Abstract
The identity and lineage potential of the cells that initiate thymopoiesis remain controversial. The goal of these studies was to determine, at a clonal level, the immunophenotype and differentiation pathways of the earliest progenitors in human thymus. Although the majority of human CD34+lin− thymocytes express high levels of CD7, closer analysis reveals that a continuum of CD7 expression exists, and 1% to 2% of progenitors are CD7−. CD34+lin− thymocytes were fractionated by CD7 expression and tested for lineage potential in B-lymphoid, T-lymphoid, and myeloid-erythroid conditions. Progressive restriction in lineage potential correlated with CD7 expression, that is, the CD7hi fraction produced T and NK cells but lacked B and myelo-erythroid potential, the CD7int (CD10+) fraction produced B, T, and NK cells, but lacked myelo-erythroid potential. The CD7− fraction produced all lymphoid and myelo-erythroid lineages and expressed HSC-associated genes. However, CD34+lin−CD7− thymocytes also expressed early T lymphoid genes Tdt, pTα, and IL-7Rα and lacked engraftment capacity, suggesting the signals that direct lymphoid commitment and corresponding loss of HSC function are rapidly initiated on arrival of HSC in the human thymus. Thus, differential levels of CD7 identify the progressive stages of lineage commitment in human thymus, initiated from a primitive CD7− lympho-myeloid thymic progenitor.
Introduction
It is generally accepted that the thymus does not contain a resident source of self-renewing cells and that progenitor cells from the bone marrow serve as the source of precursors for a lifetime of thymopoiesis.1 However, the nature of the cells that seed the thymus has been the subject of much debate. Although it is clear that lymphoid and possibly even T-lineage commitment can occur in the bone marrow, it is not known whether such commitment is required before thymic seeding. To examine this question, investigators have focused their attention on the isolation and analysis of candidate populations either in the bone marrow or in the thymus itself. The majority of the work has relied on murine transplant models to test the lineage potential and thymic engraftment of these populations. A long-held assumption that lymphoid-restricted progenitors generated in the bone marrow were responsible for thymopoiesis was supported by the identification in the murine marrow of a common lymphoid progenitor (CLP) with B-, NK-, and T-cell potential.2 Recent studies have suggested that more lineage-restricted bone marrow progenitors are also able to generate thymocytes and may be more efficient than CLP at doing so.3 Although mature B and myeloid cells can be found in low frequency in the thymus, the majority of thymocytes are T cells, further supporting the hypothesis that T-lineage commitment occurs before seeding of the thymus
Attempts to identify the earliest progenitors within the thymus have focused on the subsets within the CD4−CD8− (the so-called double negative, DN) population of thymocytes, which in mice is further divided into 4 stages based on CD44 and CD25 expression.4,5 Allman et al6 more recently identified a c-kithiIL7Rα− subset as the earliest T-lineage progenitor (ETP). This population vigorously repopulated the thymus after intrathymic transfer, yet generated some B cells and myeloid cells, suggesting that the ETP represents a less committed progenitor than the CLP.
Few studies exist that address the cell of origin in human thymopoiesis. CD34 has long been identified as a marker for hematopoietic stem and progenitor cells in human bone marrow and is also expressed on immature human thymocytes. Based on the finding that CD34 expression was higher on the CD1− fraction than on the CD1+ fraction of CD34+ thymocytes, Galy et al concluded that the CD34+CD1− cells are the most primitive population in the human thymus followed by CD34+CD1+, CD34+CD4+CD8−, and CD34+/−CD4+CD8α+cells.7 A more recent study by Weerkamp et al8 further explored the lineage potential of CD34+1a− cells finding that in addition to T cells, B-lymphoid, myeloid, and erythroid lineages could also be generated, albeit at low frequency, from this population.
CD7 has been widely used as a T-cell marker and is considered to be one of the earliest surface markers in T-cell ontogeny.9-11 Studies of the T-cell differentiation pathway in the human thymus concluded that, in contrast to bone marrow HSC, intrathymic CD34+ precursors coexpressed CD7.12,13 Schmitt et al14 found that CD3−CD34+CD7++ thymocytes were fully committed to the T-cell lineage, lacked myeloid differentiation capacity and responded to IL-7. The concept that CD7 expression and lymphoid commitment occur before migration to the thymus was further supported by the identification of CD7+ lymphoid-restricted progenitors in the fetal bone marrow15 and umbilical cord blood.16,17
The existence of thymocyte progenitors, which lack CD7 expression, has not been previously described. Here we demonstrate that, although CD7 is expressed on the majority of line-age negative human thymocytes that express CD34, a continuum of CD7 expression exists in the CD34+lin− cells, and a rare CD34+lin−CD7− subpopulation is consistently seen. The CD34+lin−CD7− cells are a subset of the more numerous CD34+CD1a− population and are highly enriched for single cells with both lymphoid (B, T, and NK) and myelo-erythroid potential. Clonal lineage specific assays and gene expression analysis reveal that CD7 expression in human CD34+lin− thymocytes can be used to identify stages of thymocyte commitment, from CD7− lympho-myeloid-erythroid progenitors to CD7intermediate CD10+(CD7intCD10+) multilymphoid (B/NK/T) progenitors and finally to CD7hi T/NK progenitors, which comprise the majority of human thymic CD34+ cells. Our data support the concept that the human thymus is directly seeded by circulating lympho-myeloid hematopoietic stem or progenitor cells, which on contact with the thymic microenvironment, rapidly lose engraftment and multilineage capacity and enter pathways committed to lymphopoiesis.
Methods
Thymus, peripheral blood, and cord blood samples were all collected as anonymous waste tissue under protocols reviewed by the Institutional Review Board at Childrens Hospital Los Angeles. Informed consent was obtained in accordance with the Declaration of Helsinki.
Cell isolation and flow cytometry
Discarded thymus tissue was obtained from patients undergoing cardiac surgery under a protocol approved by the Institutional Review Board at Childrens Hospital Los Angeles (Committee on Clinical Investigations). Patients ranged in age from 1 day to 9 years. A single-cell suspension was obtained by mechanically teasing apart the tissue in buffer (10% fetal bovine serum in phosphate-buffered saline) using 2 sterile needles, or by mechanically mincing the tissue using a pair of sterile scissors. Mononuclear cells were isolated by density gradient centrifugation over Ficoll-Paque TM Plus (Amersham BioSciences, Uppsala, Sweden) and then enriched for CD34+ cells by positive selection, using MAC MS+ separation columns and magnetic beads (Miltenyi Biotech, Auburn, CA). Cells were subsequently incubated with the following lineage specific fluorescein isothiocyanate (FITC)-labeled antibodies CD14, CD15, CD56, CD57 (BD Biosciences, San Jose, CA) and glycophorin A (Immunotech, Warrendale, PA), and allophycocyanin (APC)-labeled antibodies CD3 and CD19 (BD Biosciences) for fluorescence-activated cell sorting (FACS) of lineage negative cells.
CD34+lin− cells were then analyzed for coexpression of other cell- surface markers using CD1a FITC or R-phycoerythrin (PE; BD Pharmingen), CD4 FITC, peridiain chlorophyll protein (PerCP) or APC, CD25 PE, CD38 APC, CD44 APC, and CD10 PE-Cy7 (BD Biosciences), and IL-7Rα-PE (Immunotech). CD34+lin−CD7−, CD34+lin−CD7intCD10+ and CD34+lin−CD7hi were isolated by FACS after staining CD34+lin− cells with CD34 PerCP-Cy5.5 and CD7 PE (BD Biosciences) or CD34 PE and CD7 R-phycoerythrin-cyanine 5 (PE-Cy5; BD Biosciences, BD Pharmingen, respectively) or CD34 APC and CD7 FITC (BD Biosciences) and processed for phenotypic and/or functional analyses.
Normal (nonmobilized) peripheral blood was obtained from volunteer adult donors under a protocol approved by the Institutional Review Board at Childrens Hospital Los Angeles (Committee on Clinical Investigations). CD34+lin−CD7− cells were isolated from peripheral blood as described for thymus samples at the beginning of this section.
Analysis was performed either on a FACSCalibur or FACSVantage and cell isolation was performed on a FACSVantage or FACSAria (BD).
Gene-expression studies
RNA was extracted from FACS sorted cells using either RNA Stat-60 (Tel-Test, Friendswood, TX) or RNeasy Micro kit (Qiagen, Valencia, CA). RNA was reverse transcribed using the Omniscript RT kit (Qiagen), and the resulting cDNA was stored at −80°C for polymerase chain reaction (PCR) analysis. All PCR primer sequences were designed to span an intron. PCR primer sequences and PCR conditions used to detect expression of β2-microglobulin, Pu.1, GATA-3, pre–T alpha (pTα), Tdt, Pax-5, and IL-7Rα are described by Hao et al,16 and for SCL expression in Zhang et al.18 Primer sequences and PCR conditions for c-kit, CCR9, Notch 1, and IL2Rα are given in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). β2-microglobulin was used as a positive control for cDNA integrity in all cases.
In some instances, IL-7Rα gene expression at the single-cell level was analyzed by depositing FACS–sorted single cells from the desired population using the automated cell deposition unit of the FACSVantage directly into 1.5-mL microfuge tubes containing RNA Stat-60. RNA extraction was carried out as described by the manufacturer for larger cell numbers. cDNA produced from each cell was then split in half for the separate PCR detection of the IL-7Rα, as well as β2-microglolublin expression.
Real-time PCR for quantitative IL-7Rα and IL-2Rα expression was carried out using Taqman Gene expression assays consisting of predesigned probe and primer sets from Applied Biosystems (Foster City, CA; IL-7Rα, assay number Hs00233682_m1; and IL-2Rα, assay number Hs00907777_m1). β2-microglobulin was used as a control for cDNA integrity (assay number Hs00984230_m1).
Lineage-specific in vitro assays
Clonal myelo-erythroid assays.
To assay for the presence of clonogenic myelo-erythroid progenitors, that is, erythroid burst-forming units (BFU-Es), granulocyte-macrophage colony forming units (CFU-GMs) and granulocyte-erythroid, macrophage megakaryocyte (CFU-mix), approximately 200 to 500 freshly sorted cells were plated into 1.0-mL semisolid media (1% methylcellulose; MethoCultGFH4435; Stem Cell Technologies, Vancouver, BC). Colony forming units (CFU) were counted after 14 days and the mean of quadruplicates was recorded.
Cultures on OP9 stroma.
Cocultivation on the murine stromal line OP9 was used to test for B lymphoid, NK, and myelo-erythroid differentiation. Freshly sorted thymocytes (500-1500 cells) were seeded onto established nonirradiated OP9 stromal cells (American Type Culture Collection, Manassas, VA) in 96-well flat-bottomed plates. Cells were grown in a modified lymphoid medium (RPMI 1640, Irvine Scientific, Santa Ana, CA), 5% fetal calf serum screened for B-cell cultures (Cambrex Bio Science, Walkersville, MD), 50 μM 2-mercaptoethanol (Sigma-Aldrich, St Louis, MO), penicillin/streptomycin (Gemini Bio Products, Calabasas, CA), IL-7 (5 ng/mL, R&D Systems, Minneapolis, MN), Flt 3 ligand (FL, 5 ng/mL, R&D), and thrombopoietin (TPO, 5 ng/mL, R&D). For the first 5 to 7 days of culture, cells were stimulated with erythropoietin (EPO, 1 unit/mL, Amgen, Thousand Oaks, CA) plus or minus IL-3 (10 ng/mL; R&D). Every 7 days thereafter, half the medium was replaced with fresh medium that contained no EPO or IL3.
T-cell differentiation cultures.
Cocultivation on the OP9 stromal line engineered to express Delta-like-1 ligand (OP9-DL1, a generous gift from Dr Juan Carlos Zuniga-Pflucker, Department of Immunology, University of Toronto, Sunnybrook and Women's College Health Sciences Center, Toronto, ON)19 was used to test differentiation of progenitors to T and NK lineages. Freshly sorted thymocytes (500-1500 cells) were seeded onto established, nonirradiated OP9-DL1 stromal cells in 96-well plates. Cells were grown in a medium that was a mixture of the modified lymphoid medium containing IL-7, TPO, and FL described under “Cultures on OP9 stroma,” and conditioned medium in equal proportions. Every 7 days, half the medium was replaced with fresh medium. Conditioned medium was prepared by plating mechanically disassociated unfractionated human thymus cells in 75-cm2 flasks containing IMDM (Cambrex Bio Science), 15% horse serum, 15% fetal bovine serum (Omega Scientific, Tarzana, CA), 50 μM 2-mercaptoethanol (Sigma), penicillin/streptomycin (Gemini Bio Products, West Sacramento, CA), and glutamine (Gemini Bio Products). Nonadherent cells were discarded after 5 to 7 days, and this process was repeated for 14 to 28 days until foci of adherent cells developed. Medium was replaced every 7 days. When the cells grew to be 90% to 100% confluent, the conditioned culture medium was collected and spun at 2095g for 20 minutes at 4°C and filtered through a 0.45-μm syringe filter and frozen in aliquots at −80°C for later use. All T-cell cultures were analyzed in parallel with negative control wells that contained OP9-DL1 stroma and identical culture medium but not seeded thymocyte populations.
Transplantation studies in NOD/SCID/β2mnull mice.
The capacity of thymocyte populations to engraft bone marrow was tested in NOD/SCID/β2mnull mice (Jackson Laboratories, Bar Harbor, ME) with 3 different transplantation approaches: intravenously in nonirradiated neonates, intravenously in sublethally irradiated (300 cGy 2 hours prior) 6- to 8-week-old adults, and intrafemorally in irradiated 6- to 8-week-old week adults. The following populations were transplanted: 2 × 103 to 6 × 103 CD34+CD38− cells from cord blood (positive control), 2 × 103 to 104 CD34+lin−CD7− thymocytes, and 0.4 × 105 to 0.4 × 106 CD34+lin−CD7hi thymocytes.
After 2 to 6 weeks, mice were killed by inhalation of a mixture of 75% CO2/25% O2, and cells were harvested from marrow and thymus. Human cell engraftment was assessed by staining with human leukocyte common antigen CD45 (BD Biosciences) and analysis on the FACSCalibur.
Results
Phenotypic analysis of human thymus reveals a CD7− subset within the CD34+lin−CD1a− population
Analysis of samples of thymus from 38 infants and children up to the age of 9 years revealed that approximately 0.5% of the mononuclear fraction expressed CD34, a marker of hematopoietic stem and progenitor cells. The immunophenotype of these thymic progenitors was further analyzed after enrichment of CD34+ cells using magnetic bead isolation and flow cytometry and depletion of cells that expressed the lineage markers of T cells (CD3), NK cells (CD56, CD57), B cells (CD19), myeloid cells (CD14, CD15), and erythroid cells (glycophorin A).
Consistent with previous publications,7,8 we found that CD1a− cells constituted 30% to 50% of all CD34+ thymocytes (Figure 1A). CD7 was expressed at high levels in the majority of CD34+CD1a+ and CD34+CD1a− lineage-depleted thymocytes (> 95% of each; Table 1). However, a rare population of CD34+lin− cells expressed low or undetectable levels of CD7; these CD34+CD7− cells were approximately 10-fold enriched in the CD34+lin−CD1a− fraction compared with the CD1a+ fraction (Table 1, P < .04). Thus, the CD34+lin−CD1a− population, previously identified7 as containing the most primitive progenitor in the human thymus, can be further subdivided on the basis of CD7 expression.
CD34+ thymocytes show variable expression levels of CD1a and CD7. (A) Expression of CD1a in CD34+ enriched thymocytes. (B) The CD34 enriched, lineage-depleted fraction of thymus may be subfractionated into CD34+lin−CD7hi cells, and much rarer CD34+lin−CD7int and CD34+lin−CD7− subpopulations (shown are percentages of each gated population within CD34+lin− cells). (C) Most of the CD34+lin−CD7int population expresses CD10 and CD45. Numbers on plots are percentages of total cells.
CD34+ thymocytes show variable expression levels of CD1a and CD7. (A) Expression of CD1a in CD34+ enriched thymocytes. (B) The CD34 enriched, lineage-depleted fraction of thymus may be subfractionated into CD34+lin−CD7hi cells, and much rarer CD34+lin−CD7int and CD34+lin−CD7− subpopulations (shown are percentages of each gated population within CD34+lin− cells). (C) Most of the CD34+lin−CD7int population expresses CD10 and CD45. Numbers on plots are percentages of total cells.
CD7− thymocytes are enriched in the CD34+lin−CD1a− subset
| . | % in CD34+lin− CD1a− population (X) . | % in CD34+lin− CD1a+ population (Y) . | Ratio X/Y . |
|---|---|---|---|
| CD7− (n=6) | 0.39 ± 0.38 | 0.04 ± 0.06 | 9.8* |
| CD7int (n=6) | 1.54 ± 1.3 | 0.79 ± 0.59 | 2.0 |
| CD7hi (n=6) | 98.0 ± 0.5 | 98.9 ± 0.2 | 1.0 |
| . | % in CD34+lin− CD1a− population (X) . | % in CD34+lin− CD1a+ population (Y) . | Ratio X/Y . |
|---|---|---|---|
| CD7− (n=6) | 0.39 ± 0.38 | 0.04 ± 0.06 | 9.8* |
| CD7int (n=6) | 1.54 ± 1.3 | 0.79 ± 0.59 | 2.0 |
| CD7hi (n=6) | 98.0 ± 0.5 | 98.9 ± 0.2 | 1.0 |
Data are means plus or minus SE. The frequency of CD7− cells was 9.8-fold higher in the CD34+CD1a− population (X) compared with the frequency of CD7− cells in the CD34+CD1a+ population (Y). The frequency of CD7int and CD7hi cells was similar in X and Y.
P < .04.
As shown in Figure 1B, a continuum of CD7 expression was observed within the CD34+ thymocyte progenitors, with the CD34+lin−CD7int/neg subpopulation representing a mean plus or minus SE of 1.6% (± 0.23%) of all CD34+lin− cells, (n = 38). No correlation could be seen between the frequency of each subpopulation with the postnatal age of the thymus. It was hypothesized that the progressive development of CD7 expression may reflect the earliest differentiation stages of human thymocytes during steady-state thymopoiesis. The 3 populations, CD34+CD7−, CD34+CD7int, and CD34+CD7hi, were therefore isolated by FACS and processed for phenotypic and functional analyses.
Immunophenotypic analysis of CD34+lin−CD7− and CD34+lin−CD7hi subsets
CD38, a marker of differentiation, has been used to discriminate between hematopoietic stem and progenitor cells in human bone marrow and cord blood.20,21 The frequency of immunophenotypically primitive CD34+CD38− cells was significantly higher in the CD7− (45%) than in the CD7hi fraction (4%) of CD34+lin− thymocytes (P < .01), suggesting enrichment of primitive cells in the CD34+CD7− fraction (Table 2; Figure S1).
Coexpression of markers in CD7− vs CD7hi subpopulations of CD34+lin− thymocytes
| Phenotype . | Frequency of phenotype in CD34+lin− subpopulations, % . | P . | |
|---|---|---|---|
| CD34+lin−CD7−, mean ± SEM . | CD34+lin−CD7hi, mean ± SEM . | ||
| CD38− | 45.0 ± 12.7 | 3.9 ± 1.73 | <.01* |
| CD44+ | 98.9 ± 0.52 | 78.1 ± 15.39 | <.25 |
| IL-7Rα+ | 38.8 ± 21.0 | 54.8 ± 23.71 | <.6 |
| IL-2Rα+ | 45.3 ± 9.9 | 5.0 ± 4.6 | <.01* |
| Flt3+ | 1.5 ± 0.6 | 16.3 ± 8.5 | <.13 |
| IL-2Rβ+ | 40.0 ± 15.6 | 79.5 ± 12.1 | <.09 |
| IL-2Rγ+ | 67.3 ± 8.1 | 98.7 ± 0.6 | <.02* |
| Phenotype . | Frequency of phenotype in CD34+lin− subpopulations, % . | P . | |
|---|---|---|---|
| CD34+lin−CD7−, mean ± SEM . | CD34+lin−CD7hi, mean ± SEM . | ||
| CD38− | 45.0 ± 12.7 | 3.9 ± 1.73 | <.01* |
| CD44+ | 98.9 ± 0.52 | 78.1 ± 15.39 | <.25 |
| IL-7Rα+ | 38.8 ± 21.0 | 54.8 ± 23.71 | <.6 |
| IL-2Rα+ | 45.3 ± 9.9 | 5.0 ± 4.6 | <.01* |
| Flt3+ | 1.5 ± 0.6 | 16.3 ± 8.5 | <.13 |
| IL-2Rβ+ | 40.0 ± 15.6 | 79.5 ± 12.1 | <.09 |
| IL-2Rγ+ | 67.3 ± 8.1 | 98.7 ± 0.6 | <.02* |
There were 3 to 5 independent samples for each marker.
Statistically significant difference in the cell surface marker frequency between CD7− and CD7hi subsets.
Most of the cells in both the CD7− and CD7hi subpopulations expressed CD44, a cell adhesion/homing molecule22 (Table 2), and CD45RA (data not shown). A total of 30% to 40% of both populations also expressed low levels of CD4 (data not shown). IL-7Rα was expressed at similar levels in both CD34+lin−CD7− and CD34+lin−CD7hi cells (38% and 54%, respectively). Interestingly however, IL-2Rα expression was significantly different between the 2 populations; IL-2Rα was expressed in approximately 40% of CD34+lin−CD7− cells but was essentially absent in the CD34+lin−CD7hi fraction (P < .01, Table 2; Figure S1). IL-2Rβ and IL-2Rγ were expressed in both CD34+lin−CD7− and CD34+lin−CD7hi cells.
Gene-expression profiles of CD7− and CD7hi subsets
To explore further how CD7 expression relates to the developmental stages of thymopoiesis, expression of selected stem cell– and lymphoid-associated genes was studied by semi-quantitative RT-PCR in CD34+lin−CD7− and CD34+lin−CD7hi populations (Figure 2). Genes normally expressed in bone marrow HSC, namely, SCL, Pu.1, and c-kit, were also expressed in the CD7− thymic progenitors. SCL and c-kit were down-regulated in the CD7hi population, a pattern consistent with lineage commitment.18
Expression of stem cell– and lymphoid-associated genes in CD34+lin− subsets from thymus and blood. Equal numbers of CD34+lin−CD7hi and CD34+lin−CD7− cells from thymus and CD34+lin−CD7− cells from normal peripheral blood (PBL) were isolated by FACS and subjected to RT-PCR analysis to define the expression levels of selected stem- and lymphoid-associated genes. All results were confirmed at least twice using independently sorted cells from different thymuses.
Expression of stem cell– and lymphoid-associated genes in CD34+lin− subsets from thymus and blood. Equal numbers of CD34+lin−CD7hi and CD34+lin−CD7− cells from thymus and CD34+lin−CD7− cells from normal peripheral blood (PBL) were isolated by FACS and subjected to RT-PCR analysis to define the expression levels of selected stem- and lymphoid-associated genes. All results were confirmed at least twice using independently sorted cells from different thymuses.
Several genes associated with early T-cell development, although present in the CD7− thymic fraction, were up-regulated in the CD34+CD7hi cells. These included GATA-3, pre–T-cell receptor alpha (pTα), terminal deoxyribonucleotide transferase (TdT), and Notch-1. In contrast, expression of Pax5, a regulator of B- versus T- lineage fate, was barely detectable in both subsets. RAG-1, which plays a fundamental role in lymphocyte development by initiating the rearrangement of antigen-binding domains in B- and T-cell receptors, was undetectable in both subsets. CCR9, a chemokine receptor that plays a role in regulating both the migration of progenitor cells to the thymus and the migration of developing thymocytes within the thymus,23 was strongly expressed in both CD34+CD7− and CD34+CD7hi populations.
Consistent with the FACS analysis (Table 2), IL-7Rα message was detected in both CD7− and CD7hi subsets, whereas the IL2-Rα message was confined to the CD7− fraction. Real-time quantitative PCR demonstrated similar levels of expression of IL-7Rα in each population and down-regulation of IL-2Rα in CD7hi by 2.7-fold. To explore the heterogeneity of IL-7Rα expression within the CD7− fraction, RT-PCR of single CD34+CD7− cells was performed. IL-7Rα transcripts were detected in at least 35% of CD34+CD7− cells (n = 4 experiments each with 10 single cells analyzed per experiment).
To investigate the possibility that the CD7− fraction detected in the thymus represented contamination by circulating HSC, gene expression was also studied in CD34+lin−CD7− cells from the peripheral blood (Figure 2). CD34+lin−CD7− cells from the peripheral blood expressed SCL, c-kit, Pu.1, Notch-1, and GATA-3, similar to their thymic CD34+lin−CD7− counterparts. However, in contrast to their thymic analogs, the peripheral blood CD7− progenitors did not express the lymphoid-associated genes TdT, pre-Tα, IL-7Rα, IL-2Rα, or CCR9. Thus, although the thymic CD34+lin−CD7− population expressed stem cell–associated genes, they were nevertheless distinguishable from lympho-myeloid progenitors and stem cells in the peripheral blood by the expression of genes associated with early T-lymphoid commitment.
CD7 expression reflects the lineage potential of CD34+lin− cells
To determine whether CD7 expression defines progenitors with differing lineage potential, CD34+lin− thymocytes were isolated into 3 subsets (CD7−, CD7int, and CD7hi cells), and assayed in various lineage specific in vitro assays. It should be noted that the lineage potential of the entire CD34+lin−CD7int population was not consistent in these assays unless this immunophenotype was combined with coexpression of CD10 (Figure 1C). Thus, the full immunophenotype of the population referred to as CD7int population tested in the in vitro assays was CD34+lin−CD10+CD7int.
When cultured in methylcellulose medium, the CD7− progenitor population contained all types of clonogenic myeloid and erythroid progenitors (BFU-E, CFU-mix, and CFU-GM) at high frequency (3.3 ± 0.6%), whereas the CD7int and CD7hi populations were almost devoid of myelo-erythroid potential (P < .001, Table 3). More primitive myelo-erythroid progenitors (long-term culture-initiating cells, ie, cells able to generate CFU-C after up to 7 weeks of stromal coculture) were also detected only in the CD7− fraction (data not shown).
The CD34+lin−CD7− subpopulation contains a high frequency of myelo-erythroid progenitors
| . | % BFU-E . | % CFU mix . | % CFU-GM . | % total CFU . |
|---|---|---|---|---|
| CD34+lin−CD7−, mean ± SE (n = 24) | 0.6 ± 0.2 | 1.1 ± 0.3 | 1.6 ± 0.3 | 3.3 ± 0.6* |
| CD34+lin−CD7intCD10+, mean ± SE (n = 5) | 0 | 0.02 ± 0.02 | 0.02 ± 0.02 | 0.05 ± 0.05 |
| CD34+lin−CD7hi, mean ± SE (n = 12) | 0 | 0 | 0.02 ± 0.01 | 0.02 ± 0.01 |
| . | % BFU-E . | % CFU mix . | % CFU-GM . | % total CFU . |
|---|---|---|---|---|
| CD34+lin−CD7−, mean ± SE (n = 24) | 0.6 ± 0.2 | 1.1 ± 0.3 | 1.6 ± 0.3 | 3.3 ± 0.6* |
| CD34+lin−CD7intCD10+, mean ± SE (n = 5) | 0 | 0.02 ± 0.02 | 0.02 ± 0.02 | 0.05 ± 0.05 |
| CD34+lin−CD7hi, mean ± SE (n = 12) | 0 | 0 | 0.02 ± 0.01 | 0.02 ± 0.01 |
Cells of each immunophenotype were isolated by FACS from fresh thymus and plated in methylcellulose medium.
n indicates number of experiments.
P < .001, significant difference between frequency of total CFU in CD7− and the other two populations.
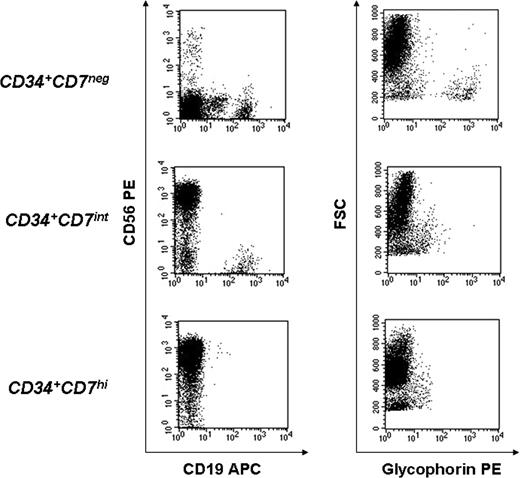
B-lymphoid and T-lymphoid potential was compared among the 3 populations by coculture on OP9 and OP9-DL1 stromal lines, respectively. On OP9 stroma, in conditions that support both B- lymphoid, NK-cell, and myelo-erythroid differentiation, only CD7− and CD7int cells were able to generate CD19+ B cells, but all 3 populations generated CD56+ NK cells (Figure 3). Myelo-erythroid potential was again shown to be exclusive to the CD7− population by the production of glycophorin A+ erythroid cells (Figure 3) and CD66b+ granulocytes (not shown) in the OP9 cultures. T cells were produced from all 3 subpopulations, when cultured on OP9-DL1 stroma, as shown by CD4 and CD8 expression (Figure 4A) and confirmed by pTα expression (Figure 4B), although T-cell production was most rapid and robust from the CD7hi subpopulation. All 3 populations were capable of generating CD7+ cells (Figure 4A). Thus, full lymphoid and myeloid potential was identified in the CD7− fraction, whereas the CD7int population gave rise to all lymphoid (B, T, and NK) cells but no myelo-erythroid cells, and the CD7hi fraction of CD34+ cells possessed only T and NK potential.
Cocultivation on OP9 stroma reveals that lineage potential of CD34+lin− thymocytes is related to CD7 expression. Approximately 1500 cells from each population (CD34+lin−CD7−, CD34+lin−CD7intCD10+, and CD34+lin−CD7hi) were isolated by FACS, cocultured on OP9 stroma in FL, TPO, IL-7, and EPO and analyzed at 2 to 4 weeks for expression of erythroid (glycophorin A) and lymphoid (CD19, CD56) markers to evaluate the lineage potential of each subset. Shown is FACS analysis of one representative experiment from a total of 4 independent experiments, using different thymus samples.
Cocultivation on OP9 stroma reveals that lineage potential of CD34+lin− thymocytes is related to CD7 expression. Approximately 1500 cells from each population (CD34+lin−CD7−, CD34+lin−CD7intCD10+, and CD34+lin−CD7hi) were isolated by FACS, cocultured on OP9 stroma in FL, TPO, IL-7, and EPO and analyzed at 2 to 4 weeks for expression of erythroid (glycophorin A) and lymphoid (CD19, CD56) markers to evaluate the lineage potential of each subset. Shown is FACS analysis of one representative experiment from a total of 4 independent experiments, using different thymus samples.
Immunophenotypic analysis of OP9-DL1 cocultures generated from CD34+lin−CD7−, CD34+lin−CD7intCD10+, and CD34+lin−CD7hi cells. Approximately 1500 cells from each population were isolated by FACS and cocultured with OP9/DL1 stroma and TPO, IL-7, and FL. Cells in culture were analyzed (A) by FACS at 2 to 4 weeks for expression of T-lineage markers (CD4, CD8) and CD7 and (B) by RT-PCR for pTα expression. Shown are data from one representative experiment of a total of 7 independent experiments for FACS analysis, and 2 independent experiments for RT-PCR, using different thymus samples.
Immunophenotypic analysis of OP9-DL1 cocultures generated from CD34+lin−CD7−, CD34+lin−CD7intCD10+, and CD34+lin−CD7hi cells. Approximately 1500 cells from each population were isolated by FACS and cocultured with OP9/DL1 stroma and TPO, IL-7, and FL. Cells in culture were analyzed (A) by FACS at 2 to 4 weeks for expression of T-lineage markers (CD4, CD8) and CD7 and (B) by RT-PCR for pTα expression. Shown are data from one representative experiment of a total of 7 independent experiments for FACS analysis, and 2 independent experiments for RT-PCR, using different thymus samples.
Clonal analysis of lineage potential
Clonal assays were performed to determine whether the lympho-myeloid potential seen in the CD7− population reflected the presence of individual multipotent, clonogenic cells. As these assays do not detect all lineages simultaneously and continuously from the same clone, a combination of “switch” assays was used to assign lineage potential to each clone. Single CD34+lin−CD7− thymic progenitors were plated into 96-well plates and cultured initially on OP9 stroma. The cloning efficiency of CD7− cells under these conditions was 19.4% plus or minus 2.8% (mean ± SE). FACS analysis of the clones generated on OP9 stroma was first used to reveal B, NK, myeloid, and erythroid potential. Where possible, clones were also replated/switched into CFU-C assay (to measure the generation of clonogenic myelo-erythroid cells), OP9-DL1 assay (to measure the generation of T and NK cells), and/or analyzed by RT-PCR for glycophorin (erythroid), pre-Tα (T-cell), and Pax-5 (B-cell) expression. The percentage of clones with both lymphoid and myeloid (and/or erythroid) potential, as determined by one or more of these assays, ranged from 13% to 39% (n = 7 experiments; Table 4). Figure 5 demonstrates the multilineage (B, T, NK, and myeloid) potential of 2 different CD34+CD7− clones shown by a combination of FACS (Figure 5A) and RT-PCR analysis at different stages of each clone's development (Figure 5B).
Multilineage potential of single CD34+lin−CD7− thymocytes
| Experiment no. . | No. of all clones/total cells plated (% plating efficiency) . | Clones with both lymphoid and myeloid potential (% of all clones) . |
|---|---|---|
| 1 | 37/384 (9.6) | 6 (16.2) |
| 2 | 13/84 (15.5) | 3 (23.0) |
| 3 | 42/192 (21.9) | 15 (35.7) |
| 4 | 18/96 (18.8) | 4 (22.2) |
| 5 | 32/96 (33.3) | 12 (37.5) |
| 6 | 15/96 (15.6) | 2 (13.3) |
| 7 | 41/192 (21.4) | 16 (39.0) |
| Mean ± SE | 19.4 ± 2.8 | 26.7 ± 4.0 |
| Experiment no. . | No. of all clones/total cells plated (% plating efficiency) . | Clones with both lymphoid and myeloid potential (% of all clones) . |
|---|---|---|
| 1 | 37/384 (9.6) | 6 (16.2) |
| 2 | 13/84 (15.5) | 3 (23.0) |
| 3 | 42/192 (21.9) | 15 (35.7) |
| 4 | 18/96 (18.8) | 4 (22.2) |
| 5 | 32/96 (33.3) | 12 (37.5) |
| 6 | 15/96 (15.6) | 2 (13.3) |
| 7 | 41/192 (21.4) | 16 (39.0) |
| Mean ± SE | 19.4 ± 2.8 | 26.7 ± 4.0 |
Data from 7 independent experiments analyzing individual CD34+lin−CD7− cells cocultured on OP9 stroma. Lympho-myeloid clones were recorded if markers of both lymphoid (ie, B and/or NK and/or T cell) and myeloid (ie, myeloid and/or erythroid) differentiation were identified by either FACS and/or RT-PCR.
Lineage analysis of clones generated from individual CD34+lin−CD7− thymocytes. Clones A and B were generated from different single CD34+lin−CD7− cells isolated by FACS, and cultured on OP9 stroma. (A) FACS of each clone harvested from OP9 stroma analyzing B (CD19) and erythroid (glycophorin; top) and NK (CD56 vs CD7) potential (middle). The same clones were replated into methylcellulose medium and generated CFU, which were analyzed by FACS for myeloid and erythroid potential (CD66b vs glycophorin; bottom). (B) RT-PCR analysis of clones A and B (at a different time point to FACS analysis) reveals the expression of T lymphoid (pTα), erythroid (glycophorin), and/or B lymphoid (Pax 5) genes. In these assays, the lineages produced from each clones fluctuate over time in culture (eg, clone B has CD19+ cells at one time but is negative for Pax 5 later in culture). Thus, lineage potential is assigned based on either FACS or RT-PCR marker expression at any time during the life of the clone.
Lineage analysis of clones generated from individual CD34+lin−CD7− thymocytes. Clones A and B were generated from different single CD34+lin−CD7− cells isolated by FACS, and cultured on OP9 stroma. (A) FACS of each clone harvested from OP9 stroma analyzing B (CD19) and erythroid (glycophorin; top) and NK (CD56 vs CD7) potential (middle). The same clones were replated into methylcellulose medium and generated CFU, which were analyzed by FACS for myeloid and erythroid potential (CD66b vs glycophorin; bottom). (B) RT-PCR analysis of clones A and B (at a different time point to FACS analysis) reveals the expression of T lymphoid (pTα), erythroid (glycophorin), and/or B lymphoid (Pax 5) genes. In these assays, the lineages produced from each clones fluctuate over time in culture (eg, clone B has CD19+ cells at one time but is negative for Pax 5 later in culture). Thus, lineage potential is assigned based on either FACS or RT-PCR marker expression at any time during the life of the clone.
CD34+lin−CD7− lympho-myeloid thymocytes do not engraft immune-deficient mice
The lympho-myeloid potential and gene expression profile of the CD34+lin−CD7− population is consistent with a hematopoietic stem-cell population present in the thymus. The capacity of this population to re-engraft the bone marrow was therefore tested by transplanting cells into immune deficient mice. Transplantation was performed using both intravenous and intra-marrow infusions into sublethally irradiated adult and neonatal NOD/SCID/β2mnull mice, a xenogeneic model that allows mostly B and some myeloid differentiation but does not permit thymic engraftment.
Although positive control cells (2 × 103 to 6 × 103 CD34+CD38− cells from umbilical cord blood) consistently engrafted the bone marrow and produced human B cells in all animals, no convincing evidence of engraftment of any lineage was found using the same or higher numbers of either CD34+CD7− (up to 5-fold higher) or CD34+CD7hi thymocytes (up to 500-fold higher than CD34+CD38− cord blood cells transplanted). Thus, although lympho-myelo-erythroid lineage potential is still present, CD34+CD7− cells within the thymus have lost marrow repopulating capacity, further evidence that they represent a stage of resident, non–self-renewing multipotent progenitors rather than circulating HSC.
Discussion
Much of the conceptual framework for how normal thymopoiesis is maintained throughout life has been developed by extrapolating from experiments that interrogate the ability of defined donor populations to generate thymocytes after transplantation in immune-deficient and/or irradiated recipients. Using these approaches, lymphoid committed progenitors such as circulating T-cell progenitors, CLP-1 and CLP-2, early lymphocyte progenitors, and multipotent progenitors in the murine bone marrow or blood have all been shown to possess T-cell potential and, to various degrees, the ability to engraft the thymus of irradiated recipients.2,3,24-28 It is assumed that these progenitor cells are first generated from HSC during normal hematopoiesis, and that, after some degree of lymphoid commitment in the bone marrow, they migrate to and seed the thymus during steady-state hematopoiesis. The experimental transplantation of HSC also generates long-term, donor-derived thymopoiesis; but because of the relative delay in T-cell production, it has been presumed that HSCs do not directly engraft the thymus but rather must first undergo some level of differentiation in the marrow. The question that remains to be answered is which of the various T-cell “competent” progenitors are the predominant source of thymopoiesis during homeostasis. Clearly, several cell types have the potential to initiate thymopoiesis in certain settings. The relative contribution of each population to thymopoiesis may be determined more by the ability of each to migrate to the thymus and their generative capacity within the normal thymic environment than the extent of prethymic lineage commitment.29
In addition to the nonphysiologic factors at play in most experimental transplantation models, these approaches are particularly problematic for human studies, which must rely on xenogeneic models in which homing and engraftment mechanisms may not be normal. In addition, primitive stem and progenitor cells are likely to lose the ability to re-engraft in a transplant setting rapidly after becoming resident in the thymic microenvironment. We thus elected to identify the cellular origins of human thymopoiesis using in vitro assays, combined with clonal analysis and gene expression, to determine the developmental relationship of discrete thymic progenitor subsets.
The combination of CD34 and CD1a expression has been previously reported to be useful for delineating the pattern of human thymic progenitor differentiation, with CD34+CD1a− cells representing the most immature thymocytes during fetal and adult life on the basis of high CD34 expression and TCR rearrangement studies.7,8,14,30,31 The present study shows that the CD34+CD1a− population is heterogeneous in terms of CD7 expression, with approximately 97% of the cells being CD34+CD7hi and the remainder being CD34+CD7 int/neg. Previous studies in fetal thymus have not described the CD34+lin−CD7− phenotype but rather have reported that all thymic progenitors express CD7.30,32 Recently, Haddad et al15 reported that the fetal bone marrow contains CD34hiCD45RAhiCD7+ cells, which represent the precursors of the CD34+CD1a− population in the fetal thymus. Although lymphoid-restricted CD7-expressing progenitors are also present in umbilical cord blood,16,17 their frequency in the bone marrow declines rapidly before birth and their contribution to postnatal thymopoiesis remains unclear.
Clonal analysis is critical to differentiate between mixtures of lineage committed cells and true multipotent progenitors. Although other studies have described multiple lineages in the murine thymus,6 specific populations have not been rigorously examined at the clonal level.33 In our experiments, cloning efficiency was at least 30-fold higher for myelo-erythroid progenitors and almost 100-fold higher for B-cell progenitors in CD34+CD7− cells than has been reported for the CD34+CD1a− human thymocyte population.8 We thus propose that the multilineage progenitors previously reported are restricted to the rare CD7− subset of the CD34+CD1a− population.
Several features mark the CD34+lin−CD7− thymocytes as direct descendants of either HSC or multipotent progenitors that have migrated from the bone marrow. Immunophenotypically, the CD7− subset of CD34+ cells was enriched by more than 10-fold for CD38− cells and most expressed c-kit and CD44. Gene expression patterns previously described in human HSC from peripheral blood and bone marrow (Pu.1, SCL, c-kit) were also found in thymic CD34+lin−CD7− cells. However, gene-expression studies also demonstrated important differences between CD34+lin−CD7− in the thymus and HSC populations in the blood, supporting the contention that at least some of the CD7− cells identified in the thymus were distinct from HSC trafficking through the thymic circulation. We propose that the expression of genes, such as IL-7Rα, pTα, and Tdt, mark the rapid induction of lymphoid commitment in bone marrow–derived HSCs or multipotent progenitor populations soon after they migrate to and seed the thymus. Immunophenotypic analysis and single-cell gene expression uncovered heterogeneity even within the CD7− population, which may reflect CD7− cells in different stages of transition after arrival in the thymus. The lack of bone marrow engraftment in NOD/SCID/ β-2mnull mice is consistent with a rapid loss of HSC function, possibly secondary to alteration of homing receptors, on migration into the thymus and under the influence of the thymic milieu. However, these conclusions must be tempered by the potential limitations of the xenogeneic system and the possibility that rare self-renewing CD7− cells circulating through the thymus were undetectable in these assays. Interestingly, Schwarz et al34 have described multipotent progenitors in murine bone marrow that efficiently generate thymopoiesis and, similar to the human CD7− cells, express CCR9. Thus, an alternative explanation for the lack of engraftment potential is that the CD34+CD7− cells in the thymus are derived directly from circulating multipotent progenitors that have lost self-renewal potential before migration from the marrow.
Previous studies of CLP in umbilical cord blood showed that onset of CD7 expression on CD34+CD38− cells heralds commitment to lymphoid lineages.16,17 CD10 expression on human bone marrow progenitors has been shown to have a similar correlation with lymphoid commitment.35 Of note, CD34+CD38−CD7+ cord blood cells coexpress CD10.16 In the studies of thymus reported here, although myelo-erythroid potential was progressively lost with CD7 expression, we found that neither CD7 nor CD10 expression alone was sufficient to define lymphoid restriction. However, by combining CD10 expression with intermediate CD7 expression, a multilymphoid progenitor with B, T, and NK but without myeloid or erythroid potential was identified. A further increase in CD7 expression on the CD34+ population was associated with the loss of B-cell potential and retention of T and NK potential. Of course, as with all immunophenotypic approaches to lineage tracking, cell-surface phenotypes do not prove that a progenitor-successor relationship exists between populations with different lineage potential. Nevertheless, to our knowledge, no other studies in mouse or human thymopoiesis have provided evidence that lymphoid commitment pathways normally confined to the bone marrow and cord blood might also occur within the thymus.
Surprisingly, IL-2Rα (CD25) was expressed in approximately 30% to 50% of CD34+CD7− cells and was down-regulated after further differentiation in the CD34+CD7hi population. Thus, based on these data, many (though not all) of the most immature progenitors in the human thymus are CD44+CD25+. In contrast, the DN1 subset, thought to represent the most primitive murine thymocytes, has a CD44+CD25− phenotype. Our data show that in human thymus the CD44+CD25− population incorporates the CD34+CD7hi population, which possesses only T and NK potential. Whether this represents a true species difference in thymocyte phenotype is unclear. As the assumption has long been that DN1 represent the most immature thymocytes, murine studies of thymic seeding cells have usually focused on this fraction. Of note, in an early study, Wu et al36 identified a primitive murine thymic population with B- and T-cell potential that was immunophenotypically similar to bone marrow HSC and that expressed moderate levels of CD4; thus, this population would not be included in a typical DN fractionation. Similarly, the lineage antibody cocktail used in the murine ETP studies did not include CD4.6,29 As the cloning efficiency for myeloid progenitors reported in ETP was very low (0.6%), it is likely that this population may also be functionally heterogeneous. Interestingly, both the CD34+lin−CD7− and CD34+lin−CD7int human thymocytes described here also expressed low but nevertheless readily detectable levels of CD4; thus, neither population can be defined as a DN subset. A functional role, if any, of CD25 (the α chain of the IL-2 receptor) on the immature thymocytes is yet to be elucidated. However, the presence of the β and γ chains of the IL-2 receptor on the CD34+CD7− population raises the possibility that the receptor is functional. The expression of CD25 in this population thus indicates either a species difference in the immunophenotypes of primitive thymocytes or the need to reexamine the application of the DN immunophenotype in staging murine thymopoiesis. At the very least, these data suggest that caution is warranted in applying the classic murine phenotypes to human studies of thymopoiesis.
In conclusion, the combination of lympho-myeloid potential and gene expression patterns normally associated with HSC suggests that human CD34+lin−CD7− thymocytes are derived directly from marrow-derived HSC. However, the coexpression of early lymphoid-specific markers on the CD34+lin−CD7− thymocytes, combined with the loss of engraftment capacity, demonstrates that differentiation is rapidly induced in these multilineage cells by the microenvironment of the human thymus. Using the differential expression of CD7 on thymic progenitors, regulation of the earliest stages of differentiation within the human thymus can now be studied.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the CHLA cardiothoracic surgery team for their invaluable assistance in supplying thymus samples for analysis and to Kaiser Permanente Sunset for their generous assistance with Cord Blood collections.
This work was supported by National Institutes of Health grants RO1-HL077912 and PO1-HL073104 (G.M.C.) and the Las Madrinas Foundation Endowment.
National Institutes of Health
Authorship
Contribution: Q.-L.H. performed FACS analysis, sorting, and culture of cells; A.A.G. performed cell isolation and RT-PCR and wrote the manuscript; J.Z. performed cell isolation and RT-PCR; L.B. performed flow cytometric sorting; E.Z. performed flow cytometric sorting; X.W. performed in vivo transplantation experiments; M.P. performed flow cytometic analysis; S.G. performed care and breeding of ID mice; and G.C. supervised the project and wrote manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Gay M. Crooks, Division of Research Immunology, Childrens Hospital Los Angeles, MS 62, 4650 Sunset Blvd, Los Angeles, CA 90027; email: gcrooks@chla.usc.edu.





This feature is available to Subscribers Only
Sign In or Create an Account Close Modal