Erlotinib, an inhibitor of the epidermal growth factor receptor (EGFR), induces differentiation, cell-cycle arrest, and apoptosis of EGFR-negative myeloblasts of patients with myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML), as well as in EGFR-negative cell lines representing these diseases (P39, KG-1, and HL 60). This off-target effect can be explained by inhibitory effects on JAK2. Apoptosis induction coupled to mitochondrial membrane permeabilization occurred independently from phenotypic differentiation. In apoptosis-sensitive AML cells, erlotinib caused a rapid (within less than 1 hour) nucleocytoplasmic translocation of nucleophosmin-1 (NPM-1) and p14ARF. Apoptosis-insensitive myeloblasts failed to manifest this translocation yet became sensitive to apoptosis induction by erlotinib when NPM-1 was depleted by RNA interference. Moreover, erlotinib reduced the growth of xenografted human AML cells in vivo. Erlotinib also killed CD34+ bone marrow blasts from MDS and AML patients while sparing normal CD34+ progenitors. This ex vivo therapeutic effect was once more associated with the nucleocytoplasmic translocation of NPM-1 and p14ARF. One patient afflicted with both MDS and non–small cell lung cancer manifested hematologic improvement in response to erlotinib. In summary, we here provide novel evidence in vitro, ex vivo, and in vivo for the potential therapeutic efficacy of erlotinib in the treatment of high-risk MDS and AML.
Introduction
Both high-risk myelodysplastic syndrome (MDS) and acute myeloid leukemia (AML) require new therapeutic approaches. On theoretical grounds, such approaches should target transformed cells and cause them to initiate at least 1 of 3 distinct yet interwoven biologic processes: (i) cell death by apoptosis, (ii) preferentially irreversible cell-cycle blockade, or (iii) terminal differentiation
A recent overall trend in oncology is the conception of so-called targeted therapies in which oncogenic events or surface markers are used to disrupt the essential transforming event and to selectively destroy tumor cells with a maximum of specificity and a minimum of side effect on nontransformed cells.1 Examples of such targeted therapies applied to hematologic malignancies include selective tyrosine kinase inhibitors (with imatinib as a ground-breaking paradigm for the treatment of chronic myeloid leukemia). For the treatment of solid tumors, quintessential examples are the small inhibitory compounds targeting the epidermal growth factor receptor (EGFR) in non–small cell lung cancer (NSCLC).2 Such small molecules directed against the EGFR include gefitinib and erlotinib.3,–5
Surprisingly, a recent study revealed the capacity of gefitinib to induce differentiation in 3 AML cell lines (U937, HL60, and Kasumi-1), which all lack expression of the EGFR,6 thus unraveling an interesting off-target effect of a compound that was believed to specifically act on EGFR-expressing cells. On the basis of this report, we studied the effects of the EGFR inhibitor erlotinib on EGFR-negative AML and MDS cells. Here, we report that erlotinib has an antineoplastic activity on MDS and AML cells that includes a proapoptotic effect. The therapeutic efficacy of erlotinib is demonstrated in vitro, ex vivo on MDS/AML-derived malignant myeloblasts, as well as in vivo. On the basis of these data, we propose the therapeutic off-target use of erlotinib for the treatment of high-risk MDS and AML.
Methods
Patients
Samples from 31 patients or healthy volunteers were assessed in our study (Table 1). Informed consent was obtained from all patients and healthy subjects in accordance with the Declaration of Helsinki. The diagnosis of AML and MDS was based on cytology of peripheral blood and bone marrow according to World Health Organization (WHO) classification and on conventional cytogenetic analysis.
Characteristics of healthy volunteers, MDS patients, and AML patients
| Sample no. . | Age, y . | Sex . | Diagnosis . | Cytogenetic analysis . | No. of cytopenias . | Risk group according to IPSS . |
|---|---|---|---|---|---|---|
| 1 | 53 | F | Normal* | 46, XX | — | — |
| 2 | 48 | M | Normal | 46, XY | — | — |
| 3 | 57 | M | Normal | 46, XY | — | — |
| 4 | 65 | M | Normal | 46, XY | — | — |
| 5 | 38 | M | Normal | 46, XY | — | — |
| 6 | 35 | M | Normal | 46, XY | — | — |
| 7 | 84 | F | RA† | 46, XX | 1 | Low |
| 8 | 76 | F | RA | del(5q) | 1 | Low |
| 9 | 78 | F | RA | 46, XX | 1 | Low |
| 10 | 51 | M | RA | del(5q) | 2 | Int-1 |
| 11 | 75 | M | RA | −y | 2 | Int-1 |
| 12 | 69 | M | RARS‡ | del(20q) | 1 | Low |
| 13 | 66 | F | RARS | 46, XX | 1 | Low |
| 14 | 77 | M | CMML | 46, XY | 1 | — |
| 15 | 77 | F | RAEB§-1 | del(11q) | 2 | Int-2 |
| 16 | 60 | F | RAEB-1 | inv(16) | 1 | Int-2 |
| 17 | 58 | M | RAEB-2 | Complex# with del(5q) | 2 | High |
| 18 | 78 | M | RAEB-2 | Complex with del(5q) | 2 | High |
| 19 | 67 | M | RAEB-2 | Complex with −7, +8 | 3 | High |
| 20 | 58 | M | RAEB-2 | −7 | 2 | High |
| 21 | 78 | M | RAEB-2 | 46, XY | 2 | Int-2 |
| 22 | 66 | M | tAML‖ | 46, XY | — | — |
| 23 | 80 | M | sAML¶ | Complex | — | — |
| 24 | 81 | M | sAML | del(9p), NPM-1 WT** | — | — |
| 25 | 80 | F | sAML | Trisomy 8 | — | — |
| 26 | 46 | F | AML | 46, XX, NPM-1 ITD†† | — | — |
| 27 | 56 | M | AML | 46, XY, NPM-1 WT | — | — |
| 28 | 52 | M | AML | 46, XY, NPM-1 WT | — | — |
| 29 | 23 | F | AML | 46, XX, NPM-1 WT | — | — |
| 30 | 73 | F | AML | 46, XX, NPM-1 WT | — | — |
| 31 | 79 | F | AML | Complex, NPM-1 WT | — | — |
| Sample no. . | Age, y . | Sex . | Diagnosis . | Cytogenetic analysis . | No. of cytopenias . | Risk group according to IPSS . |
|---|---|---|---|---|---|---|
| 1 | 53 | F | Normal* | 46, XX | — | — |
| 2 | 48 | M | Normal | 46, XY | — | — |
| 3 | 57 | M | Normal | 46, XY | — | — |
| 4 | 65 | M | Normal | 46, XY | — | — |
| 5 | 38 | M | Normal | 46, XY | — | — |
| 6 | 35 | M | Normal | 46, XY | — | — |
| 7 | 84 | F | RA† | 46, XX | 1 | Low |
| 8 | 76 | F | RA | del(5q) | 1 | Low |
| 9 | 78 | F | RA | 46, XX | 1 | Low |
| 10 | 51 | M | RA | del(5q) | 2 | Int-1 |
| 11 | 75 | M | RA | −y | 2 | Int-1 |
| 12 | 69 | M | RARS‡ | del(20q) | 1 | Low |
| 13 | 66 | F | RARS | 46, XX | 1 | Low |
| 14 | 77 | M | CMML | 46, XY | 1 | — |
| 15 | 77 | F | RAEB§-1 | del(11q) | 2 | Int-2 |
| 16 | 60 | F | RAEB-1 | inv(16) | 1 | Int-2 |
| 17 | 58 | M | RAEB-2 | Complex# with del(5q) | 2 | High |
| 18 | 78 | M | RAEB-2 | Complex with del(5q) | 2 | High |
| 19 | 67 | M | RAEB-2 | Complex with −7, +8 | 3 | High |
| 20 | 58 | M | RAEB-2 | −7 | 2 | High |
| 21 | 78 | M | RAEB-2 | 46, XY | 2 | Int-2 |
| 22 | 66 | M | tAML‖ | 46, XY | — | — |
| 23 | 80 | M | sAML¶ | Complex | — | — |
| 24 | 81 | M | sAML | del(9p), NPM-1 WT** | — | — |
| 25 | 80 | F | sAML | Trisomy 8 | — | — |
| 26 | 46 | F | AML | 46, XX, NPM-1 ITD†† | — | — |
| 27 | 56 | M | AML | 46, XY, NPM-1 WT | — | — |
| 28 | 52 | M | AML | 46, XY, NPM-1 WT | — | — |
| 29 | 23 | F | AML | 46, XX, NPM-1 WT | — | — |
| 30 | 73 | F | AML | 46, XX, NPM-1 WT | — | — |
| 31 | 79 | F | AML | Complex, NPM-1 WT | — | — |
— indicates not applicable.
Healthy volunteer.
Patient with refractory anemia.
Patient with refractory anemia with ringsideroblasts.
Patient with refractory anemia with excess of blasts (5%-9%, RAEB-1; 10%-19%, RAEB-2).
Patient with therapy-related AML.
Patient with secondary AML.
Complex karyotype was defined as 3 or more cytogenetic abnormalities.
Wild-type NPM-1.
Internal tandem duplication of NPM-1.
Cell lines and selection of CD34+ cells
Mononuclear cells (MNC) from the bone marrow (BM) were isolated by Ficoll-Paque PLUS density gradient (Amersham Biosciences, Sunnyvale, CA). To isolate CD34+ cells from MNC, a positive selection using the MiniMacs system (Miltenyi Biotec, Bergisch Gladbach, Germany) was carried out. CD34+ cells were maintained in Iscove modified Dulbecco medium (IMDM; Gibco, Carlsbad, CA) supplemented with 10% heat-inactivated fetal calf serum (FCS). The high-risk MDS cell line P39/Tsugane (kindly provided by Dr Yoshida Takeda, Japan) and the KG-1 and HL60 cell lines (kindly provided by Dr Martin Ruthardt, Germany, and Dr Bruno Cassinat, France, respectively) were cultured in FCS-supplemented RPMI 1640 (Gibco). Unless otherwise indicated, cells were seeded at a concentration of 105 cells/mL.
Assessment of cell death, cell-cycle distribution, proliferation, and differentiation
A total of 105 cells derived from patients or cell lines were resuspended in 1 mL culture medium and incubated with the indicated dosages of erlotinib (Roche, Basel, Switzerland). Control cells were incubated with the corresponding amount of dimethyl sulfoxide (DMSO). In the applied concentrations, DMSO did not affect proliferation, apoptosis, or cell-cycle distribution as compared with cells left untreated (data not shown). But because DMSO induced minor signs of differentiation, all assays assessing the differentiation-inducing capacity of erlotinib were carried out in the presence (“DMSO”) as well as in absence of DMSO (“control”). To inhibit activation of caspases, cells were incubated with 100 μM z-VAD-fmk. Apoptotic cells were detected by cytofluorometric analysis using a FACScan (Becton Dickinson, Mountain View, CA) as described previously.7,8 Cells were stained with propidium iodide (PI; 5 μg/mL; Sigma, Steinheim, Germany) and concomitantly either with 40 nM fluorochrome DiOC6(3) (3,3 dihexyloxacarbocyanine iodide; Molecular Probes, Eugene, OR) for 15 minutes at 37°C or with annexin-V-FITC (Becton Dickinson) according to the manufacturer's instructions to determine the mitochondrial transmembrane potential or phosphatidyl serine exposure, respectively.9,–11
For conventional cell-cycle analysis, cells were harvested, washed in PBS, and stained with PI (25 μg/mL, Sigma) followed by an incubation period of 30 minutes at 37°C. Subsequently, cell-cycle distribution was determined by cytofluorometric analysis using a FACS Vantage (Becton Dickinson).
Changes in proliferation were assessed by adding a predefined amount of immunofluorescent beads (AlignFlow; Invitrogen, Karlsruhe, Germany), emitting a signal at 488 nm to 300 μL of the cells cultured in suspension according to the manufacturer's instructions. In the subsequent fluorocytometric analysis, 3000 events elicited by the beads were counted with the concomitantly acquired number of cultured cells, allowing quantification of cell numbers in relation to the number of beads.
To fluorocytometrically assess differentiation, cells were harvested, washed, and stained with a phycoerythrin (PE)-conjugated anti-CD11b antibody (clone ICRF44, Becton Dickinson) indicating myelocytic differentiation as described previously.12 Isotypic mouse IgG1 (Becton Dickinson) was used to set threshold parameters.
Morphologic assessment of differentiation and apoptosis was carried out after Wright-Giemsa staining of cytospins. To quantify differentiation and apoptosis, at least 100 cells of each condition were assessed for signs of differentiation (decrease of cytoplasmic basophilia, appearance of granulation, lobulation of the nucleus) or apoptosis (chromatin condensation and nuclear fragmentation).
Analysis of the phosphoproteome
Differential expression of phosphoproteins was assessed in KG-1 cells cultured for 8 hours in the presence of 10 μM erlotinib or 0.02% DMSO. Analysis was carried out using the Kinex antibody microarray (Kinex Bioinformatics, Vancouver, BC). Samples were prepared according to the manufacturer's instructions.
Fluorocytometric assessment of STAT-5 activation
Cells were harvested, washed, and subsequently fixed and permeabilized using the BD Cytofix/Cytoperm kit (Becton Dickinson) according to the manufacturer's instructions. Cells were first stained with the primary antibody detecting activated STAT-5 (P-Tyr694, Cell Signaling, Danvers, MA), followed by staining with a secondary FITC-conjugated antibody. Isotypic controls were used to set threshold parameters.
Knockdown of proteins by small interfering RNA
Cells were transfected with the Nucleofactor system (Amaxa, Cologne, Germany) as described previously12 using 2 different small interfering RNA (siRNA) specific for NPM-1 (sense strain: 5-CAC CAC CAG UGU CUU AAG TT-3; Sigma) or JAK-2 (sense strain: 5-GAA GAG CAC CUA AGA GAC UTT-3; Eurogentec, Seraing, Belgium). Two different scramble siRNAs were used as controls. For knockdown of Src (pp60Src), PDGFRα and PDGFRβ SMARTpool siRNA together with the control siRNA and the respective antibodies were purchased from Dharmacon (Lafayette, CO).
Immunoblot analysis
Lysates from 5 × 106 cells were separated on sodium dodecyl sulfate (SDS)–polyacrylamide gels and electroblotted onto polyvinylidene difluoride membranes by using standard procedures. The membrane was blocked with 5% nonfat, dry milk and incubated with the respective primary antibody: actin (mouse monoclonal Ab; Chemicon, Temecula, CA), EGFR (rabbit polyclonal Ab; Santa Cruz Biotechnology, Santa Cruz, CA); cyclin E (mouse monoclonal Ab; Santa Cruz Biotechnology), cyclin D1 (mouse monoclonal Ab; Santa Cruz Biotechnology), retinoblastoma (mouse monoclonal Ab; Cell Signaling), retinoblastoma (P-Ser780, P-Ser795, P-Ser807/811, rabbit monoclonal Abs; Cell Signaling), STAT-5 (rabbit monoclonal Ab; Cell Signaling), activated STAT-5 (P-Tyr694; rabbit monoclonal Ab; Cell Signaling), NPM-1 (rabbit monoclonal Ab; Cell Signaling), JAK2 (rabbit polyclonal Ab; Santa Cruz Biotechnology), and activated JAK2 (P-Tyr1007/1008; rabbit monoclonal Ab; Cell Signaling). Antibodies against Src (pp60Src), PDGFRα and PDGFRβ were purchased (together with the respective SMARTpool siRNA) from Dharmacon. Blots were stained with either goat anti–rabbit peroxidase-labeled or goat anti–mouse peroxidase-labeled secondary antibody (Amersham, Arlington Heights, IL) and were revealed using an enhanced chemiluminescence detection system (Amersham).
Immunofluorescence
Cells of patients or cell lines were allowed to adhere on polylysine-L coverslips (Sigma) and were fixed in 4% paraformaldehyde at room temperature. Cells were then permeabilized with SDS 0.1% for 10 minutes, washed in PBS, and stained with the indicated primary antibodies—cytochrome c (mouse monoclonal Ab; BD Pharmingen, Heidelberg, Germany), endonuclease G (rabbit polyclonal Ab; Pro-Science, Woburn, MA), Hsp60 (mouse polyclonal Ab; Sigma), activated caspase-3 (rabbit polyclonal Ab, Cell Signaling), NPM-1 (rabbit or mouse monoclonal Ab; Santa Cruz Biotechnology), p14ARF (mouse monoclonal Ab; Sigma), NF-κB p65 (rabbit polyclonal Ab; Santa Cruz Biotechnology)—and revealed with the adequate secondary antibody coupled with Alexa 568 (red) or Alexa 488 (green) fluorochromes (Molecular Probes/Invitrogen). DNA of cells was counterstained with DAPI (Molecular Probes/Invitrogen). At least 100 cells for each slide were examined independently with a LSM 510 confocal microscope (Zeiss, Thornwood, NY) at 63× magnification. Background correction of fluorescence was performed with the LSM 5 image browser (Zeiss).
Evaluation of the therapeutic efficacy of erlotinib in a murine leukemia model
Antileukemic activity of erlotinib was evaluated in female, severe combined immunodeficient (SCID) mice (Charles River, Wilmington, MA). Animals were housed in suitable cages under specified pathogen-free conditions in rooms maintained at 23°C and 50% humidity, with a 12-hour light/12-hour dark cycle. The mice were quarantined during the acclimatization period of at least 1 week. Food (Standard 1320 and 1430; Altromin, Lange, Germany) and acidified water (pH 2.5-3.0) were available ad libitum. Mice (in groups of 4) were injected intraperitoneally with 106 KG-1 cells on day 0 as described previously.13 Mice were then randomly assigned to either the treatment or the control group. Treatment was initiated on day 7 after intraperitoneal injection of tumor cells. The treatment group received a daily dose of 100 mg/kg erlotinib (diluted in sterile water and 10% DMSO) orally on 5 consecutive days, followed by a treatment pause of 2 days. Mice in the control group received water/DMSO accordingly. Mice were controlled daily for signs of toxicity and tumor/aszites development. Xenografts were measured daily in 2 dimensions with a caliper. At the end of the studies, all animals were killed by cervical dislocation under isoflurane (1-chloro-2,2,2-trifluoroethyldifluoromethylether; CuraMED Pharma, Karlsruhe, Germany) anesthesia.
Explanted tumor and specimen investigations
Explanted tumors were fixed in 3.8% buffered formaldehyde, embedded in Paraplast (Sherwood Medical, Norfolk, NE), and prepared in 5-μm sections. For histopathologic examination, sections were stained with hematoxylin/eosin (HE) and examined by light microscopy at 200× magnification by a certified histopathologist. To assess the presence of malignant cells in the ascites, peritoneal exudate was collected by peritoneal lavage and cells were visualized on cytospins by Wright-Giemsa staining.
Statistics
Statistics were analyzed using Excel Software (Microsoft, Redmond, WA), SPSS software (SAS Institute, Cary, NC), and Scion Image 4.0 (Scion, Frederick, MD). Tumor-free survival in mice was analyzed by using the Kaplan-Meier method. Statistical significance was determined using the log-rank test. Apoptosis induced by erlotinib in ex vivo cells of the MDS and AML subgroups is depicted using a box plot graph, and statistical significance was assessed using the Student t test.
Results
Off-target effects of erlotinib on MDS/AML cell lines
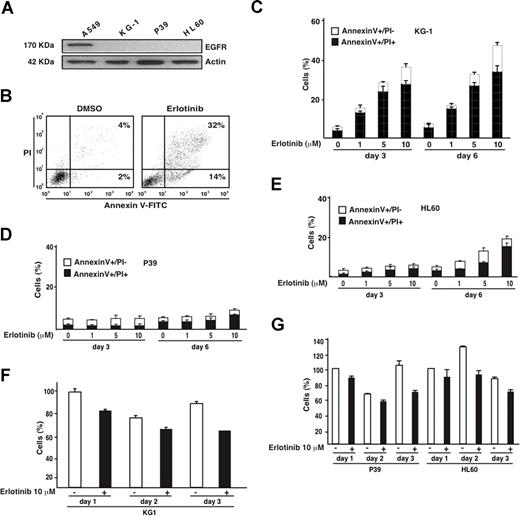
In contrast to the non–small cell lung cancer cell line A549, the AML cell lines KG-1 and HL60 and the MDS-derived cell line P39 lack EGFR expression (Figure 1A), meaning that any effect of the bona fide EGFR antagonist erlotinib on these cells must be considered as “off-target.” Erlotinib efficiently induced the apoptosis-associated phosphatidylserine exposure (detected with annexin V-FITC conjugates) followed by the loss of viability (detected with the vital dye PI) in KG-1 cells (Figure 1B,C), yet failed to do so in P39 (Figure 1D) and HL60 cells (Figure 1E). Erlotinib also reduced the absolute number of viable KG-1 (and to a lesser extent the viability of P39 or HL60 cells) cells (Figure 1F,G). Phosphoproteome analysis of KG-1 cells revealed that erlotinib influenced the phosphorylation status of numerous proteins (data not shown), including the retinoblastoma (Rb) protein, which is critical for G1/S transition of the cell cycle,14 and the platelet-derived growth factor receptors PDGFRα and PDGFRβ, which are involved in hematopoietic differentiation.15,16 We therefore assessed the effect of erlotinib on cell-cycle progression and differentiation of malignant myeloblasts.
Proapoptotic effect of erlotinib on EGFR-negative myeloid cell lines. (A) Absence of EGFR expression on KG-1, P39, and HL60 cells, as determined by immunoblot. A549 cells were included as positive control. (B) Representative annexin V-FITC/PI stainings of erlotinib-treated KG-1 cells, 3 days after treatment, as compared with DMSO-only (0.02%) treated controls. (C-E) Quantitation of dying (annexin V+/PI−) or dead (annexin V+/PI+) KG-1 (C), P39 (D), or HL60 (E) cells, 3 or 6 days after culture with the indicated concentrations of erlotinib. (F,G) Influence of erlotinib on the number of viable KG-1 (F) and P39 and HL60 (G) cells. Results are means plus or minus SD of triplicates. These experiments were repeated at least 3 times, yielding comparable results.
Proapoptotic effect of erlotinib on EGFR-negative myeloid cell lines. (A) Absence of EGFR expression on KG-1, P39, and HL60 cells, as determined by immunoblot. A549 cells were included as positive control. (B) Representative annexin V-FITC/PI stainings of erlotinib-treated KG-1 cells, 3 days after treatment, as compared with DMSO-only (0.02%) treated controls. (C-E) Quantitation of dying (annexin V+/PI−) or dead (annexin V+/PI+) KG-1 (C), P39 (D), or HL60 (E) cells, 3 or 6 days after culture with the indicated concentrations of erlotinib. (F,G) Influence of erlotinib on the number of viable KG-1 (F) and P39 and HL60 (G) cells. Results are means plus or minus SD of triplicates. These experiments were repeated at least 3 times, yielding comparable results.
Erlotinib had no effect on the total levels or Rb protein yet reduced the phosphorylation of Rb on serines 807/811, 780, and 795, coupled to a reduction in the G1/S cyclins E and D1 (Figure 2A). Accordingly, erlotinib led to a blockade in G1 with a consequent diminution of S and G2/M phases (Figure 2B,C). This cell-cycle blockade manifested before erlotinib-treated KG-1 cells became apoptotic and accumulated in the subG1 phase. Erlotinib induced cell-cycle arrest in KG-1 cells and—although to a lesser extent—in P39 and HL60 cells (Figure 2D,E).
Cell-cycle effects of erlotinib on EGFR-negative myeloid cell lines. (A) Immunoblot detection of Rb phosphorylation and G1/S cyclins. KG-1 cells were treated for the indicated period with 10 μM erlotinib, followed by lysis, SDS-PAGE, and immunochemical detection of the indicated antigens. (B) Cell-cycle analysis of KG-1 cells, 24 hours after treatment with 10 μM erlotinib as compared with cells treated with DMSO (0.02%) only. (C-E) Quantitation of erlotinib-mediated cell-cycle effects on KG-1 (C), P39 (D), or HL60 (E) cells. Results are means plus or minus SD of triplicates of 1 experiment representative for 4.
Cell-cycle effects of erlotinib on EGFR-negative myeloid cell lines. (A) Immunoblot detection of Rb phosphorylation and G1/S cyclins. KG-1 cells were treated for the indicated period with 10 μM erlotinib, followed by lysis, SDS-PAGE, and immunochemical detection of the indicated antigens. (B) Cell-cycle analysis of KG-1 cells, 24 hours after treatment with 10 μM erlotinib as compared with cells treated with DMSO (0.02%) only. (C-E) Quantitation of erlotinib-mediated cell-cycle effects on KG-1 (C), P39 (D), or HL60 (E) cells. Results are means plus or minus SD of triplicates of 1 experiment representative for 4.
When exposed to erlotinib or all-trans retinoic acid (ATRA), KG-1 cells manifested comparable signs of differentiation, namely a reduction in cytoplasmic basophilia and an increased nuclear lobulation that occurred in nonapoptotic cells (Figure 3A,B). Similar signs of differentiation could be observed in P39 and HL60 cells, which also up-regulated the myelocytic differentiation antigen CD11b (Figure 3C-H). Because these cells did not (or did only marginally) undergo apoptosis in response to erlotinib, the proapoptotic and differentiation-inducing effects of erlotinib can be dissociated (Figure 3E,H). All together, these results indicate that erlotinib can exert antineoplastic off-target effects on a range of malignant myeloid cell lines.
Erlotinib-induced differentiation of EGFR-negative myeloid cell lines. (A) Representative Wright-Giemsa staining of KG-1 cells cultured 6 days in the absence (control) or presence of DMSO (0.02%), erlotinib (10 μM), or all-trans retinoic acid (ATRA, 1 μM), revealing signs of differentiation, ie, reduced cytoplasmic basophilia and nuclear lobulation, or apoptosis, ie, chromatin condensation (pyknosis) and nuclear fragmentation (karyorrhexis). (B) Quantitation of results, as obtained in panel A, on 100 or more cells. (C-H) Erlotinib-driven differentiation in P39 (C-E) or HL60 (F-H) cells. Differentiation was quantified (X ± SD of triplicates) by immunofluorocytometric measurement of surface CD11b on day 3 (quantified in panels D,G) and day 6 (panels C,D and F,G) or by Wright-Giemsa staining of cytospins on day 6 of the incubation period (panels C,E and F,H). Results are representative of at least 3 independent experiments.
Erlotinib-induced differentiation of EGFR-negative myeloid cell lines. (A) Representative Wright-Giemsa staining of KG-1 cells cultured 6 days in the absence (control) or presence of DMSO (0.02%), erlotinib (10 μM), or all-trans retinoic acid (ATRA, 1 μM), revealing signs of differentiation, ie, reduced cytoplasmic basophilia and nuclear lobulation, or apoptosis, ie, chromatin condensation (pyknosis) and nuclear fragmentation (karyorrhexis). (B) Quantitation of results, as obtained in panel A, on 100 or more cells. (C-H) Erlotinib-driven differentiation in P39 (C-E) or HL60 (F-H) cells. Differentiation was quantified (X ± SD of triplicates) by immunofluorocytometric measurement of surface CD11b on day 3 (quantified in panels D,G) and day 6 (panels C,D and F,G) or by Wright-Giemsa staining of cytospins on day 6 of the incubation period (panels C,E and F,H). Results are representative of at least 3 independent experiments.
Mechanisms of erlotinib-mediated cell death
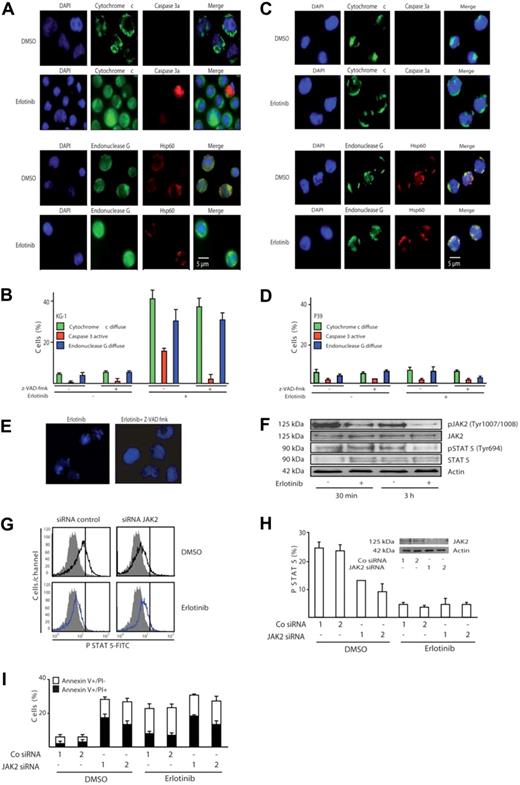
The phenotype of cell death induced by erlotinib in KG-1 was clearly apoptotic, with early phosphatidylserine exposure (Figure 1B,C), loss of nuclear DNA (Figure 2), and nuclear pyknosis and karyorrhexis (Figure 3A). Moreover, KG-1 cells incubated with erlotinib manifested biochemical hallmarks of apoptosis, namely the release of cytochrome c from mitochondria, coupled to the activation of caspase-3, as determined by immunofluorescence staining. In addition, a significant fraction of erlotinib-treated KG-1 cells exhibited the mitochondrial release of the caspase-independent death effector endonuclease G (Figure 4A,B). No such effect could be detected for P39 cells (Figure 4C,D), which did not die in response to erlotinib (Figures 1,Figure 2–3). Inhibition of caspases by Z-VAD-fmk suppressed the activation of caspase-3 in KG-1 cells treated with erlotinib yet had no effect on the mitochondrial release of cytochrome c or endonuclease G (Figure 4B), indicating that mitochondrial outer membrane permeabilization occurred independently from (and presumably upstream of) caspase activation in erlotinib-induced cell death. Z-VAD-fmk fully blocked the (caspase-dependent) pyknosis/karyorrhexis of KG-1 cells treated by erlotinib yet failed to inhibit the (caspase-independent) exposure of phosphatidylserine and cell death induced by erlotinib (Figure 4E and data not shown). Hence, caspase activation accompanies erlotinib-induced cell death yet is not required for cellular demise.
Erlotinib induces caspase-independent mitochondrial outer membrane permeabilization (MOMP) and disruption of the JAK2/STAT-5 pathway. (A) Representative fluorescence microphotographs of KG-1 cells treated with 10 μM erlotinib or DMSO as a control, after staining for the visualization of nuclear chromatin, cytochrome c, endonuclease G, and activated caspase-3 (caspase 3a). Note the mitochondrial distribution of cytochrome c and endonuclease G as cytoplasmic (nonnuclear) dots at the same location as the mitochondrial matrix marker Hsp60 or their diffuse distribution throughout the cell. (B) Quantitative assessment of the data obtained as in panel A, for KG-1 cells cultured for 3 days in the absence or presence of erlotinib and/or the pan-caspase inhibitor Z-VAD-fmk. (C,D) Immunofluorescence assessment of MOMP and caspase-3 activation in erlotinib-treated P39 cells. The same technology as in panels A and B was used on P39 cells. Results are means plus or minus SD of triplicates of 1 experiment representative of 3. (E) The pan-caspase inhibitor Z-VAD-fmk blocks pyknosis and karyorrhexis in erlotinib-treated KG-1 cells. Cells were treated as described in Figure 4A,B. (F-I) Erlotinib disrupts signaling of the JAK2/STAT-5 pathway. (F) Erlotinib decreases phosphorylation of JAK2 and STAT-5. KG-1 cells treated for 30 minutes or 3 hours with erlotinib were subjected for immunoblot detection of JAK2 and STAT-5 phosphorylation. (G,H) Impact of erlotinib and JAK2 on STAT-5 phosphorylation. After depletion of JAK2 with 2 distinct siRNAs (see panel H inset) and 24 hours of erlotinib treatment (10 μM), cells were permeabilized and subjected to the immunofluorometric quantitation of STAT-5 phosphorylation. Representative FACS histograms obtained for the first of 2 control and JAK2-specific siRNAs are shown in panel G (gray curves indicate isotype controls), and quantitative data (expressed as percentage of positive cells, X ± SD, n = 3) are depicted in panel H. Note the decrease in STAT-5 activation (shift toward the isotype) upon erlotinib treatment alone and upon down-regulation of JAK2. (I) Impact of erlotinib and JAK2 on cell death. KG-1 cells were transfected with control siRNAs or JAK2-specific siRNAs (day 0) and then cultured in the absence or presence of 10 μM erlotinib (from day 1–2 of transfection), followed by determination of the frequency (X ± SD, n = 3) of dying and dead cells using the annexin V/PI staining method used in Figure 1B. This experiment has been repeated twice, yielding similar results.
Erlotinib induces caspase-independent mitochondrial outer membrane permeabilization (MOMP) and disruption of the JAK2/STAT-5 pathway. (A) Representative fluorescence microphotographs of KG-1 cells treated with 10 μM erlotinib or DMSO as a control, after staining for the visualization of nuclear chromatin, cytochrome c, endonuclease G, and activated caspase-3 (caspase 3a). Note the mitochondrial distribution of cytochrome c and endonuclease G as cytoplasmic (nonnuclear) dots at the same location as the mitochondrial matrix marker Hsp60 or their diffuse distribution throughout the cell. (B) Quantitative assessment of the data obtained as in panel A, for KG-1 cells cultured for 3 days in the absence or presence of erlotinib and/or the pan-caspase inhibitor Z-VAD-fmk. (C,D) Immunofluorescence assessment of MOMP and caspase-3 activation in erlotinib-treated P39 cells. The same technology as in panels A and B was used on P39 cells. Results are means plus or minus SD of triplicates of 1 experiment representative of 3. (E) The pan-caspase inhibitor Z-VAD-fmk blocks pyknosis and karyorrhexis in erlotinib-treated KG-1 cells. Cells were treated as described in Figure 4A,B. (F-I) Erlotinib disrupts signaling of the JAK2/STAT-5 pathway. (F) Erlotinib decreases phosphorylation of JAK2 and STAT-5. KG-1 cells treated for 30 minutes or 3 hours with erlotinib were subjected for immunoblot detection of JAK2 and STAT-5 phosphorylation. (G,H) Impact of erlotinib and JAK2 on STAT-5 phosphorylation. After depletion of JAK2 with 2 distinct siRNAs (see panel H inset) and 24 hours of erlotinib treatment (10 μM), cells were permeabilized and subjected to the immunofluorometric quantitation of STAT-5 phosphorylation. Representative FACS histograms obtained for the first of 2 control and JAK2-specific siRNAs are shown in panel G (gray curves indicate isotype controls), and quantitative data (expressed as percentage of positive cells, X ± SD, n = 3) are depicted in panel H. Note the decrease in STAT-5 activation (shift toward the isotype) upon erlotinib treatment alone and upon down-regulation of JAK2. (I) Impact of erlotinib and JAK2 on cell death. KG-1 cells were transfected with control siRNAs or JAK2-specific siRNAs (day 0) and then cultured in the absence or presence of 10 μM erlotinib (from day 1–2 of transfection), followed by determination of the frequency (X ± SD, n = 3) of dying and dead cells using the annexin V/PI staining method used in Figure 1B. This experiment has been repeated twice, yielding similar results.
Erlotinib reduced the (auto)phosphorylation of the oncogenic JAK2 kinase on tyrosine 1007/1008, when added to KG-1 cells (Figure 4F), in accord with the previously reported capacity of erlotinib to directly inhibit JAK2.17 The transcription factor STAT-5, a direct target for phosphorylation by JAK2 on tyrosine 694,18 was hypophosphorylated after erlotinib treatment, as determined by immunoblot (Figure 4F) and confirmed by cytofluorometric analysis (Figure 4G; note the left shift of the staining after erlotinib as compared with the vehicle control). Thus both assays demonstrate that erlotinib alone is sufficient to reduce the constitutive STAT-5 activation in KG-1 cells. To assess the functional relevance of the JAK2–STAT-5 signaling in erlotinib-induced apoptosis, expression of JAK2 was down-regulated by RNA interference. As depicted in Figure 4H,I, abrogation of JAK2 expression alone was sufficient to diminish activation of STAT-5 and to concomitantly induce apoptosis in KG-1 cells. The combination of JAK2 knockdown and erlotinib did not induce more apoptosis than erlotinib induced alone. This result is compatible with the hypothesis that erlotinib causes apoptosis at least in part by inhibiting JAK2. In contrast, erlotinib was still able to increase the apoptosis induced by knockdown of the PDGFR and Src, suggesting that these kinases are not (or are less) relevant to the proapoptotic effect of erlotinib (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
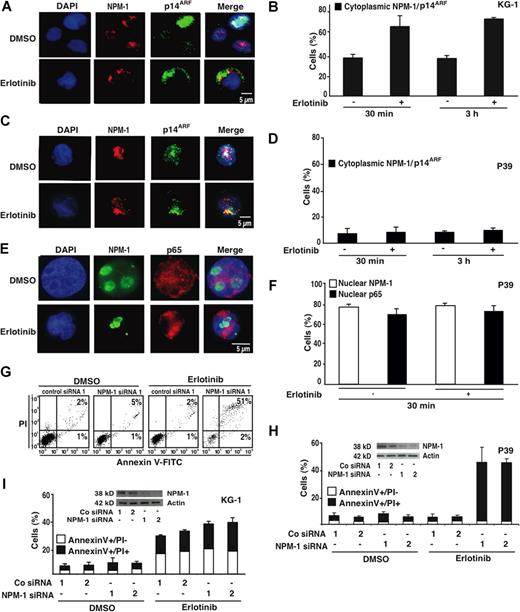
As a further attempt to explore the antineoplastic action of erlotinib, we studied its effects on the myelomonocytic master regulator nucleophosmin-1 (NPM-1). In KG-1 cells, erlotinib stimulated the rapid (within 30 minutes) redistribution of NPM-1 from the nucleus to the cytoplasm (Figure 5A,B). This translocation was always accompanied by that of the tumor suppressor protein p14ARF (Figure 5A,B), which is known to interact with NPM-1.19,,–22 Apoptosis-resistant P39 cells failed to release NPM-1, p14ARF, and NF-κB p65—which is constitutively activated in P39 cells and imparts an antiapoptotic signal12 —from their nuclei (Figure 5C-F). Depletion of NPM-1 with 2 distinct siRNAs was not sufficient to induce apoptosis, nor did it affect the apoptogenic action of erlotinib on KG-1 cells (Figure 5G-I). However, down-regulation of NPM-1 established the sensitivity of P39 cells (which normally did not undergo apoptosis; Figures 1,Figure 2–3) in response to erlotinib (Figure 5G,H). These results suggest that inactivation of NPM-1 would be essential for the lethal action of erlotinib.
Influence of nucleophosmin-1 (NPM-1) on erlotinib-mediated cytolysis. (A) Typical immunofluorescence images obtained after staining of KG-1 cells with antibodies specific for NPM-1 and p14ARF. Chromatin was counterstained using DAPI. Note the nuclear localization of NPM-1 and p14ARF that is lost after erlotinib treatment (10 μM, 30 minutes) in favor of a cytoplasmic staining. (B) Quantitation of the data obtained as in panel A, 30 minutes or 3 hours after addition of erlotinib. (C-F) Failure of erlotinib to affect the subcellular localization of NPM-1 (C-F), p14ARF (C,D) and NF-κB p65 (E,F) in erlotinib-resistant P39 cells. Representative immunofluorescence microphotographs are shown in panels C and E, and quantitative results are reported in panels D and F. (G-I) Effect of NPM-1 knockdown on the apoptosis-inducing capacity of erlotinib in P39 and KG-1 cells. NPM-1 was knocked down with 2 distinct siRNA heteroduplexes (insets in panels H and I). Twenty-four hours after transfection erlotinib (or DMSO as a solvent control) was added for 48 hours, and the frequency of dying and dead cells was measured using the annexin V/PI method. Representative FACS pictograms are shown for P39 cells in panel G, and quantitative results (X ± SD, n = 3) are depicted for both P39 and KG-1 cells in panels H and I.
Influence of nucleophosmin-1 (NPM-1) on erlotinib-mediated cytolysis. (A) Typical immunofluorescence images obtained after staining of KG-1 cells with antibodies specific for NPM-1 and p14ARF. Chromatin was counterstained using DAPI. Note the nuclear localization of NPM-1 and p14ARF that is lost after erlotinib treatment (10 μM, 30 minutes) in favor of a cytoplasmic staining. (B) Quantitation of the data obtained as in panel A, 30 minutes or 3 hours after addition of erlotinib. (C-F) Failure of erlotinib to affect the subcellular localization of NPM-1 (C-F), p14ARF (C,D) and NF-κB p65 (E,F) in erlotinib-resistant P39 cells. Representative immunofluorescence microphotographs are shown in panels C and E, and quantitative results are reported in panels D and F. (G-I) Effect of NPM-1 knockdown on the apoptosis-inducing capacity of erlotinib in P39 and KG-1 cells. NPM-1 was knocked down with 2 distinct siRNA heteroduplexes (insets in panels H and I). Twenty-four hours after transfection erlotinib (or DMSO as a solvent control) was added for 48 hours, and the frequency of dying and dead cells was measured using the annexin V/PI method. Representative FACS pictograms are shown for P39 cells in panel G, and quantitative results (X ± SD, n = 3) are depicted for both P39 and KG-1 cells in panels H and I.
In vivo and ex vivo effects of erlotinib on malignant cells of MDS and AML
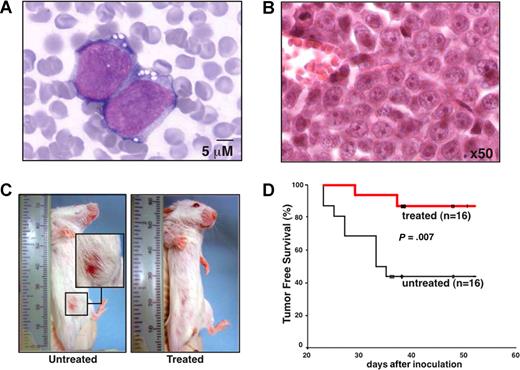
To verify that a therapeutic effect of erlotinib in EGFR-negative myeloblasts can be obtained in vivo, we inoculated KG-1 cells into the peritoneal cavity of SCID mice, a manipulation that led to the formation of malignant ascites (Figure 6A), peritoneal carcinosis, and the development of palpable tumors in the lower abdomen (Figure 6B,C). In this setting, oral treatment with erlotinib (5 days per week, 100 mg/kg per day) significantly suppressed the development of clinically detectable tumors (Figure 6C,D). Thus, erlotinib can exert antineoplastic effects in vivo at an acceptable level of toxicity.
Therapeutic efficacy of erlotinib on human AML xenografts in SCID mice. (A,B) Cytologic and histopathologic features of KG-1 cells injected into SCID mice. Forty days after intraperitoneal inoculation of 106 KG-1 cells, ascites was subjected to Wright-Giemsa staining (A) and abdominal tumors were analyzed by HE staining (B) to demonstrate the presence of AML cells in the tissues. (C) Representative mice that were vehicle-only or erlotinib-treated with the presence or absence of abdominal tumor masses are shown. (D) Kaplan-Meier plot showing tumor-free survival. Mice were inoculated with KG-1 cells on day 0, and oral erlotinib administration was started on day 7 (5 days per week, 100 mg/kg per day). This experiment has been repeated once, yielding similar results.
Therapeutic efficacy of erlotinib on human AML xenografts in SCID mice. (A,B) Cytologic and histopathologic features of KG-1 cells injected into SCID mice. Forty days after intraperitoneal inoculation of 106 KG-1 cells, ascites was subjected to Wright-Giemsa staining (A) and abdominal tumors were analyzed by HE staining (B) to demonstrate the presence of AML cells in the tissues. (C) Representative mice that were vehicle-only or erlotinib-treated with the presence or absence of abdominal tumor masses are shown. (D) Kaplan-Meier plot showing tumor-free survival. Mice were inoculated with KG-1 cells on day 0, and oral erlotinib administration was started on day 7 (5 days per week, 100 mg/kg per day). This experiment has been repeated once, yielding similar results.
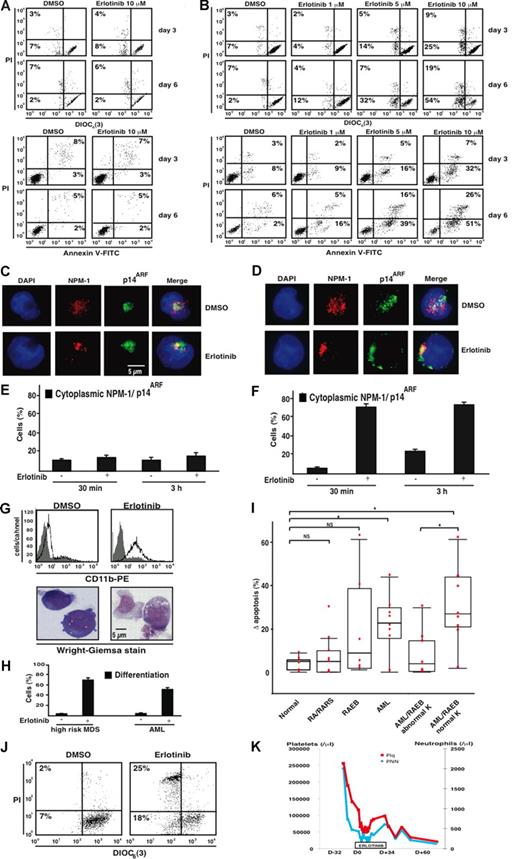
Next, we determined whether erlotinib acts on freshly purified CD34+ bone marrow blasts from AML patients, comparing them with CD34+ bone marrow progenitors from healthy volunteers. Whereas normal CD34+ cells were not harmed by erlotinib (Figure 7A), AML-derived CD34+ bone marrow blasts exhibited a dose- and time-dependent increase in apoptosis, as determined by 2 distinct methods: first, by assessing the frequency of cells that exposed phosphatidylserine on their surface (and hence stained positive with annexin V-FITC conjugates), and second, by measuring the portion of cells that lost the mitochondrial transmembrane potential (Δψm and hence manifest reduced incorporation of the Δψm-sensitive cyanine dye DiOC6(3)) before they lost viability and incorporated PI (Figure 7B). As to be expected from the results obtained with the erlotinib-sensitive AML cell line KG-1 (Figure 5A,B), primary myeloblasts from patients who responded to erlotinib ex vivo manifested a rapid nucleocytoplasmic translocalization of NPM-1 and p14ARF (Figure 7D,F), which was not seen in normal CD34+ bone marrow cells (Figure 7C,E). Moreover, the ex vivo treatment of primary myeloblasts responding to erlotinib recapitulated 2 further effects that we observed on KG-1 cells: an increase in the surface expression level of the myelocytic marker CD11b and morphologic signs of differentiation (Figure 7G,H). Ex vivo treatment of CD34+ bone marrow cells from a cohort of control subjects and patients with MDS and AML confirmed that an increasing fraction of patients—but not the healthy volunteers—manifested a strong (> 20%) apoptosis induction upon erlotinib administration (Figure 7I). We could not correlate the therapeutic response with the mutational status of NPM-1 (because only one AML patient in our cohort carried a NPM-1 mutation). During the course of our study, we had the opportunity to evaluate the in vivo effects of erlotinib in a patient diagnosed with 2 malignancies: a metastatic EGFR-positive NSCLC and a high-risk MDS (RAEB-2) with a normal karyotype. Noteworthy, the patient's CD34+ bone marrow myeloblasts responded to erlotinib ex vivo (Figure 7J). Because the NSCLC progressed after chemotherapeutic treatment (and because the patient's considerable comorbidity presented a contraindication for further chemotherapy), a single agent treatment with 150 mg/day erlotinib for the NSCLC (a disease for which this drug is approved by European Union Health Authorities) was initiated, while high-risk MDS was treated with best supportive care. Routine blood examinations carried out before and during erlotinib monotherapy revealed that the patient manifested a transient, objective hematopoietic improvement (according to the IWG-2006 criteria)23 on platelet and neutrophil counts (without achieving a complete remission considering that the initial percentage of bone marrow blasts remained stable at around 15% during the treatment). This hematopoietic improvement was maintained throughout the duration of erlotinib treatment (34 days). However, after discontinuation of erlotinib, platelet and neutrophil counts declined (Figure 7K).
Therapeutic effects of erlotinib ex vivo and in vivo. (A,B) Representative FACS diagrams of CD34+ bone marrow cells from a healthy volunteer and an AML patient with a normal karyotype. Cells were cultured with medium plus DMSO or erlotinib for 3 to 6 days, followed by the assessment of cell death with 2 distinct methods, the DiOC6(3)/PI and annexin V/PI staining of dying (DiOC6(3)low PI− or AnnV+PI−) and dead cells (PI+). Numbers in each quadrant are the percentages of total cells gated within the quadrant. (C-F) NPM-1 and p14ARF translocation induced by erlotinib in AML cells (D,F), but not in normal CD34+ bone marrow cells (C,E). Cells were treated and assessed as described in Figure 5A,D. Error bars represent SD. (G,H) Induction of myeloid differentiation by erlotinib in malignant myeloblasts. CD34+ bone marrow cells from a high-risk MDS (G,H) and an AML patient (H) were cultured for 6 days in the presence of 0.02% DMSO or 10 μM erlotinib. CD11b surface exposure was measured by FACS analysis while excluding apoptotic cells. (G) The morphology and percentage of cells exhibiting signs of erlotinib-induced differentiation were determined by Wright-Giemsa staining as described in Figure 3. (I) Quantitative comparison of apoptosis induction in a cohort of normal controls and different subgroups of MDS/AML patients. Results are depicted as a box plot, giving the increase in apoptosis induced by erlotinib (10 μM, 3 days) in vitro/ex vivo as compared with DMSO-only–treated controls in the indicated subgroups. K indicates karyotype; NS, not significant; *P < 0.01. The horizontal bar is the mean, the box the 25th percentile, and the whiskers the extremes. (J,K) Comparison of the ex vivo and in vivo effects of erlotinib on a patient with high-risk MDS. A patient diagnosed with metastatic EGFR-positive NSCLC and MDS RAEB-2 was treated with a monotherapy of erlotinib for the NSCLC. (J) Apoptosis-inducing effect of erlotinib on the patient's CD34+ bone marrow cells ex vivo (3 days, 10 μM) as assessed by staining with DiOC6(3)/PI. (K) Routine blood examinations before and during monotherapy with erlotinib demonstrated the in vivo effect of erlotinib on platelet and neutrophil counts resulting in a hematologic improvement. D-32 indicates day 32 before erlotinib treatment; D0, start of erlotinib therapy; D+34, day 34 under erlotinib treatment.
Therapeutic effects of erlotinib ex vivo and in vivo. (A,B) Representative FACS diagrams of CD34+ bone marrow cells from a healthy volunteer and an AML patient with a normal karyotype. Cells were cultured with medium plus DMSO or erlotinib for 3 to 6 days, followed by the assessment of cell death with 2 distinct methods, the DiOC6(3)/PI and annexin V/PI staining of dying (DiOC6(3)low PI− or AnnV+PI−) and dead cells (PI+). Numbers in each quadrant are the percentages of total cells gated within the quadrant. (C-F) NPM-1 and p14ARF translocation induced by erlotinib in AML cells (D,F), but not in normal CD34+ bone marrow cells (C,E). Cells were treated and assessed as described in Figure 5A,D. Error bars represent SD. (G,H) Induction of myeloid differentiation by erlotinib in malignant myeloblasts. CD34+ bone marrow cells from a high-risk MDS (G,H) and an AML patient (H) were cultured for 6 days in the presence of 0.02% DMSO or 10 μM erlotinib. CD11b surface exposure was measured by FACS analysis while excluding apoptotic cells. (G) The morphology and percentage of cells exhibiting signs of erlotinib-induced differentiation were determined by Wright-Giemsa staining as described in Figure 3. (I) Quantitative comparison of apoptosis induction in a cohort of normal controls and different subgroups of MDS/AML patients. Results are depicted as a box plot, giving the increase in apoptosis induced by erlotinib (10 μM, 3 days) in vitro/ex vivo as compared with DMSO-only–treated controls in the indicated subgroups. K indicates karyotype; NS, not significant; *P < 0.01. The horizontal bar is the mean, the box the 25th percentile, and the whiskers the extremes. (J,K) Comparison of the ex vivo and in vivo effects of erlotinib on a patient with high-risk MDS. A patient diagnosed with metastatic EGFR-positive NSCLC and MDS RAEB-2 was treated with a monotherapy of erlotinib for the NSCLC. (J) Apoptosis-inducing effect of erlotinib on the patient's CD34+ bone marrow cells ex vivo (3 days, 10 μM) as assessed by staining with DiOC6(3)/PI. (K) Routine blood examinations before and during monotherapy with erlotinib demonstrated the in vivo effect of erlotinib on platelet and neutrophil counts resulting in a hematologic improvement. D-32 indicates day 32 before erlotinib treatment; D0, start of erlotinib therapy; D+34, day 34 under erlotinib treatment.
Discussion
A previous article by Golub et al6 reported 2 possible antineoplastic effects of the EGFR antagonist gefinitib on AML cells: differentiation and proliferative arrest. On the basis of the assumption that it would be preferable to induce cell death, we investigated whether the EGFR antagonist erlotinib might induce apoptosis in myeloid malignancies. Erlotinib was unable to induce apoptosis in HL60 cells (Figures 1,Figure 2–3) and U937 cells (not shown), 2 cell lines also investigated by Golub and colleagues,6 although erlotinib efficiently induced the differentiation of HL60, U937, and P39 cells (Figure 3 and data not shown). In contrast, one particular AML cell line, KG-1, was potently killed by erlotinib, suggesting that the lethal response to this compound is dictated by a poorly understood cellular context. It is important to note that these results reveal that differentiation and cell-cycle blockade induced by EGFR antagonists exerting off-target effects on AML cells are not automatically linked to an apoptotic response. This idea is also corroborated by the fact that some experimental interventions that block erlotinib-induced differentiation (such as the knockdown of the PDGFR or Src) do not suppress the apoptotic response (Figure S1).
Erlotinib kills KG-1 cells by triggering apoptosis, based on morphologic signs (chromatin condensation and nuclear fragmentation that define apoptosis)24 and biochemical criteria (early phosphatidylserine exposure before loss of viability, nuclear DNA degradation giving rise to a peak of subdiploid cells). Apoptosis can be induced by following either the extrinsic pathway (in which ligation of death receptors causes apical caspase activation in the death-inducing signaling complex) or the intrinsic pathway (in which mitochondrial outer membrane permeabilization [MOMP] causes the liberation of the caspase activator cytochrome c). It appears clear that erlotinib stimulates the intrinsic pathway, based on the observation that mitochondria released cytochrome c (and other death effectors such as endonuclease G), before caspase-3 was activated and that caspase inhibition was unable to prevent mitochondrial outer membrane permeabilization and cell death (Figure 4). Although the particular apoptotic execution pathway that is triggered by erlotinib appears clear, it remains an ongoing conundrum through which molecular events erlotinib finally causes MOMP.
As a possibility, direct inhibition of JAK2 may account for the apoptogenic activity of erlotinib on KG-1 AML cells (Figure 4). The evidence that erlotinib functions by disrupting signaling of JAK2/STAT-5 is of particular importance if one takes into account that these molecules are considered to be crucial during leukemogenesis and deregulated in a broad spectrum of hematologic malignancies.25,,–28 Another molecular event that could be associated with the lethal effect of erlotinib was the nucleocytoplasmic translocation of the NPM-1/p14ARF complex. Thus, cell lines that were killed by erlotinib manifested this translocation, whereas others that were resistant against erlotinib failed to do so. Resistant cell lines became susceptible to erlotinib-mediated killing when NPM-1 was knocked down by specific siRNAs (Figure 5). Although this association is interesting, we lack mechanistic insights into the manner in which erlotinib inactivates NPM-1 and how the inactivation of NPM-1 makes cells permissive to the lethal action of erlotinib. These issues require further clarification in future studies.
Our study unravels a remarkable internal coherence of the in vitro, ex vivo, and in vivo results that were obtained on AML cell lines and primary, patient-derived malignant myeloblasts. Thus, normal CD34+ bone marrow cells did not respond to erlotinib, whereas blasts from a fraction of MDS and AML patients were killed by erlotinib while manifesting the nucleocytoplasmic translocation of the NPM-1/p14ARF complex and an apoptosis-independent differentiation. The antineoplastic effect of erlotinib could be confirmed in vivo, in a mouse model of xenografted human AML cells, and in one patient who presented both a high-risk MDS as well as a NSCLC (which constituted the indication for the therapeutic administration or erlotinib). Noteworthy, a recently published case report of a patient suffering concomitantly from AML and NSCLC provides further clinical evidence for the therapeutic efficacy of erlotinib in EGFR-negative myeloid malignancies. Thus this patient, who initially received a monotherapy with erlotinib, achieved a complete remission of his AML that was maintained for several months.29
From a more general point of view, the results presented here confirm an emerging concept in the field of targeted anticancer therapy: targeted therapies do not work only through an action on the molecular target for which they were originally designed. As an example, imatinib was rationally designed to antagonize the deregulated tyrosine kinase activity of Bcr-Abl (and hence to treat leukemias bearing the Philadelphia chromosome creating the Bcr-Abl fusion oncogene).30 However, imatinib later turned out to be useful for the treatment of malignancies relying on the activating mutation of c-kit (in gastrointestinal stroma cell tumors) or the PDGFR (in hypereosinophilic syndrome).31,32 In experimental models, imatinib can even cause the regression of tumors in vivo that do not respond to the drug in vitro, presumably though the inhibition of nonmutated tyrosine kinases expressed by the host immune system.33,34 From this viewpoint, the present report adds another example of a therapeutically warranted off-target effect of tyrosine kinase inhibitors. Through a hitherto unexplained molecular pathway, the EGFR antagonist erlotinib kills MDS and AML cells that lack the EGFR. Needless to say, this discovery may constitute a first step toward a novel therapeutic application of this tyrosine kinase inhibitor.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
S.B. received a scholarship from the Deutsche Forschungsgemeinschaft. L.A. received a scholarship from Assistance Publique-Hopitaux de Paris and Caisse Nationale d'Assurance Maladie des Professions Indépendantes. T.B. is supported by the Myelodysplastic Syndrome foundation. G.K. is supported by Cancéropôle Ile-de-France, Institut National du Cancer, Fondation de France, Association Laurette Fugain, Cent pour Sang la Vie, Agence National de la Recherche.
Authorship
Contribution: S.B. and L.A. designed and performed the research and analyzed the data. T.B., L.G., J.G., C.F., G.L.R., and A.M. performed some of the research. C.G., S.d.B., and P.F. provided bone marrow samples, discussed the results, and provided helpful suggestions. G.K. designed the experiments and supervised their conduct, wrote the paper, and analyzed the data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Dr Guido Kroemer, Inserm, U848, Institut Gustave Roussy, PR1, 39, rue Camille Desmoulins, F-94805 Villejuif, France; e-mail: kroemer@igr.fr.
References
Author notes
S.B. and L.A. contributed equally to this work.



This feature is available to Subscribers Only
Sign In or Create an Account Close Modal