Neuropilin-1 and -2 (NRP1 and NRP2) are the transmembrane glycoproteins interacting with 2 types of ligands: class III semaphorins and several members of the VEGF family, the main regulators of blood and lymphatic vessel growth. We show here that both NRP1 and NRP2 can also bind hepatocyte growth factor (HGF). HGF is a pleiotropic cytokine and potent proangiogenic molecule that acts on its target cells by binding to the c-met receptor. We found that the N-terminal domain of HGF is involved in the interaction with neuropilins. We demonstrated that invalidation of NRP1 or NRP2 by RNA interference in human umbilical vein endothelial cells (HUVECs) decreased HGF-induced c-met phosphorylation and VEGF-A165– and HGF-mediated intracellular signaling. Accordingly, the disruption of NRP1 or NRP2 binding to VEGF-A165 or HGF with a blocking antibody, decreased the proliferation and migration of endothelial cells. This effect may be further enhanced if VEGF-A165 or HGF binding to both NRP1 and NRP2 was disrupted. Using a mouse Matrigel model, we demonstrated that NRP1 is essential for HGF-mediated angiogenesis in vivo. Our results suggest that, in endothelial cells, both NRP1 and NRP2 function as proangiogenic coreceptors, potentiating the activity of at least 2 major proangiogenic cytokines, VEGF-A165 and HGF.
Introduction
Neuropilins (NRPs) are transmembrane glycoproteins that play an important role in various biological processes, including axonal guidance, angiogenesis, tumorigenesis, and the immunologic response.1,,–4 NRPs have been characterized as coreceptors for 2 unrelated families of extracellular secreted ligands—class III semaphorins and several members of the vascular endothelial growth factor (VEGF) family, the main regulators of blood and lymphatic vessel growth.5 NRP acts in conjunction with membrane-associated signal transducers, such as the VEGF receptor tyrosine kinases (VEGFR-1, -2, and -3)6,7 and plexins, the transmembrane receptors of the semaphorin family.8 In higher eukaryotes, 2 neuropilin genes, NRP1 and NRP2, have been identified.9 They code for proteins displaying about 44% amino-acid sequence identity, with a similar domain structure.10 Both NRP1 and NRP2 contain a large extracellular region and a short cytoplasmic tail of about 40 amino acids, lacking any enzymatic activity. The extracellular region of neuropilins contains 5 different structural domains—2 CUB motifs, a1 and a2, homologous to complement components C1r/C1s, 2 coagulation factor V/VIII homology domains b1 and b2, and one c domain (MAM, homologous to meprin, A5, μ).11 The high-affinity binding site for VEGF-A165 has been localized to the b1 and b2 domains of NRP112,13 and NRP2,14 whereas the binding of semaphorins requires both the a1a2 and b1b2 repeats.12
NRP1 and NRP2 interact selectively with different members of the VEGF and semaphorin families and have nonoverlapping expression patterns. Thus, among the VEGF members, NRP1 binds VEGF-A165, VEGF-B, VEGF-E, and placental growth factor (PlGF), whereas NRP2 binds VEGF-A165,VEGF-A145, VEGF-C, and PlGF.15 The non–heparin-binding isoforms of VEGF, such as VEGF-A121, have long been considered unable to interact with NRPs. Current evidence suggests, however, that VEGF-A121 does bind NRP1 via the C-terminal sequence of 6 amino acids encoded by exon 8.16,,–19 During the development of the cardiovascular system, NRP1 is detected primarily in the arterial endothelial cells, and NRP2 is detected in the venous and lymphatic endothelial cells.20 Genetic studies in mice have shown that both the overexpression of NRP121 and the targeted inactivation of the NRP1 gene22,23 are lethal, provoking, in addition to neuronal defects, disorganization of the vascular network and defects in heart development. Inactivation of the NRP2 gene has less severe consequences, limited to defects in the formation of small lymphatic vessels and capillaries.24 However, mice in which both neuropilin genes have been invalidated had a very severe vascular phenotype and died after embryonic day 8.5.25 Thus, neuropilins are essential for vasculogenesis, angiogenesis, and lymphangiogenesis.
The role of NRPs in the control of vascular function has been attributed principally to their ability to regulate the activities of VEGF on endothelium. In endothelial cells, NRPs are thought to increase signaling through the VEGFRs by ensuring the optimal presentation of VEGF and by stabilizing VEGF/VEGFR complexes. Thus, the interaction of VEGF-A165 with NRP1 is required for stable binding of VEGF-A165 to VEGFR-2, full activation of VEGFR-2, and downstream signaling and biological responses.17,26 Similarly, the interaction of VEGF-A or VEGF-C with NRP2 increases the VEGFR-2 phosphorylation threshold and promotes the endothelial cell survival and motility induced by VEGF-A and VEGF-C.7 In contrast, disruption of the NRP1/VEGF-A interaction with a highly specific blocking antibody reduces VEGFR-2 activation and signaling, inhibiting angiogenesis and vascular remodeling in a mouse tumor model.27 Both NRP1 and NRP2 enhance the affinity of VEGF-A121 binding to VEGFR-2 and increase VEGF-A121-induced VEGFR-2 phosphorylation, thereby regulating proliferation, migration, and sprouting of endothelial cells.19,28
Evidence has recently been obtained to suggest that, in addition to VEGF, a number of heparin-binding growth factors interact with NRPs.29 Some of these factors, including hepatocyte growth factor (HGF) and several members of the fibroblast growth factor (FGF) family, have been characterized as potent proangiogenic cytokines.30,31 At least for FGF-2, the interaction with NRP1 is thought to be physiologically relevant, as NRP1 was found to enhance the growth stimulatory activity of FGF-2 on endothelial cells.29 Two recent studies have demonstrated a novel role for NRP1 in tumor progression through enhancement of the autocrine HGF/c-met loop.32,33 These findings suggest that NRP1 may also act as functional coreceptor for HGF.
HGF is a pleiotropic cytokine that acts on target cells by binding to the c-met receptor.11 The mature factor is a heterodimer of α- and β-chains.34,35 The α-chain is primarily involved in the interaction of HGF with the c-met receptor and heparin.36,–38 After binding, HGF induces the phosphorylation of c-met, resulting in the recruitment of several downstream signaling transducers, including Grb2, Gab1, STAT3, Shc, SHIP-1, Src, and phosphatidylinositol-3 kinase.39 These events lead to privileged stimulation of the Ras–extracellular signal–regulated kinase (ERK) cascade and activation of other members of the mitogen-activated protein kinase (MAPK) family. c-met is expressed in most tissues, and HGF/c-met signaling is essential for the development and regeneration of several organs and systems,40 and for the malignant progression of various cancers.39,41 HGF is also a potent stimulator of angiogenesis.30 Endothelial cells undergoing angiogenesis have the highest levels of c-met receptor expression.42 HGF signals through c-met to regulate endothelial cell survival, proliferation, migration, matrix deposition, and degradation, together with the formation of capillary-like structures. In vivo, HGF stimulates angiogenesis in several animal models of ischemia, with an efficiency similar to, or even greater than that of VEGF-A165.43,–45
The role of NRPs in the regulation of HGF function on endothelial cells has yet to be explored. To do this, we first analyzed whether HGF could interact with NRP2 in addition to NRP1. We also carried out a structure-function analysis to map the NRP-binding region within HGF. We compared the NRP-binding properties of full-length HGF with those of recombinant proteins corresponding to the 5 HGF α-chain structural domains—the N-terminal domain (N) and 4 kringle domains.46 We then used an RNA interference approach and blocking anti-NRP antibodies to explore the role of NRPs in VEGF-A165– and HGF-induced signaling and cellular responses in human umbilical vein endothelial cells (HUVECs). Finally, we analyzed the effect of a blocking anti-NRP1 antibody on HGF-induced angiogenesis in a mouse Matrigel model.
Methods
Approval was obtained from the institutional review board at Direction Départementale des Services Vétérinaires de Paris en Charge des Affaires Vétérinaires d'Ile de France (Ministère de l'Agriculture et de la Pêche) for these studies.
Materials
Recombinant human HGF, VEGF-A165, IGF-I, NRP1/Fc chimera (NRP1 extracellular domain fused to human IgG1 Fc fragment), NRP2/Fc, VEGFR-2/Fc, a blocking goat anti-NRP1 antibody, and a nonimmune goat IgG were purchased from R&D Systems (Minneapolis, MN). Recombinant isolated fragments of the α-chain of human HGF (N-terminal domain and kringle domains 1–4) were produced and purified as previously described.46 A goat anti-human IgG Fcγ fragment-specific antibody and peroxidase-conjugated species-specific secondary IgGs were obtained from Jackson ImmunoResearch (West Grove, PA). The horseradish peroxidase–conjugated streptavidin was obtained from Pierce (Rockford, IL). A rabbit anti–von Willebrand factor antibody was purchased from Dako (Glostrup, Denmark). A rabbit anti-CD31 antibody was obtained from BD Pharmingen (San Diego, CA). A mouse antibody against phospho-p44/p42 MAPK (Thr202/Tyr204) and rabbit antibodies against Akt, phospho-Akt (Ser473), p38, and phospho-p38 MAPK (Thr180/Tyr182) were purchased from Cell Signaling Technology (Beverly, MA). Protein A Sepharose CL-4B was purchased from GE Healthcare (Chalfont St Giles, United Kingdom). A mouse antiphosphotyrosine antibody (clone 4G10) was purchased from Upstate Cell Signaling Solutions (Lake Placid, NY). The rabbit anti–c-met, anti-NRP2, and anti-ERK2 antibodies were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). The cell culture reagents, an Alexa Fluor 488 donkey anti-rat IgG and an Alexa Fluor 555 goat anti-rabbit IgG, were purchased from Invitrogen (Cergy Pontoise, France). Endothelial cell basal medium (EBM) was purchased from Clonetics (BioWhittaker, Walkersville, MD). Bovine serum albumin (BSA), lysozyme, gelatin, OPD, and biotinamidohexanoic acid 3-sulfo-N-hydroxysuccinimide ester were obtained from Sigma-Aldrich (St Louis, MO).
Cells
HUVECs were isolated from human umbilical veins by collagenase digestion and were cultured in M199 medium supplemented with 15 mM HEPES, 2 mM glutamine, 50 IU/mL penicillin, 50 μg/mL streptomycin, 2.5 μg/mL amphotericin B, 15% (vol/vol) fetal bovine serum (FBS), 5% (vol/vol) human serum, and 2 ng/mL FGF-2. Cells were grown in gelatin-coated flasks at 37°C in an atmosphere containing 5% CO2. Before stimulation, HUVECs were starved overnight in serum-free EBM. All subsequent steps were performed in this medium, unless otherwise stated. All experiments were carried out with cells from passages 2 and 3.
Solid-phase NRP1 binding assay
Microtiter plates (96-well) were coated by incubation overnight at 4°C with 25 nM HGF, isolated HGF domains, VEGF-A165, BSA, or lysozyme. Nonspecific binding sites were blocked by incubating the plates with 1% gelatin in phosphate-buffered saline (PBS) for 1 hour at 37°C. Plates were then incubated with various concentrations of NRP1/Fc or NRP2/Fc for 1 hour at 37°C. Bound NRP/Fc was detected by incubation with a goat anti-human IgG Fcγ antibody (1:500) followed by a peroxidase-conjugated anti-goat antibody (1:1000). In another experiment, 96-well microtiter plates were coated with 1 μg/mL anti–human IgG Fcγ antibody. They were incubated with 100 ng/mL of NRP1/Fc or VEGFR-2/Fc for 1 hour at 37°C and then with various concentrations of biotinylated VEGF-A165 in the presence or absence of various concentrations of N, nonbiotinylated VEGF-A165, or lysozyme. Bound biotinylated VEGF-A165 was detected by incubation with streptavidin-peroxidase. Plates were developed by incubation with Sigma Fast OPD substrate, and absorbance at 490 nm was measured with an Elx808 automated microplate reader (Bio-Tek Instruments, Winooski, VT). All experiments were carried out in triplicate and repeated at least 3 times.
Small interfering RNA transfection
For small interfering RNA (siRNA)-mediated knockdown, HUVECs were grown to 60% to 70% confluence and treated for 24 to 48 hours with 50 nM siRNA in the presence of TransIT-TKO transfection reagent (Mirus, Madison, WI) or DharmaFECT-2 (Dharmacon, Lafayette, CO) according to the manufacturer's protocol. NRP1 siRNA-A, -B, and -C targeting different sequences in the human NRP1 mRNA were obtained from Proligo (Boulder, CO) (5′-GAGAGGUCCUGAAUGUUCCTT-3′),47 Dharmacon (SMARTpool), and Invitrogen (NRP1 Stealth Select RNAi), respectively. A total of 3 different siRNAs were used as controls: a scrambled sequence from Proligo (5′-AGAGAUGUAGUCGCUCGCUTT-3′),47 the siCONTROL Non-Targeting siRNA no. 2 from Dharmacon, and the Stealth RNAi Negative Control from Invitrogen (control siRNA-A, -B, and -C, respectively). The 21-base scrambled sequence from Proligo was subjected to a BLAST search (National Center for Biotechnology Information) of the GenBank database48 to ensure the absence of gene targeting. The Silencer Validated NRP2 siRNA was purchased from Ambion (Austin, TX).
Immunoprecipitation and Western blot analysis
After treatment, cells were lysed in 50 mM Tris-HCl (pH 8.0) containing 100 mM NaCl, 1% (vol/vol) Triton X-100, 0.5% (vol/vol) NP-40, 5 mM EDTA, 1 mM PMSF, 1 μg/mL leupeptin, 1 μg/mL aprotinin, 1 mM sodium orthovanadate, 40 mM β-glycerophosphate, 50 mM NaF, and 100 μM phenylarsine oxide. Insoluble material was removed by centrifugation at 14 000g for 10 minutes at 4°C. The protein concentration in the supernatant was determined using the BCA Protein Assay Reagent (Pierce). For immunoprecipitation, 800 μg of protein was incubated with 5 μg anti–c-met antibody overnight at 4°C, and for a further 2 hours with protein A–Sepharose beads. The antigen-antibody complexes were eluted with NuPAGE LDS sample buffer (Invitrogen), separated by electrophoresis in 4% to 12% NuPAGE gels (Invitrogen), and transferred to nitrocellulose membranes. Nonspecific binding was blocked by incubating the membranes with PBS containing 5% nonfat milk powder for 1 hour at room temperature. Membranes were probed overnight at 4°C with specific primary antibody, and then with peroxidase-conjugated secondary antibody. Antigen-antibody complexes were detected with the SuperSignal West Pico chemiluminescent substrate (Pierce). Bands were visualized with an LAS-3000 Luminescent Image Analyzer (Fujifilm, Tokyo, Japan) and quantified with Multi Gauge 4.0 software (Fujifilm).
DNA biosynthesis assays
HUVECs were seeded in 24-well plates at a density of 15 000 cells per well, in complete medium. After 1 day of culture, the cells were starved by incubation for 24 hours in serum-free EBM. They were then stimulated by incubation for a further 20 hours with HGF (2 to 10 ng/mL) or VEGF-A165 (10 ng/mL) in the presence of anti-NRP1 or nonimmune IgG (20 μg/mL). Cells were incubated for the last 16 hours with 1 μCi (0.037 MBq) per well of [methyl-3H]thymidine (MP Biomedicals, Irvine, CA), and [3H]thymidine incorporation was estimated in an LS 6500 liquid scintillation β-counter (Beckman Coulter, Fullerton, CA).
Migration assay
Cell-culture inserts (8-μm pores, FluoroBlok; BD Biosciences, San Diego, CA) were coated with gelatin. HUVECs were incubated overnight with 50 μg CellTracker CM-Dil (Invitrogen). Cells were then suspended in M199 medium supplemented with 1% FBS, and incubated for 30 minutes with 20 μg/mL anti-NRP1 IgG or nonimmune IgG. A total of 5 × 104 cells were added to the upper side of each insert. The inserts were placed in 24-well plates containing medium supplemented with HGF or VEGF-A165 (10 ng/mL). Plates were incubated for 6 hours at 37°C in an atmosphere containing 5% CO2, and, after the times indicated, the number of cells that had migrated to the lower surface of the filters was evaluated by fluorescence measurements using a Wallac 1420 multilabel counter (Perkin Elmer, Turku, Finland). Each experiment (n = 3) was performed in triplicate.
In vivo assessment of angiogenesis using the Matrigel plug assay
Animals were cared for in accordance with the guidelines of the European Convention for the Protection of Vertebrate Animals used for Experimental and other Scientific Purposes, and the study protocol was approved by the local ethics committee. Briefly, Matrigel was mixed with HGF (300 ng/mL), heparin (20 IU/mL), anti-NRP1, or an isotype control IgG (20 μg/mL), and the resulting mixture was injected subcutaneously (0.5 mL) into both flanks of 7-week-old male C57BL/6 mice (Harlan, Gannat, France) under isoflurane/oxygen anesthesia. The animals were killed 8 days later, and the Matrigel plugs were removed, weighed, and photographed. For each mouse, one plug was embedded in paraffin, sectioned, and processed for haematoxylin-eosin-safran (HES) staining and immunohistochemistry analysis. The second plug was homogenized in PBS using an Ultra-Turrax T25 homogenizer (Bioblock, Illkirch, France). Debris was removed by centrifugation, the supernatant was mixed with Sulfolyzer (Sysmex; Roche, Indianapolis, IN), and its hemoglobin content was assessed by determining absorbance at 550 nm. The standard curve for hemoglobin quantification was generated with purified rat hemoglobin (Sigma) treated in the same way as the samples. Hemoglobin concentration values were normalized according to plug weight, using a correction index calculated as the ratio of the weight of plug to the weight of the smallest plug.
Immunohistochemistry
Sections (5 μm) were cut from paraffin-embedded plugs. The paraffin was then removed, and the sections were treated with the target retrieval solution (Dako) for 20 minutes at 95°C. Sections were blocked by incubation with 20% goat serum in PBS for 20 minutes, and were then incubated with a rabbit anti–von Willebrand factor or a rat anti–mouse CD31 antibody for 1 hour at room temperature, followed by an Alexa Fluor 555 goat anti–rabbit or an Alexa Fluor 488 donkey anti–rat antibody, respectively. Sections were observed with an Axioskop 2 plus fluorescence microscope (Zeiss, Jena, Germany), and images were recorded with the Archimed 4.7.0 software (Microvision Instruments, Eury, France).
Statistics
Data are presented as means plus or minus SD. Statistical analyses were performed in Excel (Microsoft, Redmond, WA) using Student t test (biochemical experiments, proliferation, and migration assays). Analysis of variance (ANOVA) was used to analyze results from Matrigel plug assays. Values were considered significantly different if P values were less than .05.
Results
NRP1 and NRP2 bind HGF
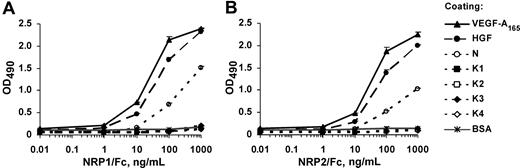
To determine whether NRP1 and NRP2 can bind HGF, we used an enzyme-linked immunosorbent assay (ELISA)–based approach. The 96-well microtiter plates were coated with HGF or with recombinant isolated HGF domains, the N-terminal domain (N), or various kringle domains (K1, K2, K3, K4), and incubated with a series of concentrations of recombinant NRP1/Fc or NRP2/Fc. Both NRP1 and NRP2 bound HGF in a concentration-dependent manner (Figure 1). The NRP/HGF binding profiles obtained were similar to those obtained with VEGF-A165, used as a positive control. The EC50 values calculated from these experiments for NRP1 binding to HGF and VEGF-A165 were 20 and 10 ng/mL, respectively, indicating that NRP1 had similar affinities for HGF and VEGF-A165 (Figure 1A). The binding affinity of HGF for NRP2 was also similar to that for VEGF-A165 (Figure 1B). The N-terminal domain of HGF (referred to as N) bound NRP1 and NRP2, albeit less efficiently than full-length HGF. In contrast, no binding of NRP1 or NRP2 was observed to any kringle domain or to BSA used as a negative control (Figure 1). These results suggest that HGF interacts with both NRP1 and NRP2 through its N-terminal domain.
HGF and the HGF N-terminal domain, but not HGF kringles, bind to NRP1 and NRP2. ELISA analysis of (A) NRP1/Fc and (B) NRP2/Fc binding to immobilized ligands. Microtiter plates were coated with VEGF-A165, HGF, N, K1, K2, K3, K4, or BSA and incubated with various concentrations of recombinant NRP1/Fc or NRP2/Fc, followed by the peroxidase-conjugated anti-Fc antibodies. Values are means plus or minus SD of 3 independent experiments.
HGF and the HGF N-terminal domain, but not HGF kringles, bind to NRP1 and NRP2. ELISA analysis of (A) NRP1/Fc and (B) NRP2/Fc binding to immobilized ligands. Microtiter plates were coated with VEGF-A165, HGF, N, K1, K2, K3, K4, or BSA and incubated with various concentrations of recombinant NRP1/Fc or NRP2/Fc, followed by the peroxidase-conjugated anti-Fc antibodies. Values are means plus or minus SD of 3 independent experiments.
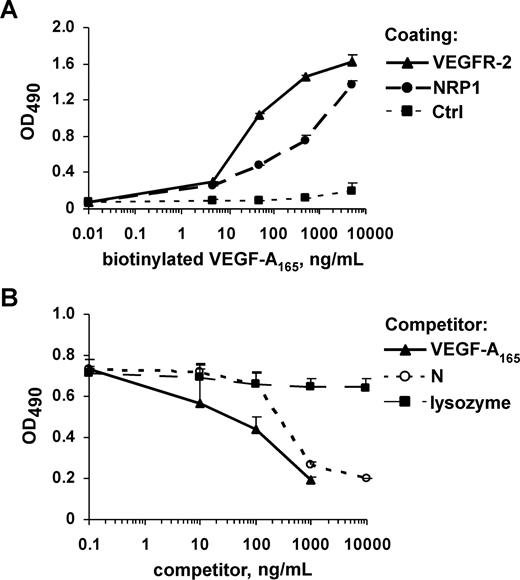
In another set of experiments, NRP1/Fc or VEGFR-2/Fc was immobilized on plates via the Fc moiety, using an anti-Fc antibody. Biotinylated VEGF-A165 bound to immobilized NRP1 and VEGFR-2 in a dose-dependent manner, whereas it did not bind to an anti-Fc antibody (Figure 2A) or to CD6/Fc, used as a negative control (data not shown). Thus, biotinylation did not alter the binding properties of VEGF-A165. We then investigated whether N could displace VEGF-A165 from its binding sites on NRP1. As expected, nonbiotinylated VEGF-A165 and N competed with biotinylated VEGF-A165 for binding to NRP1 (Figure 2B). However, the calculated IC50 values for VEGF-A165 and N were 200 and 550 ng/mL, respectively, indicating that N was a less potent competitor than VEGF-A165. In contrast, lysozyme, which has a molecular size and pI similar to those of N, did not compete with VEGF-A165 for NRP1. N therefore includes the site responsible for interaction between HGF and NRP1. Our results also suggest that the binding sites for VEGF-A165 and HGF on NRP1 overlap.
The HGF N-terminal domain competes with VEGF-A165 for binding to NRP1. (A) Solid-phase receptor assay analysis of the binding of biotinylated VEGF-A165 to VEGFR-2/Fc, and NRP1/Fc immobilized via the Fc moieties on plates coated with anti-Fc IgG. Bound VEGF-A165 was detected by peroxidase-conjugated streptavidin staining. (B) Microtiter plates coated with NRP1/Fc were incubated with biotinylated VEGF-A165 in the presence of various concentrations of N, nonmodified VEGF-A165, or lysozyme. Values are means plus or minus SD of 3 independent experiments.
The HGF N-terminal domain competes with VEGF-A165 for binding to NRP1. (A) Solid-phase receptor assay analysis of the binding of biotinylated VEGF-A165 to VEGFR-2/Fc, and NRP1/Fc immobilized via the Fc moieties on plates coated with anti-Fc IgG. Bound VEGF-A165 was detected by peroxidase-conjugated streptavidin staining. (B) Microtiter plates coated with NRP1/Fc were incubated with biotinylated VEGF-A165 in the presence of various concentrations of N, nonmodified VEGF-A165, or lysozyme. Values are means plus or minus SD of 3 independent experiments.
NRP1 and NRP2 enhance HGF-induced c-met activation and VEGF-A165– and HGF-mediated signaling
Our binding results are consistent with the hypothesis that NRP1 and NRP2 bind HGF, presumably through interaction with the HGF N-terminal domain. Therefore, NRP1 and NRP2 are putative functional coreceptors for HGF. We used an RNA interference approach to demonstrate the involvement of NRP1 and NRP2 in HGF/c-met–mediated signaling in endothelial cells.
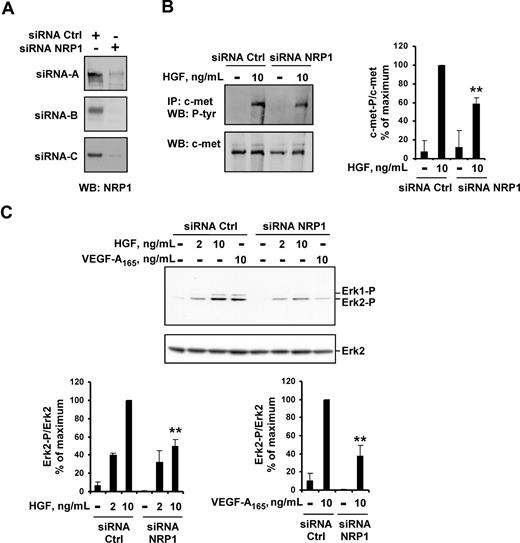
We first analyzed the impact of NRP1 knockdown on HGF function in HUVECs. Off-target effects were minimized using 3 siRNAs targeting different sequences of NRP1, and 3 control siRNAs. The transfection of HUVECs with any of the 3 NRP1 siRNAs resulted in a large decrease in NRP1 protein levels, down to the detection limit for Western blotting (Figure 3A). In HUVECs transfected with NRP1 siRNA, the levels of c-met phosphorylation induced by HGF were 40% lower than in HUVECs transfected with control siRNA (Figure 3B). Accordingly, NRP1 knockdown in HUVECs decreased the activation of several signaling pathways downstream from c-met. In HUVECs transfected with the various NRP1 siRNAs, HGF-induced ERK2 activation was 50% lower than in HUVECs transfected with any of the control siRNAs (Figure 3C). As expected, VEGF-A165–induced ERK2 activation was also reduced in these cells. Similarly, p38 kinase and Akt (Ser473) phosphorylation induced by HGF or VEGF-A165 was 50% to 60% lower in HUVECs transfected with NRP1 siRNA than in HUVECs transfected with control siRNA (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
NRP1 knockdown reduces HGF-induced c-met phosphorylation and VEGF-A165– and HGF-induced signaling. (A) HUVECs were transiently transfected with 50 nM NRP1 siRNA-A, -B, or -C, or with 50 nM control siRNA-A, -B, or -C. NRP1 protein level was evaluated by Western blotting. (B) HUVECs were transfected with 50 nM NRP1 or control siRNA-A, deprived of serum, and incubated with or without 10 ng/mL HGF for 10 minutes. c-met was immunoprecipitated from cell lysates and subjected to Western blotting with an antiphosphotyrosine or anti–c-met antibody (left). The phosphoprotein content was estimated by scanning densitometry analysis and normalized according to total c-met levels (right). (C) HUVECs were transfected with 50 nM NRP1 or control siRNA-A, -B, or -C, deprived of serum, and incubated with or without HGF (2 or 10 ng/mL), or with VEGF-A165 (10 ng/mL). Cell lysates were analyzed by Western blotting with anti–phospho-ERK1/2 and anti–total ERK2 antibody (top; shown for siRNA-C). Bands were quantified by scanning densitometry, and results were normalized according to total ERK2 content (bottom). Data are expressed as a percentage of the maximal phosphorylation obtained in HUVEC transfected with control siRNAs. Values are means plus or minus SD of 3 independent experiments. **P < .01 (Student t test).
NRP1 knockdown reduces HGF-induced c-met phosphorylation and VEGF-A165– and HGF-induced signaling. (A) HUVECs were transiently transfected with 50 nM NRP1 siRNA-A, -B, or -C, or with 50 nM control siRNA-A, -B, or -C. NRP1 protein level was evaluated by Western blotting. (B) HUVECs were transfected with 50 nM NRP1 or control siRNA-A, deprived of serum, and incubated with or without 10 ng/mL HGF for 10 minutes. c-met was immunoprecipitated from cell lysates and subjected to Western blotting with an antiphosphotyrosine or anti–c-met antibody (left). The phosphoprotein content was estimated by scanning densitometry analysis and normalized according to total c-met levels (right). (C) HUVECs were transfected with 50 nM NRP1 or control siRNA-A, -B, or -C, deprived of serum, and incubated with or without HGF (2 or 10 ng/mL), or with VEGF-A165 (10 ng/mL). Cell lysates were analyzed by Western blotting with anti–phospho-ERK1/2 and anti–total ERK2 antibody (top; shown for siRNA-C). Bands were quantified by scanning densitometry, and results were normalized according to total ERK2 content (bottom). Data are expressed as a percentage of the maximal phosphorylation obtained in HUVEC transfected with control siRNAs. Values are means plus or minus SD of 3 independent experiments. **P < .01 (Student t test).
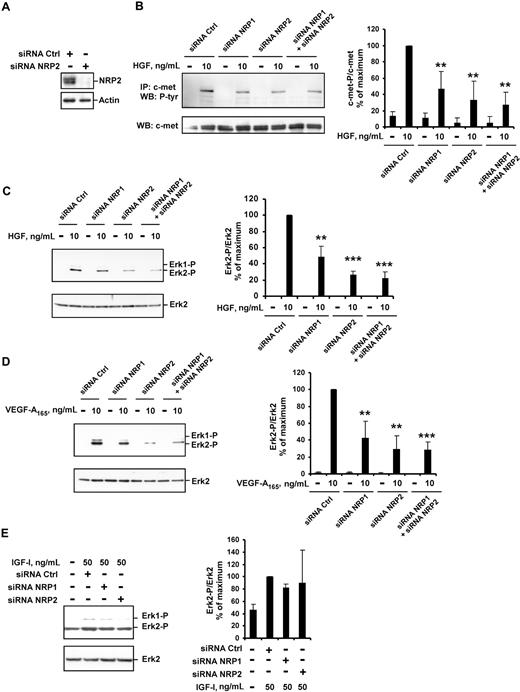
We further transfected HUVECs with NRP2 siRNA. In these conditions, NRP2 protein levels were very much lower than in HUVECs transfected with control siRNA (Figure 4A). In HUVECs transfected with NRP2 siRNA, the phosphorylation of c-met and activation of ERK2 following stimulation with HGF were reduced (by 60% and 70%; Figure 4B,C, respectively). As expected,7 VEGF-A165–induced ERK2 activation was also reduced in these cells (Figure 4D). The transfection of HUVECs with both NRP1 and NRP2 siRNAs led to the marked inhibition of both VEGF-A165– and HGF-induced signaling (Figure 4B-D). Thus, both NRP1 and NRP2 contribute to the enhancement of VEGF-A165 and HGF signaling in HUVECs.
NRP1 and NRP2 double knockdown leads to effective inhibition of VEGF-A165– and HGF-mediated signaling. HUVECs were transfected with 50 nM NRP1 siRNA-C, NRP2 siRNA, or both, or with a control siRNA-C, deprived of serum, and incubated with or without 10 ng/mL of HGF, 10 ng/mL of VEGF-A165, or 50 ng/mL of IGF-I for 10 minutes. (A) NRP2 protein level was evaluated by Western blotting. (B) c-met was immunoprecipitated from cell lysates and subjected to Western blotting with an antiphosphotyrosine or anti–c-met antibody (left). The phosphoprotein content was estimated by scanning densitometry analysis and normalized according to total c-met levels (right). (C-E) Cell lysates were analyzed by Western blotting with anti–phospho-ERK1/2 and anti–total ERK2 antibody (left). Bands were quantified by scanning densitometry, and results were normalized according to total ERK2 content (right). Data are expressed as percentages of the maximal phosphorylation obtained in HUVECs transfected with control siRNA. Values are means plus or minus SD of 3 independent experiments. **P < .01; ***P < .001 (Student t test).
NRP1 and NRP2 double knockdown leads to effective inhibition of VEGF-A165– and HGF-mediated signaling. HUVECs were transfected with 50 nM NRP1 siRNA-C, NRP2 siRNA, or both, or with a control siRNA-C, deprived of serum, and incubated with or without 10 ng/mL of HGF, 10 ng/mL of VEGF-A165, or 50 ng/mL of IGF-I for 10 minutes. (A) NRP2 protein level was evaluated by Western blotting. (B) c-met was immunoprecipitated from cell lysates and subjected to Western blotting with an antiphosphotyrosine or anti–c-met antibody (left). The phosphoprotein content was estimated by scanning densitometry analysis and normalized according to total c-met levels (right). (C-E) Cell lysates were analyzed by Western blotting with anti–phospho-ERK1/2 and anti–total ERK2 antibody (left). Bands were quantified by scanning densitometry, and results were normalized according to total ERK2 content (right). Data are expressed as percentages of the maximal phosphorylation obtained in HUVECs transfected with control siRNA. Values are means plus or minus SD of 3 independent experiments. **P < .01; ***P < .001 (Student t test).
Finally, we analyzed the effect of NRP down-regulation on signaling induced by insulin-like growth factor type I (IGF-I), a non–heparin-binding angiogenic stimulator that signals through a receptor tyrosine kinase and has not been characterized as an NRP-interacting protein. ERK1/2 activation in HUVECs stimulated with IGF-I did not differ significantly between cells transfected with control siRNA and those transfected with NRP1 or NRP2 siRNA (Figure 4E). Thus, the enhancement of intracellular signaling by NRPs concerns specifically the HGF- or VEGF-A165–induced effects.
NRP1 and NRP2 enhance VEGF-A165– and HGF-induced cellular responses in HUVECs
During the first few hours following transfection, NRP1 siRNA had no significant effect on HUVEC morphology or viability. However, consistent with reported data,49,50 the prolonged (more than 24 hours) inhibition of NRP1 expression was harmful for HUVECs, decreasing cell adhesion and survival. Note that the short half-lives of the nonmodified or nonstabilized siRNAs in vivo render them unsatisfactory for animal studies. We therefore used a blocking anti-NRP1 antibody as another approach to studying the involvement of NRP1 in the regulation of VEGF-A165– and HGF-induced cellular responses.
We first confirmed, by Western blot analysis, the blocking activity of this antibody in an ERK1/2 activation assay: VEGF-A165– and HGF-induced ERK2 activation was decreased by 30% in the presence of 20 μg/mL anti-NRP1 IgG (Figure 5A). As ERK1/2 has been shown to be essential for endothelial cell proliferation,51 we used a [3H]thymidine incorporation assay to analyze the effect of anti-NRP1 antibody on HUVEC proliferation. At a concentration of 20 μg/mL, anti-NRP1 antibodies significantly decreased HGF- and VEGF-A165–induced proliferation (by 47% and 40%, respectively), whereas nonimmune IgG had no effect (Figure 5B). We also checked that nonimmune IgG had no effect on cell proliferation when used alone (data not shown).
The blocking anti-NRP1 and anti-NRP2 antibodies decreases VEGF-A165– and HGF-induced cellular responses in HUVECs. Serum-deprived HUVEC were preincubated with 20 μg/mL anti-NRP1 IgG or nonimmune IgG and then stimulated for 10 minutes with HGF or VEGF-A165. (A) ERK1/2 phosphorylation was analyzed by Western blotting. Figures (bottom) indicate phospho-ERK1/2 content as estimated by scanning densitometry with normalization according to ERK2 content. Data are expressed as a percentage of the maximal phosphorylation obtained in HUVECs stimulated with 10 ng/mL HGF in the presence of nonimmune IgG. (B) DNA synthesis was determined by [3H]thymidine incorporation. Data are expressed as fold stimulation of basal incorporation observed in unstimulated cells in the presence of nonimmune IgG. Values are means plus or minus SD of 3 independent experiments performed in triplicate. (C) Migration was analyzed by Transwell assay. CM-Dil–stained HUVECs were preincubated with 20 μg of anti-NRP1 IgG or nonimmune IgG and dispensed into the upper chamber of the Fluoroblok inserts. Migration was induced by adding 10 ng/mL of HGF or VEGF-A165 to the lower chamber. The number of migrating cells was quantified by spectrofluorimetry. The results shown are means plus or minus SD of 3 independent experiments performed in triplicate. (D) HUVEC were preincubated with 20 μg/mL of anti-NRP1 IgG, anti-NRP2 IgG, or both, or with a nonimmune IgG and stimulated with HGF or VEGF-A165. DNA synthesis was determined by [3H]thymidine incorporation. Data are expressed as fold stimulation of basal incorporation observed in unstimulated cells in the presence of non immune IgG. Values are means plus or minus SD of 3 independent experiments performed in triplicate. *P < .05; **P < .01; ***P < .001 (Student t test).
The blocking anti-NRP1 and anti-NRP2 antibodies decreases VEGF-A165– and HGF-induced cellular responses in HUVECs. Serum-deprived HUVEC were preincubated with 20 μg/mL anti-NRP1 IgG or nonimmune IgG and then stimulated for 10 minutes with HGF or VEGF-A165. (A) ERK1/2 phosphorylation was analyzed by Western blotting. Figures (bottom) indicate phospho-ERK1/2 content as estimated by scanning densitometry with normalization according to ERK2 content. Data are expressed as a percentage of the maximal phosphorylation obtained in HUVECs stimulated with 10 ng/mL HGF in the presence of nonimmune IgG. (B) DNA synthesis was determined by [3H]thymidine incorporation. Data are expressed as fold stimulation of basal incorporation observed in unstimulated cells in the presence of nonimmune IgG. Values are means plus or minus SD of 3 independent experiments performed in triplicate. (C) Migration was analyzed by Transwell assay. CM-Dil–stained HUVECs were preincubated with 20 μg of anti-NRP1 IgG or nonimmune IgG and dispensed into the upper chamber of the Fluoroblok inserts. Migration was induced by adding 10 ng/mL of HGF or VEGF-A165 to the lower chamber. The number of migrating cells was quantified by spectrofluorimetry. The results shown are means plus or minus SD of 3 independent experiments performed in triplicate. (D) HUVEC were preincubated with 20 μg/mL of anti-NRP1 IgG, anti-NRP2 IgG, or both, or with a nonimmune IgG and stimulated with HGF or VEGF-A165. DNA synthesis was determined by [3H]thymidine incorporation. Data are expressed as fold stimulation of basal incorporation observed in unstimulated cells in the presence of non immune IgG. Values are means plus or minus SD of 3 independent experiments performed in triplicate. *P < .05; **P < .01; ***P < .001 (Student t test).
VEGF-A165– and HGF-induced activation of p38 kinase, a key kinase for HUVEC migration,52 was decreased by NRP1 knockdown. We analyzed the involvement of NRP1 in HUVECs chemotaxis by carrying out a Transwell migration assay. HGF-induced HUVEC migration was 39% lower in the presence of anti-NRP1 IgG than in the presence of nonimmune IgG (Figure 5C). Similarly, VEGF-A165–induced HUVEC migration was reduced by 35% in the presence of anti-NRP1 IgG (Figure 5C).
Our results indicated that the disruption of HGF/NRP1 or VEGF-A165/NRP1 interaction caused only partial inhibition of the HGF- or VEGF-A165–induced cellular responses in HUVECs. We analyzed whether this effect could be enhanced by concomitant blocking of both NRP1 and NRP2 binding activities. [3H]thymidine incorporation assays showed that disruption of the VEGF-A165/NRP2 or HGF/NRP2 interaction with a blocking anti-NRP2 antibody inhibited VEGF-A165 and HGF stimulatory activity by 40% (Figure 5D). A stronger effect was observed if HUVEC proliferation was stimulated by VEGF-A165 and HGF in the presence of both anti-NRP1 and anti-NRP2 antibodies (about 65% inhibition).
NRP1 is essential for HGF-induced angiogenesis in vivo
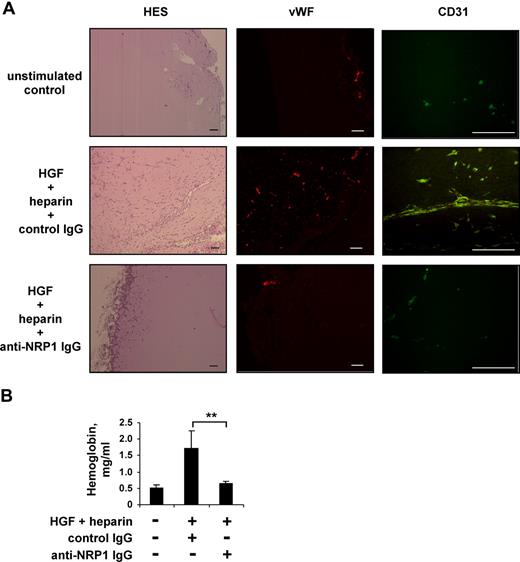
To study the role of NRP1 in the regulation of HGF function in vivo, we analyzed the effect of blocking anti-NRP1 antibody on HGF-induced angiogenesis in Matrigel plugs formed in mice after the subcutaneous injection of Matrigel. Neovascularization was induced by mixing Matrigel with angiogenic cocktail containing 300 ng/mL of HGF and 20 IU/mL of heparin. At this concentration, heparin does not itself stimulate angiogenesis, but instead enhances the angiogenic response induced by HGF.53 The HGF/NRP1 interaction was disrupted by adding a blocking anti-NRP1 antibody to some plugs, whereas the control plugs contained a nonimmune IgG. Mice were killed 8 days after Matrigel injection. Macroscopic analysis of the recovered plugs showed that, in the absence of HGF and heparin (PBS control group with nonimmune IgG), the implants remained white, with no visible vessels. In contrast, plugs containing HGF and heparin (HGF/heparin group with nonimmune IgG) were reddish in color and appeared to be vascularized. Histologic analysis after HES staining revealed that many cells had invaded the Matrigel implant of the HGF/heparin/nonimmune IgG group. Most of these cells were organized into tubular structures with lumina lined by a single cell layer, and some erythrocytes were visible within the tubules. These cells stained positive for von Willebrand factor and CD31, and were therefore of endothelial origin (Figure 6A). The addition of anti-NRP1 IgG strongly inhibited cellular invasion of the Matrigel and the formation of vessels (HGF/heparin/anti-NRP1 IgG group; Figure 6A).
Blocking anti-NRP1 antibody inhibits HGF-induced angiogenesis in a mouse Matrigel model. Mice were injected with Matrigel with or without an angiogenic cocktail and 20 μg/mL anti-NRP1 or nonimmune IgG. (A) Matrigel sections were analyzed by histology after HES staining and by immunochemical staining with an anti–von Willebrand factor or anti-CD31 antibody, using an Axioskop 2 plus fluorescence microscope (Carl Zeiss) equipped with Achroplan 4×/0.10 (left panel), Plan Neofluar 5×/0.15 (middle panel), or Plan Neofluar 40×/0.75 (right panel) objectives. Scale bar equals 100 μm. (B) Quantification of the hemoglobin content of plugs by spectrophotometry. **P < .01 (Student t test).
Blocking anti-NRP1 antibody inhibits HGF-induced angiogenesis in a mouse Matrigel model. Mice were injected with Matrigel with or without an angiogenic cocktail and 20 μg/mL anti-NRP1 or nonimmune IgG. (A) Matrigel sections were analyzed by histology after HES staining and by immunochemical staining with an anti–von Willebrand factor or anti-CD31 antibody, using an Axioskop 2 plus fluorescence microscope (Carl Zeiss) equipped with Achroplan 4×/0.10 (left panel), Plan Neofluar 5×/0.15 (middle panel), or Plan Neofluar 40×/0.75 (right panel) objectives. Scale bar equals 100 μm. (B) Quantification of the hemoglobin content of plugs by spectrophotometry. **P < .01 (Student t test).
We then quantified Matrigel vascularization by determining the hemoglobin content of the implants. Plugs recovered from the animals of PBS group contained 0.52 mg/mL hemoglobin. The hemoglobin content of the Matrigel plugs of the HGF/heparin/nonimmune IgG group was much higher (up to 334%), consistent with the formation of a larger number of functional blood vessels. The hemoglobin content of plugs from the HGF/heparin/anti-NRP1 IgG group was similar to that of plugs from the PBS group (Figure 6B), demonstrating that the anti-NRP1 antibody had inhibited the vascularization induced by the angiogenic cocktail.
Discussion
In endothelial cells, NRPs serve as coreceptors for members of the VEGF family, regulating VEGFR-dependent angiogenic events. The interaction of different VEGFs with neuropilins is thought to be mediated primarily by VEGF heparin-binding domains.1 The core NRP1-binding region of VEGF-A165 has been mapped to the carboxy-terminal 23 residues within the VEGF-A165 heparin-binding site.54 The structure of this region55 is remarkably similar to the structure of the hairpin-loop region within the N-terminal domain of HGF,56 the only protein for which such a fold has been described. It therefore appeared possible that VEGF-A165 and HGF might interact via their heparin-binding regions with the same molecular partners, including NRPs. Indeed, our binding studies showed that HGF interacted with both NRP1 and NRP2. Our results also suggested that HGF interacts with NRPs via the HGF N-terminal domain. Moreover, we found that the N-terminal domain of HGF competes with VEGF-A165 for binding to NRP1, indicating that the binding sites for HGF and VEGF-A165 on NRP1 probably overlap. The heparin-binding domain of VEGF-A165 differs from that of HGF in surface charge distribution, suggesting that VEGF-A165 and HGF interact with heparin in different ways.55 We also found that lysozyme, a highly basic protein, did not displace VEGF-A165 from its binding sites on NRP1, indicating that the VEGF-A165/NRP1 interaction is not simply charge dependent. These observations are not consistent with NRP1 behaving simply as a heparin mimetic.29 Instead, they suggest that NRP1 is involved in a specific interaction that may be strengthened by heparin or proteoglycans.
Our findings suggest that NRP1 and NRP2 act as functional coreceptors, regulating the activity not only of VEGF-A165, but also of HGF, in endothelial cells. Indeed, invalidation of NRP1 or NRP2 in HUVECs by RNA interference or disruption of the NRP1/HGF or NRP2/HGF interaction with blocking antibody decreased HGF-induced c-met activation and cellular responses, but only moderately. This effect was enhanced when HGF binding to both NRP1 and NRP2 was disrupted. Nevertheless, even in these conditions, the inhibition of HGF activity remained incomplete (about 65% inhibition). These results indicate that HGF can activate the c-met receptor independently of NRPs, albeit less efficiently. In contrast, NRPs are unlikely to signal autonomously following interaction with HGF, and most probably function as enhancers potentiating the signaling initiated by the binding of HGF to c-met.
Our findings are consistent with the idea that NRPs do not signal alone and act essentially in conjunction with receptor tyrosine kinases, such as plexins or VEGFRs. The cytoplasmic region of NRPs is too small to function as catalyst. Although highly conserved across species,1 it is not required for the effects of semaphorins on axon guidance.57 NRP1, however, regulates the adhesion of endothelial cells independently of VEGFR-2.50 This suggests that the intracellular domain of NRPs may serve as a binding site for docking proteins involved in the recruitment of signaling kinases. Indeed, it contains the binding sequence for PDZ domain–containing proteins,10 which are believed to assemble signal transduction components and to ensure their proper subcellular compartmentalization.58 This would explain how NRPs convey signals during angiogenesis, in both VEGFR-2–dependent and VEGFR-2–independent manners.59
We found that NRP1 invalidation inhibited the VEGF-A165– and HGF-induced activation of ERK1/2, Akt, and p38 kinase, and the proliferation and migration of HUVECs to similar extents. Our results are partly consistent with reported data on the consequences of NRP1 targeting for VEGF-A165–induced activities in HUVECs. Pan et al recently reported the production and biological activity of 2 blocking monoclonal antibodies raised against the semaphorin and VEGF binding sites of NRP1.27 Both antibodies inhibit the VEGF-A165–induced migration and sprouting of endothelial cells and vascular remodeling. The antibody blocking the VEGF binding site on NRP1 had a small effect on VEGFR-2 phosphorylation, p38 kinase activation, and HUVEC proliferation, with no effect on ERK1/2 and Akt activity. The authors suggested that NRP1 might act independently of the VEGFR-2 receptor, and might principally control endothelial cell migration. Another study, analyzing the biological activity of a bicyclic peptide NRP1 antagonist, EG3287, showed that disruption of the VEGF/NRP1 interaction inhibits the VEGF-A165–induced activation of ERK1/2 and Akt, but has no effect on endothelial cell proliferation.17 Finally, using RNA interference–mediated silencing of NRP1, Murga et al demonstrated that NRP1 is necessary for both VEGF-induced ERK1/2 phosphorylation and proliferation in HUVECs.50 These various results suggest that the amplitude and spectrum of effects induced by NRP inhibition may vary with cellular context.
Our findings provide new insight into the molecular mechanisms underlying the cooperation between VEGF-A and HGF in induction of the angiogenic response.43,60 They suggest that these 2 cytokines act through common coreceptors—the NRPs. NRPs, given their multiple interactions with various partners, are probably part of multicomponent complexes containing other receptors, effectors, adhesion molecules, and adaptor proteins. These complexes, known as signalosomes, are currently believed to be the principal signal-transducing elements regulating receptor activation, endocytosis, recycling, degradation, and the recruitment of downstream effectors.61 NRPs may have a general scaffold function in signalosome formation. By independent recruitment of NRPs, and activating associated molecules, VEGF and HGF may be involved in modulating signalosome functioning, thereby influencing signaling through unrelated receptors.
In conclusion, we demonstrate that, in addition to VEGF-A165, both NRP1 and NRP2 bind to and enhance the activity of HGF, a powerful proangiogenic cytokine. Recent studies have indicated that other heparin-binding proangiogenic cytokines may also interact with NRPs.29 Thus, the role of NRPs in vascular biology may be more extensive than simple regulation of the activity of VEGFs. Both NRP1 and NRP2 are up-regulated during pathological angiogenesis,4 a context in which NRPs probably act as multifunctional enhancers coordinating the action of diverse proangiogenic stimuli. The pharmacologic targeting of NRPs is therefore a promising therapeutic approach for the treatment of angiogenesis-associated disorders.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We would like to thank the Obstetrics Department of Lariboisière Hospital (Paris) for providing umbilical cord specimens.
This study was supported in part by research funding from Servier Laboratories.
Authorship
Contribution: E.S. designed and performed the research, analyzed the data, and cowrote the paper; J.P. designed the research, analyzed and interpreted data, and edited the paper; M.B. and D.A. carried out research and data analysis; G.T. designed the research, analyzed and interpreted data, and edited the paper; and T.M-R. designed the research, collected, analyzed, and interpreted data, and cowrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interest.
Correspondence: Tatyana Merkulova-Rainon, Institut des Vaisseaux et du Sang, Hôpital Lariboisière, 8 rue Guy Patin, 75475 Paris Cedex 10, France; e-mail: merkoulova@larib.inserm.fr.

![Figure 5. The blocking anti-NRP1 and anti-NRP2 antibodies decreases VEGF-A165– and HGF-induced cellular responses in HUVECs. Serum-deprived HUVEC were preincubated with 20 μg/mL anti-NRP1 IgG or nonimmune IgG and then stimulated for 10 minutes with HGF or VEGF-A165. (A) ERK1/2 phosphorylation was analyzed by Western blotting. Figures (bottom) indicate phospho-ERK1/2 content as estimated by scanning densitometry with normalization according to ERK2 content. Data are expressed as a percentage of the maximal phosphorylation obtained in HUVECs stimulated with 10 ng/mL HGF in the presence of nonimmune IgG. (B) DNA synthesis was determined by [3H]thymidine incorporation. Data are expressed as fold stimulation of basal incorporation observed in unstimulated cells in the presence of nonimmune IgG. Values are means plus or minus SD of 3 independent experiments performed in triplicate. (C) Migration was analyzed by Transwell assay. CM-Dil–stained HUVECs were preincubated with 20 μg of anti-NRP1 IgG or nonimmune IgG and dispensed into the upper chamber of the Fluoroblok inserts. Migration was induced by adding 10 ng/mL of HGF or VEGF-A165 to the lower chamber. The number of migrating cells was quantified by spectrofluorimetry. The results shown are means plus or minus SD of 3 independent experiments performed in triplicate. (D) HUVEC were preincubated with 20 μg/mL of anti-NRP1 IgG, anti-NRP2 IgG, or both, or with a nonimmune IgG and stimulated with HGF or VEGF-A165. DNA synthesis was determined by [3H]thymidine incorporation. Data are expressed as fold stimulation of basal incorporation observed in unstimulated cells in the presence of non immune IgG. Values are means plus or minus SD of 3 independent experiments performed in triplicate. *P < .05; **P < .01; ***P < .001 (Student t test).](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/4/10.1182_blood-2007-04-084269/3/m_zh80040815350005.jpeg?Expires=1769095764&Signature=gdxnB1tKnAn-roZClSphRh7S1~Tlo-FUtc~fGPI0BQJHzC3FdJxM2NajU-IRqBAXLUWvILtyyRvIpoYWEnRAlSvhowFPg8hT1Fx~FDdJ2YPVyhXSAFghHZcTmteorRwNWs1avxb5hGJRIE1NHyUhc3Ht7jGcYv4~uv9A73~0LL8p0FGS69mYv7Z080vi3tQfn94BUu~VpP4oWe85nU1pvO8O2hiHYsE1WIPB8QrnF7fMbOnfI1isZ0vhX~wgLQGq0ealCqqDonN0dQqQHQ06ddEao03dVppj2ON5shoxN-dY4k5W5m23Vb1zEqIMbyy9mKsJ9aYJOLoXCF7bXSxE1A__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)