We characterized the mutational status of the FLT3 tyrosine kinase domain (FLT3-TLD) in 3082 patients with newly diagnosed AML. FLT3-TKD mutations were detected in 147 of 3082 (4.8%) patients. Similar to the FLT3 juxtamembrane domain mutations (FLT3-LM), there was a high correlation of FLT3-TKD mutations with normal karyotype (88 of 1472; 6.0%). FLT3-TKD mutations were most frequent in the AML FAB subtypes M5b (15 of 114; 13.2%), M3v (6 of 51; 11.8%), and M4 (39 of 484; 8.1%). Similar to FLT3-LM, the FLT3-TKD mutations show elevated peripheral leukocytes compared with FLT3wt AML. FLT3-TKD had a high incidence in cases with NPM1 mutations (23 of 262; 8.8%), CEBPA mutations (6 of 76; 7.9%), and NRAS mutations (6 of 78; 7.7%). FLT3-TKD in combination with FLT3-LM (17 of 594 patients; 2.9%) and KITD816 (1 of 44; 2.3%) was rare. Unlike the FLT3-LM, which are associated with inferior survival, prognosis was not influenced by FLT3-TKD in the total cohort of 1720 cases, where follow-up data were available (97 FLT3-TKD; 1623 FLT3-WT). In t(15;17)/PML-RARA with FLT3-TKD mutations, in FLT3-LM/TKD double-mutated, and in MLL-PTD/TKD double-mutated cases prognosis was unfavorably influenced by FLT3-TKD mutations. In contrast, we found an additional favorable impact of FLT3-TKD on EFS in prognostically favorable AML with NPM1- or CEBPA mutations.
Introduction
Actual therapy concepts try to modify the intensity of therapy of acute myeloid leukemia (AML) in accordance to the individual risk of relapse. Karyotype represents the strongest prognostic parameter in AML. However, 45% of all AML patients show a normal karyotype and thus an intermediate prognosis.1,–3 Because AML with normal karyotype is considered a heterogeneous disease with respect to molecular aspects, the definition of suitable molecular markers for the determination of prognosis and for minimal residual disease (MRD) detection is of increasing importance
Mutations of the fms-tyrosine kinase (FLT3) were first described in 19974 and account for the most frequent molecular mutations in AML.5,6 The FLT3 gene is a member of the class III receptor tyrosine kinase family, including c-kit, c-fms, and the platelet-derived growth factor receptors.6,,–9 In normal bone marrow, FLT3 expression is restricted to immature hematopoietic progenitor cells and mediates stem-cell proliferation.6,8,10,11
FLT3 length mutations (FLT3-LM or FLT3-ITD for “internal tandem duplication”) represent one of the most frequent genetic alterations in AML. They show a frequency of 20% to 27% in AML in adults9,12,,,–16 and of 10% to 16% in childhood cases17,18 and are associated with progression of myelodysplastic syndrome (MDS) to secondary AML (s-AML).19 FLT3-LM mostly are represented by internal tandem duplications and/or insertion-deletion mutations in exons 11 and 12 of the human FLT3 gene on chromosome 13q12, which codes for the juxtamembrane domain of the FLT3 protein. The mutations are heterogeneous and consist of internal tandem duplications of 6 to 30 amino acids, resulting in an elongated FLT3 protein with constitutive PTK activity.20 These mutations lead to autophosphorylation of the receptor and result in increased proliferation of AML cells in vitro and to a decrease of apoptosis. Constitutively active FLT3 mutants have transforming potential in interleukin-3–dependent cells and activate several signal transduction pathways, including STAT-5 and MAPK.21,,–24
FLT3-LM are highly associated with normal karyotype, t(15;17)/PML-RARA and t(6;9)/DEK-CAN,15,16 and are prognostically unfavorable in adults12,,,–16 and in pediatric patients.17,25,26
In addition to the juxtamembrane domain mutations, mutations in the tyrosine kinase domain (FLT3-TKD mutations) have been described in AML.7,9,16,27 FLT3-TKD mutations are small mutations in the activation loop of FLT3, mostly representing point mutations in codon D835 or deletions of codon I836. They induce constitutive tyrosine phosphorylation leading to activation of the receptor tyrosine kinase and are supposed to represent gain-of-function mutations.7,22,28,,,–32 Corresponding activation loop mutations have been reported at position D816 of KIT and also in other receptor tyrosine kinases, eg, RET.21
Studies on the frequency of FLT3-TKD in AML were performed by Abu-Duhier et al (n = 97),7 Yamamoto et al (n = 429),8 Thiede et al (n = 979),16 Moreno et al (n = 208),27 and Fröhling et al (n = 224),33 with AML showing a normal karyotype. According to these studies, FLT3-TKD show an incidence of 5.8% to 7.7% in AML and thus are less frequent than FLT3-LM.7,9,16,34
Because of the high frequency of FLT3 mutations in AML, concentration focuses on the development of FLT3 inhibitors for treatment of this AML subgroup. Several selective FLT3 tyrosine kinase inhibitors have demonstrated in vitro and in vivo activity.35,,,–39
Because of the low frequency, the prognostic significance of FLT3-TKD mutations is still unclear. In the present study, we analyzed the frequency of FLT3-TKD mutations in 3082 patients with AML at diagnosis and did correlations to other biologic factors, such as karyotype, molecular mutations, cytomorphologic subtypes, and leukocyte count. The influence of FLT3-TKD on prognosis was evaluated in detail.
Methods
Patients
Bone marrow samples or blood samples from 3082 consecutive patients with AML at diagnosis were screened for FLT3-TKD mutations.
The range of age was 17.5 to 91.8 years (median, 63.1 years). Of the 3082 patients, 2546 (82.6%) had de novo AML, 334 (10.8%) had secondary AML after myelodysplastic syndrome (MDS) (s-AML), and 202 (2.0%) had AML after treatment of a previous malignant disease (t-AML). Patients were treated according to protocols of the AML Cooperative Group (AMLCG) study group (80%)40 or according to other intensive AML therapy protocols. The study was approved by the Ethics Committee of participating institutions, and informed consent was obtained in accordance with the Declaration of Helsinki.
Samples were evaluated by cytomorphology, cytochemistry, multiparameter flow cytometry, cytogenetics, fluorescence in situ hybridization, and molecular genetics in parallel.41,,–44 The cytomorphologic classification of AML was performed according to the French-American-British (FAB) classification45,46 (Table 1).
Characterization of the patient cohort (n = 3082)
| . | No. of patients . | Percentage of patients . |
|---|---|---|
| History of AML | ||
| De novo AML | 2546 | 82.6 |
| Secondary AML (s-AML) | 334 | 10.8 |
| t-AML | 202 | 2.0 |
| Total | 3082 | 100.0 |
| Cytogenetic subgroup | ||
| Normal | 1472 | 47.8 |
| t(15;17)* | 130 | 4.2 |
| t(8;21) | 88 | 2.9 |
| Trisomy 8 | 129 | 4.2 |
| 11q23/MLL | 109 | 3.5 |
| 5q-/-5 | 31 | 1.0 |
| 7q-/-7 | 60 | 1.9 |
| inv(16)/t(16;16)† | 119 | 3.9 |
| inv(3)/t(3;3) | 44 | 1.4 |
| t(6;9)(p23;q34) | 6 | 0.2 |
| Complex aberrant | 430 | 14.0 |
| Other aberrations | 400 | 15.0 |
| Total | 3018 | 100.0 |
| FAB subtype | ||
| M0 | 132 | 4.8 |
| M1 | 491 | 17.8 |
| M2 | 1054 | 38.2 |
| M3 | 85 | 3.1 |
| M3v | 51 | 1.8 |
| M4 | 484 | 17.5 |
| M4eo | 118 | 4.3 |
| M5a | 107 | 3.9 |
| M5b | 114 | 4.1 |
| M6 | 112 | 4.1 |
| M7 | 14 | 0.5 |
| Total | 2762 | 100.0 |
| . | No. of patients . | Percentage of patients . |
|---|---|---|
| History of AML | ||
| De novo AML | 2546 | 82.6 |
| Secondary AML (s-AML) | 334 | 10.8 |
| t-AML | 202 | 2.0 |
| Total | 3082 | 100.0 |
| Cytogenetic subgroup | ||
| Normal | 1472 | 47.8 |
| t(15;17)* | 130 | 4.2 |
| t(8;21) | 88 | 2.9 |
| Trisomy 8 | 129 | 4.2 |
| 11q23/MLL | 109 | 3.5 |
| 5q-/-5 | 31 | 1.0 |
| 7q-/-7 | 60 | 1.9 |
| inv(16)/t(16;16)† | 119 | 3.9 |
| inv(3)/t(3;3) | 44 | 1.4 |
| t(6;9)(p23;q34) | 6 | 0.2 |
| Complex aberrant | 430 | 14.0 |
| Other aberrations | 400 | 15.0 |
| Total | 3018 | 100.0 |
| FAB subtype | ||
| M0 | 132 | 4.8 |
| M1 | 491 | 17.8 |
| M2 | 1054 | 38.2 |
| M3 | 85 | 3.1 |
| M3v | 51 | 1.8 |
| M4 | 484 | 17.5 |
| M4eo | 118 | 4.3 |
| M5a | 107 | 3.9 |
| M5b | 114 | 4.1 |
| M6 | 112 | 4.1 |
| M7 | 14 | 0.5 |
| Total | 2762 | 100.0 |
A total of 136 cases of FAB M3/M3v morphology were associated with 130 cases of t(15;17) in chromosome banding analyses because of additional 6 cryptic PML-RARA rearrangements.
A total of 118 cases of FAB M4eo morphology were associated with 119 cases of inv(16)/t(16;16) because of atypical morphology in one case.
Paired samples of 152 patients were compared with respect to the FLT-TKD mutation status at diagnosis and at relapse.
Molecular analysis
Ficoll, mRNA extraction, cDNA synthesis, mutation analysis for FLT3-LM and control polymerase chain reaction (PCR) were performed as previously described.15 Analysis for TKD mutations was performed by LightCycler based melting curve analysis with forward primer 17F: 5′-CCGCCAGGAACGTGCTTG-3′), reverse primer 17R: 5′-ATGCCAGGGTAAGGATTCACACC-3′ and hybridization probes FLT3-sensor 5′-ACTCATGATATCTCGAGCCAATCC-FL-3′ and FLT3-anchor 5′- LCred640-AAGTCACATATCTTCACCACTTTCCCGT-3′-PH. The PCR reaction was carried out in a 20-μL reaction volume with each 0.5 μM of forward and reverse primer, 0.75 μM Hyb-Probes, 4 mM MgCl2 and 2 μL LightCycler-FastStart DNA Master Hybridization Probes (Roche Diagnostics, Mannheim, Germany). LightCycler data were analyzed using the LightCycler 3.0 software (Roche Diagnostics) and the second derivative maximum method. Each 20-μL reaction contains 2 μL cDNA, an equivalent of approximately 3000 cells. Amplification was performed with 45 cycles using 60°C annealing temperature. Final melting curve analysis was started at 40°C up to 95°C with slope of 0.2°C/sec and continuous detection with channel F2/F1 (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article).
Evaluation of this assay was performed in 100 samples by parallel assessment with standard PCR, EcoRV restriction enzyme digestion, and agarose gel electrophoresis, as previously described.9
Screening for FLT3-LM, FLT3-TKD, NRAS mutations, and NPM1 mutations was performed as described elsewhere.15,16,43,47,48 Mutations of the CEBPA gene were analyzed by denaturing high performance liquid chromatography (WAVE, Transgenomics, Omaha, NE) with subsequent direct sequencing of aberrant fragments.
Sequencing analysis
Approximately 100 ng purified PCR products were directly sequenced with 3.3 pmol primers as described with BigDye Terminator Cycle Sequencing Kit (Applera, Weiterstadt, Germany). After initial denaturation at 95°C for 5 minutes, 25 cycles at 94°C for 15 seconds and 60°C for 4 minutes were performed. Sequence analysis was performed on an ABI 310 or 3100 Sequence Detection system.
Statistical analysis
Overall survival (OS) and event-free survival (EFS) were performed according to Kaplan-Meier. OS was calculated from time of diagnosis to death and EFS from time of diagnosis to death, documentation of persistent leukemia, or relapse. The correlation of OS and EFS with other parameters was assessed by Cox regression. The comparison of survival curves was performed using double-sided log-rank test. Comparisons of dichotomous variables between different groups were performed by use of 2-sided Fisher exact test. For statistical analysis, SPSS (version 12.4) software (SPSS, Chicago, IL) was used.
Complete remission was defined by less than 5% blasts in a normocellular bone marrow with normal levels of peripheral neutrophil and platelet counts.
Results
Frequency of the FLT3-TKD mutation
In total, 3082 samples were analyzed at diagnosis. Of these, 147 were positive for a FLT3-TKD mutation (4.8%). As estimated from the LightCycler curves and sequence data, 137 had a mutation/weight ratio corresponding to 20% to 100% of mutated cells. Ten cases had a TKD/wildtype (TKDwt) ratio of approximately 10%. Lower ratios were not observed. On the other hand, 2935 cases (95.2%) tested negative for a FLT3-TKD mutation.
Sequencing analysis
To further characterize the detected mutations, PCR products of cases with low melting temperatures profiles were directly sequenced. In 10 cases, sequencing with a sensitivity of approximately 10% did not reveal a mutation because the mutated clone as derived from the melting peak was below 10% of the whole cell population. Of 147 mutated cases, 137 (93.2%) could be further characterized. Changes of D835 were most frequent (122 of 137; 89.1%). In detail, a change from asparagine to tyrosine (D835Y) was found in 68 of 147 cases (49.6%). In 118 of 122 cases with FLT3-TKD mutations (96.7%), only one allele was mutated. Compound heterozygotes were found in 4 of 122 cases with FLT3-TKD mutations (3.3%) (asp > ala/asp > tyr; asp > tyr/asp > glu; asp > tyr/asp > his; and asp > tyr/asp > glu). In one case, we found mutations of both D835 and a ΔI836 (1 of 137; 0.7%). ΔI836 was observed in 14 cases (10.2%) (Table 2).
Distribution of point mutations in 137 cases with FLT3-TKD mutations
| Amino acid exchange . | No. of patients . | Percentage of patients . |
|---|---|---|
| D835Y | 68 | 49.6 |
| D835H | 20 | 14.6 |
| D835V | 14 | 10.2 |
| D835E | 11 | 8.0 |
| D835A | 3 | 2.2 |
| D835S | 1 | 0.7 |
| D835N | 1 | 0.7 |
| Δ I836 | 14 | 10.2 |
| D835 compound heterozygous | ||
| D835Y + D835E | 2 | 1.4 |
| D835Y + D835A | 1 | 0.7 |
| D835Y + D835H | 1 | 0.7 |
| D835 total | 122 | 89.1 |
| D835V and Δ I836 | 1 | 0.7 |
| Total | 137 | 100.0 |
| Amino acid exchange . | No. of patients . | Percentage of patients . |
|---|---|---|
| D835Y | 68 | 49.6 |
| D835H | 20 | 14.6 |
| D835V | 14 | 10.2 |
| D835E | 11 | 8.0 |
| D835A | 3 | 2.2 |
| D835S | 1 | 0.7 |
| D835N | 1 | 0.7 |
| Δ I836 | 14 | 10.2 |
| D835 compound heterozygous | ||
| D835Y + D835E | 2 | 1.4 |
| D835Y + D835A | 1 | 0.7 |
| D835Y + D835H | 1 | 0.7 |
| D835 total | 122 | 89.1 |
| D835V and Δ I836 | 1 | 0.7 |
| Total | 137 | 100.0 |
Biologic parameters of patients with FLT3-TKDmut compared with FLT3-TKDwt
The 147 FLT3-TKD mutated cases included 72 males (49.0%) and 75 females (51.0%), thus revealing no sex difference. The median age of the FLT3-TKD mutated patients was 59.5 years (range, 18.7-84.6 years) compared with 63.2 years (range, 17.5-91.8 years) in the unmutated group (not significant). Thus, FLT3-TKD mutations were not significantly correlated with sex or age. The median white blood count (WBC) was 40.0 × 109/L (range, 1.2 × 109/L to 332.0 × 109/L) in the cohort with FLT3-TKD mutations compared with 50 × 109/L (range, 1.0-840.0 × 109/L) in the cohort with FLT3-LM vs 7.6 × 109/L (range, 115.0-462.0 × 109/L) in the patients with FLT3- wild-type. Thus, WBC was significantly higher in FLT3-TKD–mutated than in FLT3wt AML (P < .001), and lower than in FLT3-LM (P = .04).
Correlation of the FLT3-TKD mutation status with AML history
The distribution of FLT3-TKD mutations in de novo AML, s-AML, and t-AML revealed the highest incidence in de novo AML (135 of 2546; 5.3%) compared with s-AML (8 of 334; 2.4%) and t-AML (4 of 202; 2.0%). Thus, FLT3-TKD mutations were significantly more frequent in de novo than in s-AML after MDS or precedent chemotherapy (P = .01).
Correlation of the FLT3-TKD mutation status to cytomorphology
The FAB subtype was available in 2762 of 3082 patients (89.6%) and was correlated to the FLT3-TKD status. Overall FLT3-TKD mutations were detected in all morphologic subgroups. They were significantly overrepresented in AML M3v (6 of 51; 11.8%; P = .02), M4 (39 of 484; 8.1%; P < .001), and M5b (15 of 114; 13.2%; P < .001). In contrast, they were significantly under-represented in the subtypes M2 (25 of 1054) and M6 (M6: 0 of 112; 0%; P = .015) (Table 3).
Correlation of FLT3-TKD mutations with the FAB subtype in 2762 patients with AML at diagnosis
| FAB . | Total, no. . | FLT TKDmut, no. . | FLT TKDmut, % . | P . |
|---|---|---|---|---|
| M0 | 132 | 3 | 2.3 | .158 |
| M1 | 491 | 31 | 6.3 | .096 |
| M2 | 1054 | 25 | 2.4 | <.001 |
| M3 | 85 | 4 | 4.7 | .949 |
| M3v | 51 | 6 | 11.8 | .020 |
| M4 | 484 | 39 | 8.1 | <.001 |
| M4eo | 118 | 5 | 4.2 | .751 |
| M5a | 107 | 6 | 5.6 | .711 |
| M5b | 114 | 15 | 13.2 | <.001 |
| M6 | 112 | 0 | 0.0 | .015 |
| M7 | 14 | 0 | 0.0 | .397 |
| Total | 2762 | 134 | 4.9 | — |
| FAB . | Total, no. . | FLT TKDmut, no. . | FLT TKDmut, % . | P . |
|---|---|---|---|---|
| M0 | 132 | 3 | 2.3 | .158 |
| M1 | 491 | 31 | 6.3 | .096 |
| M2 | 1054 | 25 | 2.4 | <.001 |
| M3 | 85 | 4 | 4.7 | .949 |
| M3v | 51 | 6 | 11.8 | .020 |
| M4 | 484 | 39 | 8.1 | <.001 |
| M4eo | 118 | 5 | 4.2 | .751 |
| M5a | 107 | 6 | 5.6 | .711 |
| M5b | 114 | 15 | 13.2 | <.001 |
| M6 | 112 | 0 | 0.0 | .015 |
| M7 | 14 | 0 | 0.0 | .397 |
| Total | 2762 | 134 | 4.9 | — |
— indicates not applicable.
Correlation of the FLT3-TKD mutation status with cytogenetics
Karyotype was available in 3018 of 3082 cases (97.9%). In 64 of 3082 cases (2.1%), karyotype was not available because of a low number of analyzed metaphases. Cases were grouped into 12 categories: normal karyotype, t(15;17)/PML-RARA, t(8;21)/AML1-ETO, inv(3)/t,3,3 inv(16)/t(16;16)/CBFB-MYH11, 11q23/MLL, complex aberrant karyotype (defined by 3 or more chromosomal abnormalities), trisomy 8, 5q- or loss of chromosome 5, 7q- or loss of chromosome 7, and all others, the so-called other aberrations.
FLT3-TKD mutations were observed in all cytogenetic subgroups. However, some specific associations with distinct cytogenetic subgroups were observed. Compared with the total cohort, they were significantly overrepresented in normal karyotype (88 of 1384; 6.0%; P = .003) compared with the overall cohort. They showed an elevated frequency in t(15;17)/PML-RARA (10 of 130; 7.7%), but this association did not reach significance. In complex aberrant karyotype, they were significantly underrepresented (4 of 430; 0.9%; P < .001; Table 4).
Correlation of FLT3-TKD mutations with cytogenetics in 3018 patients
| Cytogenetic subgroup . | Total, no. . | FLT TKDmut, no. . | FLT TKDmut, % . | P . |
|---|---|---|---|---|
| Normal | 1472 | 88 | 6.0 | .003 |
| t(15;17)/PML-RARA | 130 | 10 | 7.7 | .116 |
| t(8;21) | 88 | 2 | 2.3 | .260 |
| Trisomy 8 | 129 | 6 | 4.7 | .934 |
| 11q23/MLL | 109 | 7 | 6.4 | .421 |
| 5q-/-5 | 31 | 0 | 0.0 | .209 |
| 7q-/-7 | 60 | 1 | 1.7 | .251 |
| inv(16)/t(16;16) | 119 | 6 | 5.0 | .902 |
| inv(3)/t(3;3) | 44 | 2 | 4.5 | .936 |
| t(6;9)(p23;q34) | 6 | 0 | 0.0 | .582 |
| Complex aberrant | 430 | 4 | 0.9 | <.001 |
| Other aberrations | 406 | 19 | 4.7 | .900 |
| Total | 3018 | 145 | 4.8 | — |
| Cytogenetic subgroup . | Total, no. . | FLT TKDmut, no. . | FLT TKDmut, % . | P . |
|---|---|---|---|---|
| Normal | 1472 | 88 | 6.0 | .003 |
| t(15;17)/PML-RARA | 130 | 10 | 7.7 | .116 |
| t(8;21) | 88 | 2 | 2.3 | .260 |
| Trisomy 8 | 129 | 6 | 4.7 | .934 |
| 11q23/MLL | 109 | 7 | 6.4 | .421 |
| 5q-/-5 | 31 | 0 | 0.0 | .209 |
| 7q-/-7 | 60 | 1 | 1.7 | .251 |
| inv(16)/t(16;16) | 119 | 6 | 5.0 | .902 |
| inv(3)/t(3;3) | 44 | 2 | 4.5 | .936 |
| t(6;9)(p23;q34) | 6 | 0 | 0.0 | .582 |
| Complex aberrant | 430 | 4 | 0.9 | <.001 |
| Other aberrations | 406 | 19 | 4.7 | .900 |
| Total | 3018 | 145 | 4.8 | — |
— indicates not applicable.
Correlation of the FLT3-TKD mutation status with other molecular markers in normal karyotype AML
The correlation of FLT3-TKD with other molecular markers was analyzed for FLT3-LM, MLL-PTD, and mutations in NPM1, NRAS, KITD816, and CEPBA. FLT3-TKD mutations were significantly over-represented in the patients with NPM1 mutations (23 of 262; 8.8%) (P < .012). In patients with FLT3-LM, they were significantly under-represented (17 of 594; 2.9%; P = .001; Table 5).
Correlation of FLT3TKD mutations with other molecular mutations in normal karyotype AML
| Molecular mutation, mutation status . | No. . | FLT3-TKDmut+ . | Percentage . | P . |
|---|---|---|---|---|
| FLT3-LM, N = 1596 | ||||
| + | 594 | 17 | 2.9 | — |
| − | 1002 | 72 | 7.1 | .001 |
| MLL-PTD, N = 1044 | ||||
| + | 84 | 6 | 7.1 | — |
| − | 960 | 64 | 6.7 | .867 |
| NPM1, N = 517 | ||||
| + | 262 | 23 | 8.8 | — |
| − | 255 | 7 | 2.7 | <.012 |
| CEBPA, N = 455 | ||||
| + | 76 | 6 | 7.93 | — |
| − | 379 | 26 | 6.9 | .408 |
| NRAS, N = 1266 | ||||
| + | 78 | 6 | 3.8 | — |
| − | 1156 | 68 | 6.4 | .221 |
| KITD816, N = 1427 | ||||
| + | 44 | 1 | 2.3 | — |
| − | 1383 | 12 | 0.9 | .487 |
| Molecular mutation, mutation status . | No. . | FLT3-TKDmut+ . | Percentage . | P . |
|---|---|---|---|---|
| FLT3-LM, N = 1596 | ||||
| + | 594 | 17 | 2.9 | — |
| − | 1002 | 72 | 7.1 | .001 |
| MLL-PTD, N = 1044 | ||||
| + | 84 | 6 | 7.1 | — |
| − | 960 | 64 | 6.7 | .867 |
| NPM1, N = 517 | ||||
| + | 262 | 23 | 8.8 | — |
| − | 255 | 7 | 2.7 | <.012 |
| CEBPA, N = 455 | ||||
| + | 76 | 6 | 7.93 | — |
| − | 379 | 26 | 6.9 | .408 |
| NRAS, N = 1266 | ||||
| + | 78 | 6 | 3.8 | — |
| − | 1156 | 68 | 6.4 | .221 |
| KITD816, N = 1427 | ||||
| + | 44 | 1 | 2.3 | — |
| − | 1383 | 12 | 0.9 | .487 |
+ indicates positive mutation status; −, negative mutation status; and —, not applicable.
Stability of the FLT3-TKD in paired samples from diagnosis and relapse
The stability of FLT3-TKD was analyzed by evaluation of 152 paired samples at diagnosis and relapse. The mutation status was TKDwt/TKDwt at both time points in 139 cases. Four cases revealed an FLT3-TKD mutation at diagnosis as well as at relapse. Thus, the TKD status was unchanged at diagnosis and relapse in 143 cases (94.1%). In contrast, 9 cases (5.9%) that were FLT3-TKD mutated at diagnosis did not reveal this mutation at relapse. This meant that, of 13 cases that were positive at diagnosis, 9 (69%) lost the mutation at relapse. Although the limited number of cases has to be taken into account, this may suggest that FLT3-TKD is a rather unstable mutation not valid for follow-up studies.
Prognostic significance of the FLT3-TKD mutation
When all patients were taken into account, OS and EFS were not significantly influenced by the presence of FLT3-TKD mutations (Figure S2A,B). This pattern remained unchanged if all cases with FLT3-LM were excluded (Figure S2C,D). We then compared OS and EFS with respect to FLT3-TKD mutations in different cytogenetic subgroups. In t(8;21), inv16/t(16;16) 11q23/MLL rearrangements, inv(3)/t(3;3), +8, and in complex aberrant karyotype, the influence of FLT3-TKD could not be determined because of the rare occurrence in these cytogenetic subgroups. In normal karyotype, OS and EFS did not differ significantly in dependence on FLT3-TKD mutations (Figure S3A-E). In patients with karyotypes associated with an intermediate prognosis, FLT3-TKD had no prognostic impact as well (Figure S4A-D). In addition, when this analysis was restricted to the de novo AML case, no prognostic significance could be worked out (Figures S2E,F and S3E,F).
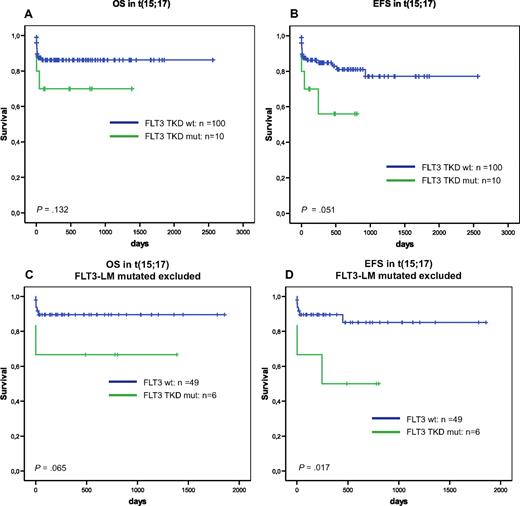
In the subgroup with t(15;17), EFS was significantly deteriorated by FLT3-TKD mutations (P = .051) (Figure 1B). This became even more evident when the analysis was restricted to the cases without FLT3-LM (Figure 1C,D).
Prognostic influence of FLT3-TKD in APL. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) vs FLT3-TKD mutated cases (FLT3-TKDmut) in t(15/17)/PML-RARA positive AML. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in t(15/17)/PML-RARA positive AML exclusive all cases that are FLT3-LM mutated.
Prognostic influence of FLT3-TKD in APL. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) vs FLT3-TKD mutated cases (FLT3-TKDmut) in t(15/17)/PML-RARA positive AML. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in t(15/17)/PML-RARA positive AML exclusive all cases that are FLT3-LM mutated.
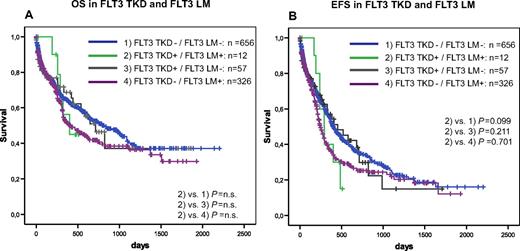
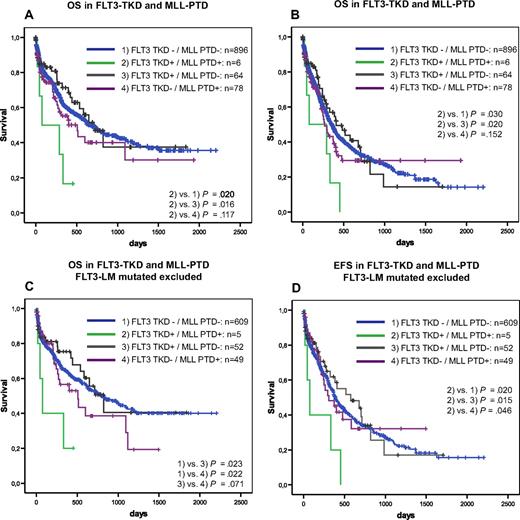
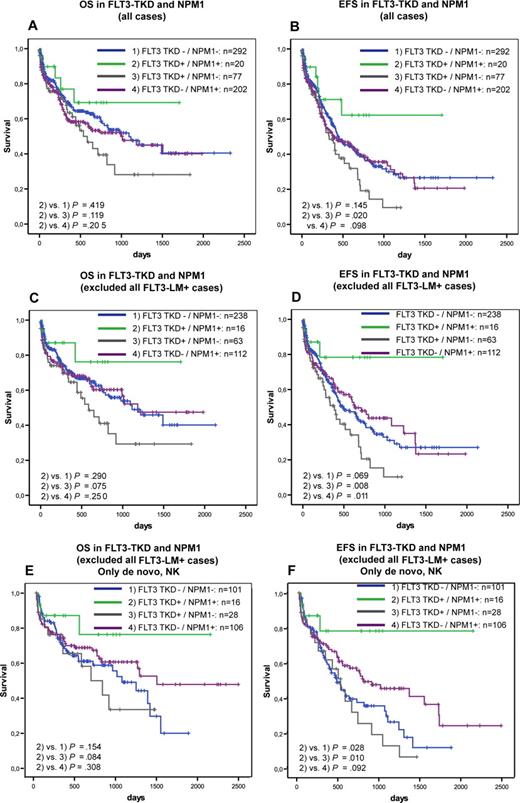
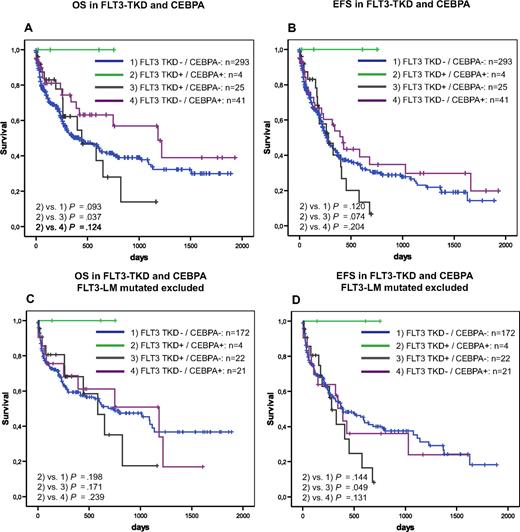
Furthermore, we analyzed the prognostic impact of FLT3-TKD mutations in dependence on FLT3-LM, MLL-PTD, NPM1, and CEBPA. In the NRAS–mutated patients, the influence of FLT3-TKD mutations could not be analyzed because of the low incidence in this subgroup. Kaplan-Meier plots showed an unfavorable impact of FLT3-TKD in prognostically unfavorable groups, such as FLT3-LM (Figure 2) and MLL-PTD (Figure 3A,B). This cooperative adverse effect of FLT3-TKD and MLL-PTD was even stronger after the FLT3-LM positive cases had been excluded (Figure 3C,D). In contrast, there seems to be a favorable impact in the 2 subgroups with the favorable markers NPM1 (Figure 4A,B) and CEBPA (Figure 5). For NPM1 this cooperative positive effect was even stronger when the FLT3-LM mutated cases were excluded. If this analysis was further broken down to de novo AML, normal karyotype, and exclusion of FLT3-LM mutated cases, FLT3-TKD/NPM1 double–mutated cases had a highly significantly better EFS than sole FLT3-TKD mutated (P = .010) or FLT3wt/NPM1wt cases (P = .028) (Figure 4E,F). Because of the limited sample numbers in subgroups, a comparable detailed analysis was not possible for all other mutations.
Prognostic correlations of two different types of FLT3 mutations. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on FLT3-LM.
Prognostic correlations of two different types of FLT3 mutations. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on FLT3-LM.
Prognostic correlations between MLL-PTD and FLT3-TKD. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on MLL PTD. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on MLL PTD exclusive all cases that are FLT3-LM mutated.
Prognostic correlations between MLL-PTD and FLT3-TKD. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on MLL PTD. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on MLL PTD exclusive all cases that are FLT3-LM mutated.
Prognostic correlations between NPMI and FLT3-TKD. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FL3T-TKDmut) in dependence on NPM1 mutations. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FL3T-TKDmut) in dependence on NPM1 mutations all cases that are FLT3-LM mutated. Overall survival (E) and event-free survival (F) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FL3T-TKDmut) in dependence on NPM1 mutations, exclusive all cases that are FLT3-LM mutated in de novo normal karyotype AML.
Prognostic correlations between NPMI and FLT3-TKD. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FL3T-TKDmut) in dependence on NPM1 mutations. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FL3T-TKDmut) in dependence on NPM1 mutations all cases that are FLT3-LM mutated. Overall survival (E) and event-free survival (F) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FL3T-TKDmut) in dependence on NPM1 mutations, exclusive all cases that are FLT3-LM mutated in de novo normal karyotype AML.
Prognostic correlations between CEBPA and FLT3-TKD. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on CEPBA mutations. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on CEPBA mutations exclusive all cases that are FLT3-LM mutated.
Prognostic correlations between CEBPA and FLT3-TKD. Overall survival (A) and event-free survival (B) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on CEPBA mutations. Overall survival (C) and event-free survival (D) of FLT3-TKD wild-type (FLT3-wt) versus FLT3-TKD mutated cases (FLT3-TKDmut) in dependence on CEPBA mutations exclusive all cases that are FLT3-LM mutated.
Discussion
Molecular mutations, especially in normal karyotype AML, are of increasing importance for the definition of prognosis in AML. Some mutations have been shown to be prognostically favorable, such as NPM148,49 and CEBPA,50,51 Others, such as the MLL-PTD, are unfavorable.52,53 Although for the FLT3-LM an association with an unfavorable prognosis within normal karyotype was clearly established, the prognostic impact of FLT3-TKD remained unclear, so we did a detailed analysis on 3082 patients with AML. Although we performed a very specific and sensitive assay, which was potentially able to detect not only the codon 835 of 836 mutations but covered a total of 17 codons in the TKD domain, the frequency of FLT3-TKD in our study was only 4.8%. This slightly lower frequency compared with previous studies9,12,16,27,54 (Table 6) might be explained by differences of age as the median age of patients in this study was 63 years in comparison to comparative studies (<50 years).55,–57 Likewise to the FLT3-LM, the FLT3-TKD seem to be more frequently associated with de novo AML rather than with s-AML or t-AML, as suggested here and in some previous reports.15,33 Only one analysis found an equal distribution of FLT3-TKD in de novo and in s-AML.27
Frequency of FLT3-TKD mutations in this study and in previous studies
| Reference (year) . | Total, n . | Positive FLT-TKD mutation status, n . | FLT-TKD mutations, % . |
|---|---|---|---|
| Current study | 3082 | 147 | 4.8 |
| Abu-Duhier et al7 (2001) | 97 | 7 | 7.2 |
| Yamamoto et al9 (2001) | 429 | 30 | 7.0 |
| Thiede et al16 (2002) | 979 | 75 | 7.7 |
| Moreno et al27 (2003) | 208 | 20 | 7.7 |
| Andersson et al55 (2004) | 109 (>60 y) | 11 | 10.1 |
| Aurewarakul et al56 (2005) | 256 | 15 | 5.8 |
| Wang et al57 (2005) | 143 | 92 | 6.3 |
| Mead et al54 (2007) | 1107 | 127 | 11 |
| Overall | 5303 | 397 | 7.5 |
| Reference (year) . | Total, n . | Positive FLT-TKD mutation status, n . | FLT-TKD mutations, % . |
|---|---|---|---|
| Current study | 3082 | 147 | 4.8 |
| Abu-Duhier et al7 (2001) | 97 | 7 | 7.2 |
| Yamamoto et al9 (2001) | 429 | 30 | 7.0 |
| Thiede et al16 (2002) | 979 | 75 | 7.7 |
| Moreno et al27 (2003) | 208 | 20 | 7.7 |
| Andersson et al55 (2004) | 109 (>60 y) | 11 | 10.1 |
| Aurewarakul et al56 (2005) | 256 | 15 | 5.8 |
| Wang et al57 (2005) | 143 | 92 | 6.3 |
| Mead et al54 (2007) | 1107 | 127 | 11 |
| Overall | 5303 | 397 | 7.5 |
Like previous studies,9,33,54 we found the D835Y mutation to be the most frequent amino acid exchange followed by the D835V and the D835E mutations. Thus, the mutations are not randomly distributed within the TK domain. The occurrence of 2 different TKD mutations in combination was rare in only 3% of all cases at all. This suggests that a compound heterozygous mutation status does not provide a further advantage for the leukemic clone.
Similar to AML with FLT3-LM, the FLT3-TKD is clearly associated with higher WBC compared with FLT3-wt AML. However, median WBC in FLT3-TKD was lower than in FLT3-LM, which is in accordance with some previous studies,33,54 although others were not able to show any influence of the FLT3-TKD on peripheral leukocytes.9,27,54
A preliminary analysis of paired samples from diagnosis and relapse suggests low stability of this mutation. Loss of the FLT3-TKD mutation at relapse was found in 69% of cases, which were positive at diagnosis (6% of all cases). This was in the range of 2 previous studies that observed loss in 3%58 and in 8% of the cases, respectively.13 Therefore, the FLT3-TKD mutations might be less suitable as MRD markers compared with other markers. However, this is a matter for future studies. Efforts were already made to establish a real-time PCR assay for the follow-up of FLT3-TKD mutations.59
We found a relatively high frequency of FLT3-TKD in t(15;17)/PML-RARA compared with the total cohort. This association between the TKD mutations and acute promyelocytic leukemia confirmed the results of our previous analysis which included part of the patients of this study.60 Likewise to the FLT3-LM,60 we found the FLT3-TKD significantly associated with the FAB subtype M3v (11.8%), whereas in M3 the frequency (4.7%) was comparable to the overall frequency of FLT3-TKD mutations in AML. In conclusion, this association of the FLT3-LM and of the TKD mutations with PML-RARA in parallel with the low frequency of other molecular markers in acute promyelocytic leukemia suggests a cooperation of both subtypes of FLT3 mutations with PML-RARA to cause AML with preference for the cytomorphologic M3v subtype.61 Such cooperation is further emphasized by the previous observation of a higher PML-RARA expression in FLT3-LM positive patients in comparison to FLT3-LM negative patients.57
In contrast to the FLT3-LM, which are significantly under-represented in the subgroup with inv(16)/CBFB-MYH11 (Schnitt-ger et al,15 0%; Thiede et al,16 0.6%), the frequency of FLT3-TKD in inv(16)/CBFB-MYH11 was only slightly lower than the overall frequency in this study and in previous analyses (this study, 5.0%; Care et al,62 4.8%; Thiede et al,16 5.0%). A recent large study found the TKD mutations even significantly associated with inv(16).54 Thus, the relationship between the FLT3-TKD mutations and CBFB-MYH11 is not yet clear. In contrast to FLT3-LM, which were observed in 90% of t(6;9)/DEK-CAN cases,16 the FLT3-TKD mutations were not detected in this rare entity. In addition, the FLT3-TKD mutations were significantly underrepresented in AML with complex aberrant karyotype in our study, which was in correspondence with the FLT3-LM also being rare in this cytogenetic subgroup according to our precedent analysis.15 In conclusion, the FLT3-LM and the TKD mutations show many parallels with respect to their distribution within the different cytogenetic AML subgroups, as demonstrated by their association to normal karyotype, to t(15;17)/PML-RARA preferentially in AML M3v, and by their low occurrence in complex aberrant karyotype.
Another focus of this analysis was the distribution of the FLT3-TKD mutations within AML with different molecular mutations. According to the 2 hit hypothesis, at least 2 different mutation types are needed to induce AML. Type I mutations encode tyrosine kinases and increase proliferation, whereas type II mutations encode transcription factors and block differentiation.21 The over-representation of FLT3-TKD, being interpreted as type I mutations, in cases with NPM1, CEBPA, and NRAS mutations (representing type II mutations) in this analysis might support this model.
FLT3-TKD and FLT3-LM seem to differ with respect to their molecular cooperation partners, as the FLT3-LM were found underrepresented in NRAS mutated cases in contrast to the over-representation of TKD mutations in this subgroup.12 Second, the FLT3-TKD did not differ from the overall frequency in patients with MLL-PTD, whereas the FLT3-LM were overrepresented in MLL-PTD positive cases.63
Our data support previous suggestions that FLT3-TKD and FLT3-LM show no64 or rare coincidence.9,16,33,65 It was suggested that the FLT3-LM and FLT3-TKD are caused by totally different underlying mutational mechanisms and, in addition, do not cooperate well to amplify any leukemogenic effect. The same seems to be true for FLT3-TKD and KIT mutations because these 2 targets were also very rare in this study
The correlation of the FLT3-TKD mutations with cytomorphology allowed further speculations with respect to their role in leukemogenesis. The association with the monocytic subtype FAB M5 in this and in previous studies,9,16 such as the FLT3-LM, which shows a similar distribution,9,15,16 is indicative for a participation of FLT3 mutations in monocyte differentiation in AML.16,66 It had previously been shown that FLT3 was continuously expressed during monocyte differentiation and that FLT3 was needed for optimal differentiation of monocytes from CD34 positive stem cells.67 In addition, the underrepresentation of FLT3-TKD mutations in the FAB subtype M6 (present study)16 might be the result of the low rate of FLT3-TKD mutations in complex aberrant karyotype, as AML M6 is correlated with this cytogenetic subgroup.68
With respect to prognosis in the present study, which included 3082 AML cases, there was no influence on OS or EFS in the overall AML population, in de novo AML, nor in the normal or intermediate karyotype subgroups in accordance with most previous studies in adults,33,54 in elderly patients,65 and in children.26 However, there are also reports of a worsened outcome,27 and the meta-analysis of Yanada et al69 on 1160 patients resulted in a significantly worse outcome. This is in contrast to a large recent study, which found a significantly improved survival in the patients with FLT3-TKD mutations.54 Another study showed an inferior outcome in the intermediate cytogenetic subgroup, whereas there was no impact of the FLT3-TKD mutations on survival in the poor cytogenetic subgroup.16 These different findings may result from selection within the cohorts. We could show the prognostic effect of the FLT3-TKD is dependent on additional mutations in single patients, which came out by detailed subgroup analyses, which have not been performed previously.
Accordingly, if subgroups with defined genetic profiles are regarded, a significant prognostic impact could be worked out. The only cytogenetic subgroup with an obvious prognosis effect of the FLT3-TKD (a negative one in this case) was t(15;17)/PML-RARA. Other negative effects were detected in AML with FLT3-LM or MLL-PTD, which are themselves prognostically unfavorable. Paradoxically, in NPM1 and CEBPA mutated cases, which per se have a favorable prognosis, FLT3-TKD mutations had a further favorable prognostic impact. These effects were even stronger when the analysis was restricted to de novo AML or AML without FLT3-LM. As the case numbers in some of the double-mutated groups were rather small, especially in the FLT3-TKD/CEBPA double-mutated group, these finding have to be taken with caution and should serve mainly as a working hypothesis for further studies within clinical trials.
Different signaling properties of both mutation subtypes were suggested by Choudhary et al29 who showed that FLT3-LM gain a function over ligand-activated FLT3-wild-type, which is not gained by FLT3-TKD mutations.29 Gene-expression profiling with microarray analysis showed different gene-expression patterns for FLT3-LM and for FLT3-TKD positive AML.70 The differences between FLT3-TKD and FLT3-LM regarding the distribution within distinct cytogenetic subgroups, the coincidence with other molecular markers, and the differences with respect to the prognostic impact in this and in the precedent studies emphasize the diversity within both molecular subtypes. In conclusion, FLT3-LM mutations and FLT3-TKD mutations should be regarded as 2 biologically and prognostically different mutations within a single gene.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors appreciate the excellent technical help of Gudrun Mellert, Claudia Tschulik, Madlen Fuchs, Theresa Förster, and Nina Leopold, and the authors thank all participants of the AMLCG study group for sending bone marrow or blood samples to our laboratory for reference diagnosis and for submitting clinical data, as most of the patients were treated within the AMLCG study group.
This investigation was performed in part in the Labora-tory for Leukemia Diagnostics, Medical Department III (Head, Prof Dr W. Hiddemann), Ludwig-Maximilians-University Munich.
Authorship
Contribution: S.S. was the principal investigator; U.B. and S.S. analyzed data and wrote the manuscript; W.K. was responsible for immunophenotyping and contributed in statistical evaluation; T.H. was responsible for cytomorphology; C.H. was responsible for cytogenetics; and S.S. did the molecular genetic analysis.
Conflict-of-interest disclosure: C.H., W.K., T.H., and S.S. in part own the MLL Munich Leukemia Laboratory, which is offering comprehensive leukemia diagnostics. U.B. declares no competing financial interests.
Correspondence: Susanne Schnittger, Munich Leukemia Laboratory, Max-Lebsche-Platz 31, 81377 Munich, Germany; e-mail: Susanne.Schnittger@mll-online.com.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal