIC-53 immunoprecipitates (IPαERGIC-53) were probed by Western blotting (WB) using anti-ERGIC-53 and anti-HA antibodies. Because ERGIC-53 antibodies were covalently coupled to immunoprecipitation beads, only the antibody light chain is detectable (*). (D) ERGIC-53 coprecipitated HA-MCFD2 WT and ΔSLQ were quantified in 4 independent experiments and relative values are indicated. (E) Far UV CD spectra of recombinant MCFD2 (—) and MCFD2 ΔSLQ ( ). Each spectrum shows the average of 4 scans over the range 200 to 250 nm with step size of 0.1 nm and a bandwidth of 1.0 nm.
). Each spectrum shows the average of 4 scans over the range 200 to 250 nm with step size of 0.1 nm and a bandwidth of 1.0 nm.
MCFD2 ΔSLQ displays impaired binding to ERGIC-53 due to a structural defect. (A) Amino acid sequence of MCFD2. EF-hand motifs are underlined and the ΔSLQ mutation is boxed. (B) Schematic representation of HA-tagged constructs. The endogenous signal sequence of MCFD2 was replaced by the signal sequence of calreticulin (SScal). (C) Cell lysates (5% input) and anti-ERGIC-53 immunoprecipitates (IPαERGIC-53) were probed by Western blotting (WB) using anti-ERGIC-53 and anti-HA antibodies. Because ERGIC-53 antibodies were covalently coupled to immunoprecipitation beads, only the antibody light chain is detectable (*). (D) ERGIC-53 coprecipitated HA-MCFD2 WT and ΔSLQ were quantified in 4 independent experiments and relative values are indicated. (E) Far UV CD spectra of recombinant MCFD2 (—) and MCFD2 ΔSLQ ( ). Each spectrum shows the average of 4 scans over the range 200 to 250 nm with step size of 0.1 nm and a bandwidth of 1.0 nm.
). Each spectrum shows the average of 4 scans over the range 200 to 250 nm with step size of 0.1 nm and a bandwidth of 1.0 nm.
To date, 30 distinct mutations4-6,12-15 have been identified in LMAN1, accounting for approximately 75% of F5F8D patients, compared with 11 mutations identified in the MCFD2 gene.7,14-16 In LMAN1, no missense mutations have been identified which affect binding of either MCFD2 or factors V and VIII. In contrast, 2 missense mutations (Asp129Glu and Ile136Thr) in the second EF-hand domain of MCFD2 were shown to abolish binding to ERGIC-53 and cause F5F8D.7 No structural analysis was performed with these mutants. Here we show that the ΔSLQ missense mutation modifies the α-helical content of MCFD2 and impedes binding to ERGIC-53. Thus, our structural observations explain why ΔSLQ leads to a bleeding phenotype similar to a MCFD2 null mutation although ΔSLQ truncates only 3 residues near the putative calcium-binding domain of the second EF-hand. Collectively, our data clearly emphasize the importance of the second EF-hand of MCFD2 for protein conformation, interaction with ERGIC-53, and factor V and factor VIII transport.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Dr Michael Morris for helpful discussions. We are grateful to Luciana Palumbo for expert technical assistance.
This study was supported by a Swiss National Science Foundation Professorship to M.N.A. B.N. is supported by the Roche Research Foundation. Y.K. is a recipient of a Japan Society for the Promotion of Science Research Fellowship for Young Scientists. K.K. was supported in part by CREST project from the Japan Science and Technology Agency and by Grants-in-Aid from the Ministry of Education, Culture, Sports, Science, and Technology of Japan. H-P.H. is supported by the Swiss National Science Foundation and the University of Basel, Switzerland.
National Institutes of Health
Authorship
Contribution: M.N.A. directed the study and designed and performed the genetic analysis. B.N. and H-P.H designed and performed the study of the mutant MCFD2 in HeLa cells. Y.K., K.Y., and K.K. designed and performed the circular dichroism measurements of the recombinant MCFD2. F.B. and P. d.M. performed the clinical study. All authors contributed to writing the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marguerite Neerman-Arbez, Department of Genetic Medicine and Development, 1 Rue Michel Servet, 1211 Geneva, Switzerland; e-mail: Marguerite.Arbez@medecine.unige.ch.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal