Abstract
We searched for JAK2 exon 12 mutations in patients with JAK2 (V617F)-negative myeloproliferative disorders. Seventeen patients with polycythemia vera (PV), including 15 sporadic cases and 2 familial cases, carried deletions or duplications of exon 12 in circulating granulocytes but not in T lymphocytes. Two of the 8 mutations detected were novel, and the most frequent ones were N542-E543del and E543-D544del. Most patients with PV carrying an exon 12 mutation had isolated erythrocytosis at clinical onset, unlike patients with JAK2 (V617F)-positive PV, most of whom had also elevations in white blood cell and/or platelet counts. Both patients with familial PV carrying an exon 12 mutation had an affected sibling with JAK2 (V617F)-positive PV. Thus, several somatic mutations of JAK2 exon 12 can be found in a myeloproliferative disorder that is mainly characterized by erythrocytosis. Moreover, a genetic predisposition to acquisition of different JAK2 mutations is inherited in families with myeloproliferative disorders.
Introduction
The identification of a unique gain-of-function mutation of the Janus kinase 2 (JAK2) gene in patients with chronic myeloproliferative disorders1-5 has provided clinicians with a very useful diagnostic tool. In fact, the JAK2 (V617F) mutation can be detected in approximately 95% of patients with polycythemia vera (PV), and in more than half of those with essential thrombocythemia (ET) or primary myelofibrosis (PMF).1,6-8 While only a minority of sporadic patients with PV do not carry JAK2 (V617F), the proportion of familial cases of PV that are negative is definitely higher.9,10 Moreover, most patients with idiopathic erythrocytosis do not carry JAK2 (V617F) in circulating granulocytes.11
The JAK2 (V617F) mutation involves the exon 14, which encodes a part of the JH2 auto-inhibitory domain of the JAK2 kinase. Scott and coworkers12 recently described mutations of JAK2 exon 12 in JAK2 (V617F)-negative patients with PV or idiopathic erythrocytosis. Their findings have been confirmed by other studies.13-17 Using cellular models, it has been shown that exon 12 mutations can activate pathways associated with erythropoietin signaling, and in a murine model one of these mutations resulted in a myeloproliferative phenotype.12
In this work, we searched for exon 12 mutations in patients with JAK2 (V617F)-negative myeloproliferative disorders.
Methods
We studied 147 patients with JAK2 (V617F)-negative myeloproliferative disorders. These investigations were approved by the local ethics committees of Pavia, Basel, and Vienna: the procedures followed were in accordance with the Helsinki Declaration of 1975, as revised in 2000, and samples were obtained from subjects after written informed consent was obtained in accordance with the Declaration of Helsinki. The 2001 World Health Organization (WHO) criteria18 were applied for the diagnosis of PV and ET, while the Italian consensus criteria were used for the diagnosis of PMF.19 Of the 147 patients reported in this work, 26 had sporadic PV, 11 had familial PV,9,10 75 had sporadic ET, and 35 had sporadic PMF.
The isolation of granulocytes and T lymphocytes was performed as previously described.6,20 Quantitative real-time polymerase chain reaction (qRT-PCR)–based allelic discrimination assays6,7 were used for the detection of the JAK2 (V617F) mutation.
For exon 12 mutation screening, primers were designed to amplify a region of 453 base pairs (bp) containing the 128 bp of the exon 12 as described in Document S1 (available on the Blood website; see the Supplemental Materials link at the top of the online article). Direct sequencing was performed using the Big Dye Terminator v1.1 Cycle Sequencing kit (Applied Biosystems, Foster City, CA) and an ABI Prism 3130 Genetic Analyzer (Applied Biosystems).
Results and discussion
Of the 147 patients with JAK2 (V617F)-negative myeloproliferative disorders, 17 carried mutations of exon 12 in circulating granulocytes. Specifically, exon 12 mutations were detected in 15/26 sporadic cases and in 2/11 familial cases of PV. All positive patients met the revised WHO criteria21 for PV. Demographic and hematologic characteristics of these patients, and details of their mutations are reported in Table 1. A comparison between patients with sporadic PV who carried a JAK2 exon 12 mutation and those who did not is reported in Table S1.
Clinical and hematologic characteristics at diagnosis for the 17 patients carrying a JAK2 exon 12 mutation.
| UPN . | Sex/age . | Clinical diagnosis . | Hemoglobin level, g/L . | WBC count, 109/L . | PLT count, 109/L . | Serum Epo, mU/mL . | Palpable splenomegaly . | JAK2 exon 12 granulocytes . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA alteration . | Predicted amino acid change . | Pattern on sequencing* . | ||||||||
| PaS39A3V | M/80 | PV | 185 | 11.8 | 514 | <2 | Yes | 1627-1632del6 | E543-D544del | Het |
| PaS33A4V | M/40 | PV | 238 | 6.8 | 308 | <2 | No | 1606-1638dup33 | V536-I546dup11 | Het |
| PaS90A6V | F/47 | PV | 191 | 7.3 | 261 | <2 | No | 1624-1629del6 | N542-E543del | Het |
| PaS25A4V | F/35 | PV | 170 | 10.0 | 274 | <2 | No | 1608-1640dup33 | F537-I546dup10+F547L | Hom |
| PaS116A6V | M/53 | PV | 220 | 5.8 | 366 | 3.2 | No | 1624-1629del6 | N542-E543del | Het |
| PaS1A7V | F/76 | PV | 172 | 6.1 | 788 | 3.5 | No | 1622-1627del6 | R541-E543delinsK | Het |
| PaS23A5V | F/78 | PV | 175 | 4.6 | 339 | <2 | No | 1627-1632del6 | E543-D544del | Het |
| PaS6A95V | M/47 | PV | 207 | 6.0 | 360 | 3.0 | No | 1624-1629del6 | N542-E543del | Het |
| PaS1A5V | F/53 | Familial PV | 167 | 4.2 | 306 | 3.6 | No | 1622-1627del6 | R541-E543delinsK | Het |
| PaS12A0V | M/58 | Familial PV | 230 | 5.7 | 144 | <2 | No | 1624-1629del6 | N542-E543del | Het |
| Ba002 | F/46 | PV | 226 | 6.0 | 215 | <2 | Yes | 1620-1621del2,1626-1629del4 | I540-E543delinsMK | Het |
| Ba041 | M/43 | PV | 214 | 7.5 | 297 | 17.2 | No | 1611-1616del6 | F537-K539delinsL | Het |
| Ba138 | F/69 | PV | 194 | 8.6 | 102 | <2 | No | 1613-1615del3, A1616T | H538-K539delinsL | Het |
| Ba166 | F/46 | PV | 170 | 9.0 | 956 | <2 | No | 1627-1632del6 | E543-D544del | Het |
| Ba221 | F/36 | PV | 171 | 9.3 | 689 | 3.0 | Yes | 1627-1632del6 | E543-D544del | Het |
| Vi0064 | F/64 | PV | 199 | 11.2 | 700 | <2.5 | No | 1627-1632del6 | E543-D544del | Het |
| Vi0327 | F/70 | PV | 177 | 5.7 | 253 | ND | No | 1627-1632del6 | E543-D544del | Het |
| UPN . | Sex/age . | Clinical diagnosis . | Hemoglobin level, g/L . | WBC count, 109/L . | PLT count, 109/L . | Serum Epo, mU/mL . | Palpable splenomegaly . | JAK2 exon 12 granulocytes . | ||
|---|---|---|---|---|---|---|---|---|---|---|
| DNA alteration . | Predicted amino acid change . | Pattern on sequencing* . | ||||||||
| PaS39A3V | M/80 | PV | 185 | 11.8 | 514 | <2 | Yes | 1627-1632del6 | E543-D544del | Het |
| PaS33A4V | M/40 | PV | 238 | 6.8 | 308 | <2 | No | 1606-1638dup33 | V536-I546dup11 | Het |
| PaS90A6V | F/47 | PV | 191 | 7.3 | 261 | <2 | No | 1624-1629del6 | N542-E543del | Het |
| PaS25A4V | F/35 | PV | 170 | 10.0 | 274 | <2 | No | 1608-1640dup33 | F537-I546dup10+F547L | Hom |
| PaS116A6V | M/53 | PV | 220 | 5.8 | 366 | 3.2 | No | 1624-1629del6 | N542-E543del | Het |
| PaS1A7V | F/76 | PV | 172 | 6.1 | 788 | 3.5 | No | 1622-1627del6 | R541-E543delinsK | Het |
| PaS23A5V | F/78 | PV | 175 | 4.6 | 339 | <2 | No | 1627-1632del6 | E543-D544del | Het |
| PaS6A95V | M/47 | PV | 207 | 6.0 | 360 | 3.0 | No | 1624-1629del6 | N542-E543del | Het |
| PaS1A5V | F/53 | Familial PV | 167 | 4.2 | 306 | 3.6 | No | 1622-1627del6 | R541-E543delinsK | Het |
| PaS12A0V | M/58 | Familial PV | 230 | 5.7 | 144 | <2 | No | 1624-1629del6 | N542-E543del | Het |
| Ba002 | F/46 | PV | 226 | 6.0 | 215 | <2 | Yes | 1620-1621del2,1626-1629del4 | I540-E543delinsMK | Het |
| Ba041 | M/43 | PV | 214 | 7.5 | 297 | 17.2 | No | 1611-1616del6 | F537-K539delinsL | Het |
| Ba138 | F/69 | PV | 194 | 8.6 | 102 | <2 | No | 1613-1615del3, A1616T | H538-K539delinsL | Het |
| Ba166 | F/46 | PV | 170 | 9.0 | 956 | <2 | No | 1627-1632del6 | E543-D544del | Het |
| Ba221 | F/36 | PV | 171 | 9.3 | 689 | 3.0 | Yes | 1627-1632del6 | E543-D544del | Het |
| Vi0064 | F/64 | PV | 199 | 11.2 | 700 | <2.5 | No | 1627-1632del6 | E543-D544del | Het |
| Vi0327 | F/70 | PV | 177 | 5.7 | 253 | ND | No | 1627-1632del6 | E543-D544del | Het |
All T-cell DNA was wild-type. Reference ranges are as follows Hb, 130 to 170 g/L (males), 120 to 160 g/L (females); WBC count, 4 to 11 × 109/L; PLT, 100 to 400 × 109/L; serum Epo, 5 to 30 mU/mL.
UPN indicates unique patient number (Pa, Pavia; Ba, Basel; Vi, Vienna); PV, polycythemia vera; Epo, erythropoietin; ND, not determined; Het, heterozygous; and Hom, homozygous.
Analysis of heights of wild-type and mutant peaks showed proportions of mutant alleles ranging from approximately 10% to approximately 90% of total alleles.
Twelve of the 17 patients with PV carrying a JAK2 exon 12 mutation had isolated erythrocytosis at presentation, with WBC counts less than or equal to 12 × 109/L, and with platelet counts less than or equal to 400 × 109/L. This frequency (12/17 or 71%) was significantly higher than that observed in a series of 92 patients diagnosed with JAK2 (V617F)-positive PV at the Department of Hematology, Pavia, Italy (20/92 or 22%, P < .001). All patients but one carrying a JAK2 exon 12 mutation had low serum erythropoietin levels, indicating a combination of absolute erythrocytosis and suppressed endogenous erythropoietin production.22
Overall, 8 different mutations were found in this study, and the 2 duplications are novel at the time of this writing. All mutations were detected in circulating granulocytes but not in T lymphocytes, indicating that these were acquired somatic mutations. The pattern on sequencing was heterozygous in 16 of 17 patients (Table 1): representative patterns are reported in Figure S1.
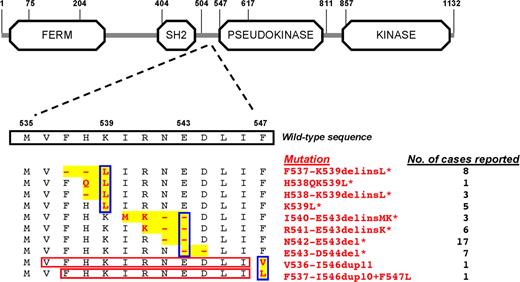
Mutations spanned from base 1606 to 1640, and the 2 duplications modified the rest of the sequence by adding 33 bp (Figure 1, Figure S2). In terms of protein, deletions predicted amino acid changes spanning from phenylalanine 537 to aspartic acid 544, while duplications predicted changes from phenylalanine 547 onward within the JH2 pseudokinase domain. Three categories of molecular lesions were identified (Figure 1): (i) those involving a K539L substitution; (ii) those involving a deletion of glutamic acid 543 (E543del); and (iii) amino acid duplications involving a substitution of phenylalanine 547. Based on data of this study and findings of previous reports,12-17 the most frequent exon 12 mutations detected so far are N542-E543del, F537-K539delinsL, and E543-D544del.
Schematic representation of the JAK2 gene with the wild-type sequence of amino acids encoded by exon 12, and the exon 12 mutations detected so far. Regions of rearrangement are highlighted in yellow, while mutations are shown in red; the red rectangles indicate the duplicated sequences. All mutations so far identified in patients with PV or idiopathic erythrocytosis are illustrated: those previously reported12-17 are marked with an asterisk. Blue rectangles indicate the 3 types of mutations: (i) those involving a K539L substitution; (ii) those involving a deletion of glutamic acid 543 (E543del); (iii) and duplications involving a substitution of phenylalanine 547. On the right, for each mutation the number of cases reported so far is indicated. Information on JAK2 domains is from URL: http://smart.embl-heidelberg.de.
Schematic representation of the JAK2 gene with the wild-type sequence of amino acids encoded by exon 12, and the exon 12 mutations detected so far. Regions of rearrangement are highlighted in yellow, while mutations are shown in red; the red rectangles indicate the duplicated sequences. All mutations so far identified in patients with PV or idiopathic erythrocytosis are illustrated: those previously reported12-17 are marked with an asterisk. Blue rectangles indicate the 3 types of mutations: (i) those involving a K539L substitution; (ii) those involving a deletion of glutamic acid 543 (E543del); (iii) and duplications involving a substitution of phenylalanine 547. On the right, for each mutation the number of cases reported so far is indicated. Information on JAK2 domains is from URL: http://smart.embl-heidelberg.de.
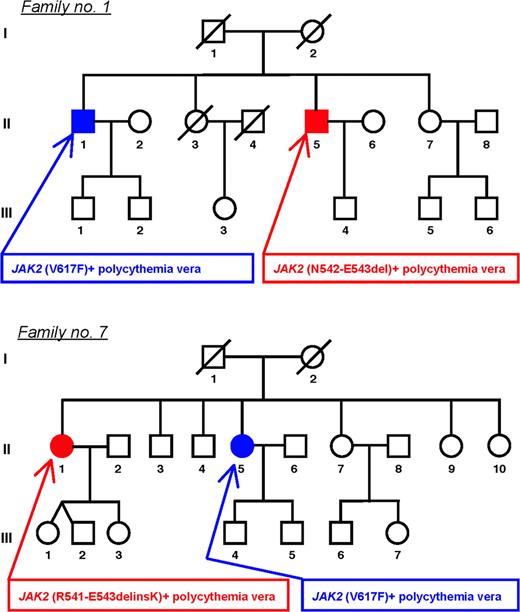
Two of 11 patients with JAK2 (V617F)-negative familial PV carried a mutation of exon 12 (Figure 2). Both patients had an affected sibling with JAK2 (V617F)-positive PV. While subjects carrying an exon 12 mutation showed isolated erythrocytosis, their JAK2 (V617F)-positive siblings had also thrombocytosis.
Pedigrees of 2 families with myeloproliferative disorders characterized by intra-family discordance of JAK2 mutation type. Each family member is identified by a pedigree number. Circles represent female family members; squares male family members; filled symbols members with phenotypically expressed myeloproliferative disorder; slashes members who had died. In family 1, 2 brothers (II-1 and II-5) had PV. Subject II-1 presented at the age of 57 with ischemic stroke, and at the age of 63 he was diagnosed with PV (erythrocytosis and thrombocytosis). This patient was later found to be JAK2 (V617F)-positive, and his granulocyte mutant alleles increased from 6% to 20% over time. He died at the age 69 of pulmonary embolism despite low-dose aspirin and cytoreductive treatment. His brother (subject II-5) was found to have erythrocytosis on routine blood counts at the age of 58; a diagnosis of PV was made, and venesection therapy was started. He was found to carry the N542-E543del mutation in circulating granulocytes. In family 7, 2 sisters (II-1 and II-5) had PV. Subject II-5 presented at the age of 59 with erythrocytosis and thrombocytosis. When she was admitted to the Department of Hematology, Pavia, Italy, at the age of 78, she was found to have post-PV myelofibrosis: she had splenomegaly (below the umbilical line), elevated circulating CD34+ cell count (244 × 106/L), and 91% granulocyte JAK2 (V617F) mutant alleles. Her sister (subject II-1) presented at the age of 53 with erythrocytosis, was diagnosed with PV, and started venesection therapy. She was found to carry the R541-E543delinsK mutation in circulating granulocytes.
Pedigrees of 2 families with myeloproliferative disorders characterized by intra-family discordance of JAK2 mutation type. Each family member is identified by a pedigree number. Circles represent female family members; squares male family members; filled symbols members with phenotypically expressed myeloproliferative disorder; slashes members who had died. In family 1, 2 brothers (II-1 and II-5) had PV. Subject II-1 presented at the age of 57 with ischemic stroke, and at the age of 63 he was diagnosed with PV (erythrocytosis and thrombocytosis). This patient was later found to be JAK2 (V617F)-positive, and his granulocyte mutant alleles increased from 6% to 20% over time. He died at the age 69 of pulmonary embolism despite low-dose aspirin and cytoreductive treatment. His brother (subject II-5) was found to have erythrocytosis on routine blood counts at the age of 58; a diagnosis of PV was made, and venesection therapy was started. He was found to carry the N542-E543del mutation in circulating granulocytes. In family 7, 2 sisters (II-1 and II-5) had PV. Subject II-5 presented at the age of 59 with erythrocytosis and thrombocytosis. When she was admitted to the Department of Hematology, Pavia, Italy, at the age of 78, she was found to have post-PV myelofibrosis: she had splenomegaly (below the umbilical line), elevated circulating CD34+ cell count (244 × 106/L), and 91% granulocyte JAK2 (V617F) mutant alleles. Her sister (subject II-1) presented at the age of 53 with erythrocytosis, was diagnosed with PV, and started venesection therapy. She was found to carry the R541-E543delinsK mutation in circulating granulocytes.
The present study confirms previous observations on JAK2 exon 12 mutations in myeloproliferative disorders, describes 2 novel mutations, and provides evidence for intrafamily discordance of JAK2 mutation type in familial disorders.
The seminal study by Scott and coworkers12 led to the conclusion that JAK2 exon 12 mutations define a distinctive myeloproliferative syndrome that affects patients who currently receive a diagnosis of PV or idiopathic erythrocytosis. Our findings confirm that these patients indeed have a myeloproliferative syndrome presenting mainly with isolated erythrocytosis associated with suppressed erythropoietin production. Exon 12 mutations were shown to be associated with peculiar biochemical abnormalities of JAK2 signaling,12 which likely favor erythrocytosis.
Of our 17 patients carrying exon 12 mutations, all but one had heterozygous patterns on sequencing (Table 1 and Figure S1). In our original report on the unique JAK2 (V617F) mutation,4 48 of 83 (58%) of JAK2 (V617F)-positive patients with PV showed a heterozygous pattern on sequencing, while the remaining 35 of 83 (42%) had a homozygous pattern. These observations may suggest that mitotic recombination (ie, the mechanism leading to homozygosity,4 ) is more likely to occur in PV patients carrying JAK2 (V617F) than in those with exon 12 mutations.
Variable proportions of patients with JAK2 (V617F)-negative PV that carry an exon 12 mutation have been reported.12,13,15-17 We used direct sequencing, which allowed us to identify novel mutations but is unlikely to detect percentages of mutant alleles lower than 10%. At least a portion of our negative PV patients (Table S1) might therefore carry low percentages of mutant alleles in granulocytes. The inability of direct sequencing to detect them may justify the discrepancy between our findings and those of previous reports.12,13
Studies of families with myeloproliferative disorders9,10,23,24 showed that JAK2 (V617F) represents an acquired somatic mutation in familial cases as it does in sporadic cases,25 and that a genetic predisposition to acquisition of JAK2 (V617F) is inherited. The current study indicates that this predisposition regards any type of JAK2 mutations, not just JAK2 (V617F). It might be hypothesized that these individuals have defective mechanisms for repairing or neutralizing spontaneous mutations, so that those involving gain-of-function emerge and lead to clonal proliferation. In addition, if one considers the 2 families reported in Figure 2, only few individuals in these families have a disease phenotype. This makes the distinction between familial and sporadic cases more subtle, and raises the question whether a genetic predisposition is present also in the so-called sporadic cases.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Tiina Berg, Sabrina Boggi, and Roland Jäger for their technical assistance.
This study was supported by grants from Associazione Italiana per la Ricerca sul Cancro (AIRC), Fondazione Cariplo, Fondazione Istituto di Ricovero e Cura a Carattere Scientifico (IRCCS) Policlinico San Matteo, Ministry of University and Research, and Alleanza Contro il Cancro, Italy (M.C.); grants from the Swiss National Science Foundation (310 000-108 006/1), the Swiss Cancer League (OCS-01742-08-2005), and the Krebsliga beider Basel (R.S.); and grants from the Initiative for Cancer Research of the Medical University of Vienna and the Privatstiftung Unruhe (R.K.).
Authorship
Contribution: M.C. and R.S. designed research, analyzed data, and wrote the paper; F.P., E.R., A.T., and H.G. collected and analyzed clinical data; and D.P., S.L, A.B., M.F., R.K., and L.C. performed molecular investigations.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Prof Mario Cazzola, Department of Hematology, Fondazione IRCCS Policlinico San Matteo, 27100 Pavia, Italy; e-mail: mario.cazzola@unipv.it.
References
Author notes
D.P. and S.L. contributed equally to this work.