Dendritic cells (DCs) are considered critical for the induction of graft-versus-host disease (GVHD) after bone marrow transplantation (BMT). In addition to their priming function, dendritic cells have been shown to induce organ-tropism through induction of specific homing molecules on T cells. Using adoptive transfer of CFSE-labeled cells, we first demonstrated that alloreactive T cells differentially up-regulate specific homing molecules in vivo. Host-type dendritic cells from the GVHD target organs liver and spleen or skin- and gut-draining lymph nodes effectively primed naive allogeneic T cells in vitro with the exception of liver-derived dendritic cells, which showed less stimulatory capacity. Gut-derived dendritic cells induced alloreactive donor T cells with a gut-homing phenotype that caused increased GVHD mortality and morbidity compared with T cells stimulated with dendritic cells from spleen, liver, and peripheral lymph nodes in an MHC-mismatched murine BMT model. However, in vivo analysis demonstrated that the in vitro imprinting of homing molecules on alloreactive T cells was only transient. In conclusion, organ-derived dendritic cells can efficiently induce specific homing molecules on alloreactive T cells. A gut-homing phenotype correlates with increased GVHD mortality and morbidity after murine BMT, underlining the importance of the gut in the pathophysiology of GVHD.
Introduction
Allogeneic hematopoietic stem-cell transplantation is an important therapeutic option for a variety of malignant and nonmalignant diseases.1 However, the occurrence of acute graft-versus-host disease (GVHD), a serious systemic illness primarily involving the intestine, liver, and skin, constitutes one of the limitations for its widespread use.2,3 GVHD occurs when alloreactive donor T cells recognize alloantigens on normal host tissues. Pathophysiologically, 3 distinct phases (as defined by Ferrara and Reddy) can be distinguished, which are characterized by tissue injury from the conditioning regimen, donor T-cell activation, and efferent effector functions.4 The gastrointestinal tract hereby seems to play a crucial role in the amplification of systemic disease by permitting release of inflammatory stimuli from the gut.5
To mount an efficient alloresponse, antigens need to be presented by professional antigen presenting cells (APCs) with host APCs being crucial for the initiation of GVHD.6,,,–10 Depletion of APCs in the liver and the spleen significantly inhibits the recruitment and proliferation of donor T cells,11 and the elimination of Langerhans cells seems to be efficient in preventing skin GVHD.12 However, there is also increasing evidence implicating cross-presenting donor APCs in acute13 and chronic14 GVHD. The importance of antigen expression by epithelial, nonhematopoietic GVHD target organ cells is still being disputed.15,16
Recent studies have shown that dendritic cells not only are required for the activation of T cells but also influence their migratory behavior.17,–19 Dendritic cells from mesenteric lymph nodes (MLNs),20,21 Peyer patches,22 and peripheral lymph nodes (PLNs)23 have all been shown to imprint tissue-specific homing molecules on T cells. However, this process is flexible and allows reprogramming after exposure to different dendritic cells.23,24 Whereas gut homing is induced by retinoic acid,25 skin homing seems to be dependent on vitamin D3.26 More recently, dendritic cells have also been shown to affect B-cell homing.27
This process of imprinting and the tight control of T-cell migration is of particular relevance for a number of physiologic and pathologic conditions.28 Since alloreactive T cells can exert their pathologic effect only after infiltration into GVHD target organs, interfering with this process is an attractive approach in preventing or treating GVHD. The feasibility of this method has been ascertained in several studies with various molecules.29,30
In this study, we examined the effect of different organ-derived host-type dendritic cells on alloreactive T cells and found that dendritic cells derived from the liver had less allostimulatory capacity than any other organ-derived dendritic cell. However, dendritic cells from the gut-associated lymphoid tissue specifically induced a gut-homing phenotype resulting in significantly higher mortality and morbidity after bone marrow transplantation. In addition, microarray analysis performed on those stimulated T cells revealed novel differentially induced target genes.
Methods
Antibodies and flow cytometry
Most fluorochrome- or biotin-labeled antimurine antibodies were obtained from BD Biosciences Pharmingen (San Diego, CA) with the exception of rat antimouse CD16/CD32 (2.4G2), produced by the Monoclonal Antibody Core Facility at the Memorial Sloan-Kettering Cancer Center. Foxp3 (FJK-16s) and integrin α4 chain (R1-2) antibodies were purchased from eBioscience (San Diego, CA) and antimurine integrin β1 chain (Hmβ1-1) was from BioLegend (San Diego, CA). Staining for E- and P-selectin ligands was performed using chimeric proteins consisting of the extracellular domains of E- and P-selectin fused to the human Fc region (from BD Biosciences Pharmingen and R&D Systems, Minneapolis, MN, respectively), which was detected with a secondary phycoerythrin-conjugated goat antihuman FcR antibody (Jackson ImmunoResearch, West Grove, PA). Fluorescence-activated cell sorting (FACS) staining was performed as previously described.31 Samples were acquired on a LSR I cytometer (Becton Dickinson, San Jose, CA) with CellQuest software (Becton Dickinson) and analyzed with FlowJo (Tree Star, Ashland, OR).
Analysis of T-cell proliferation in vivo
T cells were positively selected from red blood cell (RBC)–lysed splenocytes using CD5 MicroBeads (Miltenyi Biotec, Auburn, CA) and subsequently stained with 5 μM carboxyfluorescein diacetate succinimidyl ester (CFSE; Molecular Probes, Eugene, OR) for 20 minutes at 37°C. Between 15 and 30 × 106 stained cells were transplanted into lethally irradiated allogeneic recipients. Organs were harvested after 72 hours and analyzed with FACS.
Isolation of dendritic cells
To expand dendritic cells, C57BL/6 mice were injected subcutaneously with 10 × 106 B16 melanoma cells transduced with murine FLT3L (kindly provided by U. von Andrian, the CBR Institute for Biomedical Research, Harvard Medical School, Boston, MA). After 12 to 17 days, mice were killed and spleen, PLNs (axillary and inguinal), and MLNs were harvested, homogenized between frosted slides, and incubated for 15 minutes at 37°C in digest medium containing 1 mg/mL collagenase D and 100 μg/mL Dnase I (both from Roche, Indianapolis, IN). Livers were digested in Liver Digest Medium (Gibco, Carlsbad, CA) and nonparenchymal cells isolated by percoll separation (Amersham Biosciences, Piscataway, NJ). In some cases, recombinant human FLT3L (generously provided by Amgen, Thousand Oaks, CA) was given subcutaneously for 10 consecutive days before harvest of organs. Dendritic cells were purified immunomagnetically by positive selection with CD11c (N418) MicroBeads (Miltenyi Biotec) according to the manufacturer's instructions. Purity was analyzed by FACS and was between 90% and 95%.
T-cell stimulation
Donor-type T cells (B10.BR) were positively selected using CD5 MicroBeads (Miltenyi Biotec) and cocultured with host-type dendritic cells for 4 days at a ratio of 2:1 in T-cell medium consisting of RPMI 1640 (Mediatech, Herndon, VA), 10% heat-inactivated fetal bovine serum (FBS), 0.01 M HEPES, 2 mM l-glutamine, gentamicin (20 μg/mL), and β-mercaptoethanol. To increase maturation of dendritic cells, E coli O111:B4 lipopolysaccharide (LPS; Calbiochem, San Diego, CA) was added at 100 ng/mL at the start of the culture. In some cases, LPS was substituted with recombinant murine sCD40L from PeproTech at 1 μg/mL (Rocky Hill, NJ).
Cell proliferation and suppression assay
Various doses of organ-derived allogeneic dendritic cells were cocultured with 105 splenic donor-type T cells (at ratios of 1:100, 1:10, and 1:1) in triplicate wells of 96-well plates for 4 days in the presence of 100 ng/mL LPS. Tritiated thymidine ([3H]thymidine, 1 μCi [0.037 MBq]/well) was added for the last 18 hours of stimulation, and counts per minute were measured with a Topcount NXT microplate scintillation counter (Packard, Meridan, CT). For the suppression assay, 105 C57BL/6-derived T cells (responder) were cultured for 4 days with 2 × 105 irradiated (2000 cGy) BALB/c-derived splenocytes as stimulators and 6 × 105 B10.BR-derived CD4+CD25+ regulatory T cells that were isolated using magnetic beads after 4 days of T-cell stimulation with dendritic cells as described in the previous paragraph.
Enzyme-linked immunosorbent assay
Concentration of cytokines in cell culture supernatants or sera of mice that underwent transplantation was determined with commercially available enzyme-linked immunosorbent assay (ELISA) kits (R&D Systems) according to the manufacturer's protocol. Serum was obtained by retro-orbital bleeding and stored at −80°C until analysis.
Intracellular cytokine staining
Intracellular cytokine production of in vitro–activated donor T cells was measured by staining with (surface) fluorochrome-conjugated antibodies, fixing, and permeabilizing with the Cytofix/Cytoperm Kit (Pharmingen). Cells were subsequently stained with intracellular cytokine (IFN-γ and TNF-α) PE-conjugated antibodies. In some experiments, T cells were stimulated with PMA (10 ng/mL) and ionomycin (2 μM) for the final 4 hours, and brefeldin A (10 μg/mL) was added for the last 3 hours of incubation.
RNA isolation, labeling, microarray hybridization, and data analysis
RNA was extracted from fresh cultures by homogenization in TRIzol reagent (Gibco). Total RNA (2 μg) was used for cDNA synthesis using an oligo(dT)-T7 primer and the SuperScript Double-Stranded cDNA Synthesis Kit (Invitrogen Life Technologies, Carlsbad, CA). Synthesis, linear amplification, and labeling of cRNA were accomplished by in vitro transcription using the MessageAmp aRNA Kit (Ambion, Austin, TX) and biotinylated nucleotides (Enzo Diagnostics, Farmingdale, NY). Labeled and fragmented cRNA (10 μg) was then hybridized onto a GeneChip Mouse Genome 430A 2.0 Array (Affymetrix, Santa Clara, CA) representing approximately 14 000 mouse genes and scanned with a high-numeric aperture and flying objective (FOL) lens in a GS3000 scanner (Affymetrix). The image was quantified using Microarray Suite 5.1 (Affymetrix). Three independent RNA samples were analyzed for each of the 4 culture conditions. Using GCOS 1.3 (Affymetrix), we performed a cross comparison of each experimental sample to each reference. Cutoff values for fold change and P values were plus or minus 1.25 and .001, respectively. All array data have been deposited in GEO32 (accession number GSE5503).
Mice and bone marrow transplantation
The bone marrow transplantation (BMT) procedure was performed as described previously.31 Female C57BL/6 (B6) (H-2b) and B10.BR (H-2k) mice were obtained from The Jackson Laboratory (Bar Harbor, ME). All bone marrow transplantation protocols were approved by the Memorial Sloan-Kettering Cancer Center Institutional Animal Care and Use Committee.
Assessment of GvHD
Survival was monitored daily, and mice were individually scored weekly for 5 clinical parameters (weight loss, hunched posture, activity level, fur ruffling, and skin lesions) on a scale from 0 to 2, as previously described.33 A clinical GVHD score was generated by the summation of the 5 criteria, and mice scoring 5 or greater were killed.
Histopathologic analysis of target organ GVHD
Mice were killed after BMT for histopathologic analysis of GVHD target organs (small and large bowel, liver, and skin). Organs were harvested, formalin-preserved, paraffin-embedded, sectioned, and hematoxylin/eosin stained. Cutaneous GVHD was assessed by determining the number of apoptotic cells per millimeter of epidermis (G.F.M).34 A semiquantitative score consisting of 19 to 22 different parameters associated with GVHD was calculated for liver and gut samples (C.L.).35 Immunohistochemical staining for CD3 was performed using standard protocols.
Assessment of organ infiltration
For assessment of organ infiltration with donor T cells, recipient mice were killed after BMT and their target organs (skin, spleen, liver, PLNs, and MLNs) harvested. Mononuclear cells were isolated using standard procedures.
Statistics
All values shown in graphs represent the mean plus or minus SEM. Groupwise comparisons except for survival and GVHD score analysis were done with the one-way ANOVA test. The Mann-Whitney test or the Wilcoxon matched pairs test was used where appropriate. Survival data were analyzed with the Mantel-Cox log-rank test. For GVHD scores and the change in weight from baseline, the statistical analysis performed to test whether a differential change occurred between treatment groups was the difference in the area under curve (AUC) between groups, using all possible pairwise contrasts. Every mouse could not be followed for the full length of the study. To account for informative dropouts, the AUCs were calculated up to the minimum follow-up time for each pairwise difference. A P less than .05 was considered statistically significant.
Results
Homing molecules are differentially up-regulated on allogeneic donor T cells in vivo
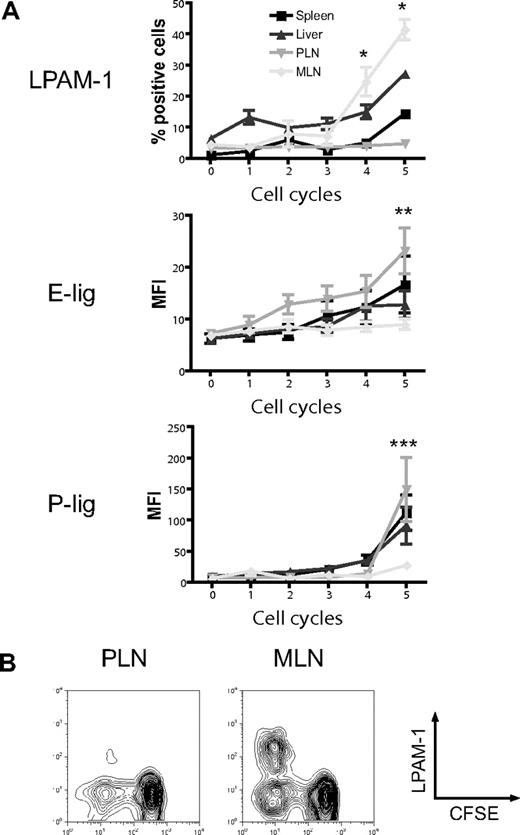
The induction of particular homing molecules on T cells seems to occur rapidly in secondary lymphoid organs in vivo.36 To determine whether alloreactive T cells display differential expression of homing molecules depending on their site of activation, we analyzed the expression of the gut-homing molecule LPAM-1 (α4β7 integrin) and the skin-homing molecules E- and P-selectin ligands (E-lig and P-lig), primarily the carbohydrate epitope cutaneous lymphocyte antigen (CLA) and P-selectin glycoprotein ligand 1 (PSGL1), respectively, in the spleen, liver, PLNs, and MLNs. Three days after adoptive transfer of CFSE-labeled naive allogeneic T cells (B10.BR) into lethally irradiated hosts (C57BL/6), we found significant up-regulation of LPAM-1 on rapidly dividing alloreactive T cells in MLNs compared with PLNs (Figure 1A,B).37 The expression of E-lig and P-lig was up-regulated in PLNs but not in MLNs. We conclude that alloreactive T cells in MLNs up-regulate LPAM-1, whereas alloreactive T cells in PLNs express more E-lig and P-lig.
Differential induction of homing molecules on allogeneic donor T cells in vivo. CFSE-labeled naive allogeneic donor T cells (B10.BR) were adoptively transferred into lethally irradiated hosts (C57BL/6), and expression of LPAM-1, E-lig, and P-lig on donor T cells was determined in spleen, liver, PLNs, and MLNs after 3 days. (A) Whereas the gut-homing molecule LPAM-1 is induced in MLNs, E-lig and P-lig are induced in PLNs (PLNs vs MLNs, *P = .002, **P < .005, ***P = .001). Expression of all molecules is intermediate in spleen and liver. Error bars represent SEM. (B) Representative example for LPAM-1 up-regulation on dividing (CFSElo) donor T cells in MLNs.
Differential induction of homing molecules on allogeneic donor T cells in vivo. CFSE-labeled naive allogeneic donor T cells (B10.BR) were adoptively transferred into lethally irradiated hosts (C57BL/6), and expression of LPAM-1, E-lig, and P-lig on donor T cells was determined in spleen, liver, PLNs, and MLNs after 3 days. (A) Whereas the gut-homing molecule LPAM-1 is induced in MLNs, E-lig and P-lig are induced in PLNs (PLNs vs MLNs, *P = .002, **P < .005, ***P = .001). Expression of all molecules is intermediate in spleen and liver. Error bars represent SEM. (B) Representative example for LPAM-1 up-regulation on dividing (CFSElo) donor T cells in MLNs.
Expanded organ-derived dendritic cells are heterogeneous and phenotypically immature but rapidly mature in the presence of LPS in vitro
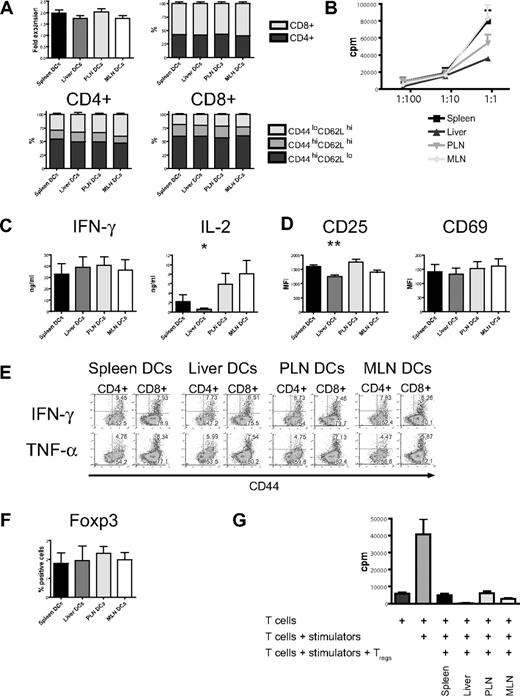
We isolated dendritic cells to better define their contribution to this induction of homing molecules.17 Administration of FMS-like tyrosine kinase 3 ligand (FLT3L) was necessary to generate sufficient numbers of host-type (C57BL/6) dendritic cells for use in bulk cultures and was given either through implantation of a murine FLT3L-transduced melanoma cell line (B16-FLT3L) as previously described22,23,38 or as a recombinant human protein. Dendritic cells were isolated from (1) the secondary lymphoid and GVHD target organ spleen, (2) the liver as a direct GVHD target organ, (3) PLNs, and (4) MLNs as secondary lymphoid organs for the GVHD target organs skin and gut, respectively. The subset composition of dendritic cells varied considerably among different organs, except for a remarkable similarity between lymph nodes (Figure 2A). All dendritic cells were phenotypically immature and had high expression of MHC II (I-A/I-E) and low levels of CD80, CD86, or CD40. Liver-derived dendritic cells had significantly fewer MHC II–positive and CD40+ cells (Figure 2B,C). However, adding LPS to our culture conditions allowed all dendritic cells to rapidly acquire a fully mature phenotype irrespective of their origins (Figure 2C).
Organ-derived dendritic cells are heterogeneous and phenotypically immature. (A) Subset composition of organ-derived dendritic cells (CD11c+) defined as lymphoid (CD8+), myeloid (CD11b+), or plasmacytoid (B220+) shows a large proportion of liver-derived dendritic cells being negative for all 3 subset markers and a high number of plasmacytoid dendritic cells in both lymph node groups (n = 14-15). (B) Organ-derived dendritic cells are phenotypically immature, express mostly MHC II (I-A/I-E), but only partly CD80, CD86, or CD40. Liver-derived dendritic cells have significantly fewer MHC II–positive and CD40+ cells (n = 6-8, *P < .05). Error bars represent SEM. (C) All organ-derived dendritic cells acquire a mature phenotype after overnight culture in the presence of LPS (100 ng/mL) with further up-regulation of MHC II and full induction of CD80, CD86, and CD40 (representative experiment).
Organ-derived dendritic cells are heterogeneous and phenotypically immature. (A) Subset composition of organ-derived dendritic cells (CD11c+) defined as lymphoid (CD8+), myeloid (CD11b+), or plasmacytoid (B220+) shows a large proportion of liver-derived dendritic cells being negative for all 3 subset markers and a high number of plasmacytoid dendritic cells in both lymph node groups (n = 14-15). (B) Organ-derived dendritic cells are phenotypically immature, express mostly MHC II (I-A/I-E), but only partly CD80, CD86, or CD40. Liver-derived dendritic cells have significantly fewer MHC II–positive and CD40+ cells (n = 6-8, *P < .05). Error bars represent SEM. (C) All organ-derived dendritic cells acquire a mature phenotype after overnight culture in the presence of LPS (100 ng/mL) with further up-regulation of MHC II and full induction of CD80, CD86, and CD40 (representative experiment).
Liver-derived dendritic cells have less allostimulatory capacity
In vitro stimulation with host-type dendritic cells (C57BL/6) efficiently activated naive allogeneic donor-type T cells (B10.BR) as evidenced by the increase of absolute cell numbers and the acquisition of an effector memory phenotype (CD44hiCD62Llo) with a predominance of CD8+ T cells (Figure 3A). This was similar among all 4 groups. However, proliferation as determined by 3H-thymidine incorporation was lower in the liver group (Figure 3B) and was associated with lower levels of interleukin 2 (IL-2) in cell culture supernatants, whereas IFN-γ and IL-12p40 secretion was similar (Figure 3C; Figure S3, available on the Blood website; see the Supplemental Materials link at the top of the online article). Expression of the activation marker CD25 was significantly lower, whereas expression levels of CD69 did not differ from the other organs (Figure 3D). Production of IFN-γ and TNF-α when determined by intracellular cytokine staining was similar in all 4 groups (Figure 3E). After additional stimulation with PMA and ionomycin, expression of IFN-γ and CD69 was comparable (Figure S5). Regulatory T cells detected by staining for Foxp3 had an activated phenotype (CD44hiCD62Llo, not shown) and were found in similar numbers in all groups (Figure 3F). To determine their suppressive capacity, we performed a suppression assay and found that regulatory T cells from all 4 groups strongly suppressed proliferation (Figure 3G). And although “liver” regulatory T cells showed more suppression than the other 3 groups, this was not statistically significant. We conclude that liver-derived dendritic cells are less effective in the activation of allogeneic T cells.
Organ-derived dendritic cells activate naive allogeneic T cells in vitro, but liver-derived dendritic cells are less efficient. (A) Four days after coculture with different organ-derived host-type dendritic cells, absolute cell numbers of donor-type T cells are almost doubled with preferential stimulation of CD8+ cells (n = 15-17). CD4+ and CD8+ T cells express mostly an effector memory phenotype (CD44hiCD62Llo). (B) Liver-derived dendritic cells induce less proliferation of allogeneic T cells, as assessed by 3H-thymidine incorporation (1 representative of 4 experiments shown). (C) Determination of cytokines in cell culture supernatant shows comparable IFN-γ but significantly less IL-2 secretion by T cells stimulated with liver-derived dendritic cells (n = 6, *P < .05). (D) Expression of the activation marker CD25 is significantly lower in the liver group (n = 8-10, **P < .001), whereas CD69 is comparable. (E) Intracellular staining for IFN-γ and TNF-α shows similar expression in all 4 groups. (F) The induction of activated Foxp3+ regulatory T cells by all organ-derived dendritic cells is comparable. (G) The suppressive capacity of isolated CD4+CD25+ regulatory T cells is similar in a suppression assay using C57BL/6 responder and BALB/c stimulator. Panels D and F are gated on CD44hiCD62Llo T cells. Error bars represent SEM.
Organ-derived dendritic cells activate naive allogeneic T cells in vitro, but liver-derived dendritic cells are less efficient. (A) Four days after coculture with different organ-derived host-type dendritic cells, absolute cell numbers of donor-type T cells are almost doubled with preferential stimulation of CD8+ cells (n = 15-17). CD4+ and CD8+ T cells express mostly an effector memory phenotype (CD44hiCD62Llo). (B) Liver-derived dendritic cells induce less proliferation of allogeneic T cells, as assessed by 3H-thymidine incorporation (1 representative of 4 experiments shown). (C) Determination of cytokines in cell culture supernatant shows comparable IFN-γ but significantly less IL-2 secretion by T cells stimulated with liver-derived dendritic cells (n = 6, *P < .05). (D) Expression of the activation marker CD25 is significantly lower in the liver group (n = 8-10, **P < .001), whereas CD69 is comparable. (E) Intracellular staining for IFN-γ and TNF-α shows similar expression in all 4 groups. (F) The induction of activated Foxp3+ regulatory T cells by all organ-derived dendritic cells is comparable. (G) The suppressive capacity of isolated CD4+CD25+ regulatory T cells is similar in a suppression assay using C57BL/6 responder and BALB/c stimulator. Panels D and F are gated on CD44hiCD62Llo T cells. Error bars represent SEM.
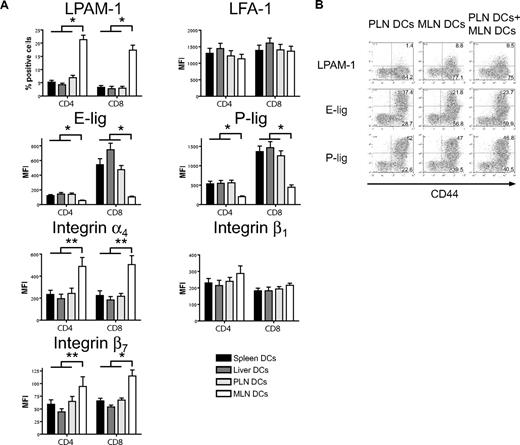
MLN dendritic cells induce high LPAM-1 but low E- and P-selectin ligand expression on allogeneic T cells in vitro
We next addressed whether host-derived dendritic cells from spleen, liver, PLNs, and MLNs could differentially induce the same homing molecules in vitro that we had previously observed to be induced in vivo (Figure 1). Only dendritic cells from MLNs induced LPAM-1 expression on donor CD4+ and CD8+ cells, whereas they were significantly less capable of inducing expression of E-lig and P-lig (Figure 4A). The induction of LPAM-1 in the MLN group was due to significantly higher expression of both subunits, the α4 and β7 chains. LFA-1, which has recently been implicated in liver homing,39 was not differentially induced by liver-derived dendritic cells. All differences were detected on T cells with an effector memory phenotype only (data not shown). As previously described for Peyer patch–derived dendritic cells,23 and in accordance with an active mechanism,25 MLN dendritic cells (DCs) dominantly induced LPAM-1 and partially suppressed the induction of E-lig and P-lig when competing with PLN DCs at equivalent concentrations (Figure 4B). Thus, all organ-derived dendritic cells activate allogeneic T cells but only MLN DCs up-regulate LPAM-1 and only weakly induce E- and P-lig compared with all other dendritic cells, thereby inducing a gut-homing phenotype.
Strong induction of LPAM-1, but less induction of E-lig and P-lig by MLN DCs. (A) Four days after coculture with different organ-derived dendritic cells, expression of LPAM-1, LFA-1, E-lig, P-lig, integrin α4, integrin β1, and integrin β7 allogeneic T cells was analyzed. Effector memory cells (CD44hiCD62Llo) in the MLN DC group express LPAM-1 and significantly less E-lig or P-lig (*P < .001, n = 8-15). There is also statistically significantly higher expression of integrin α4 and integrin β7 in the MLN DC group (**P < .05, n = 5-6). Error bars represent SEM. (B) Coculture of PLN and MLN DCs at equivalent concentrations shows that MLN DCs induce LPAM-1 irrespectively of PLN DCs, and negatively affect the induction of E-lig and P-lig (1 representative of 3 independent experiments).
Strong induction of LPAM-1, but less induction of E-lig and P-lig by MLN DCs. (A) Four days after coculture with different organ-derived dendritic cells, expression of LPAM-1, LFA-1, E-lig, P-lig, integrin α4, integrin β1, and integrin β7 allogeneic T cells was analyzed. Effector memory cells (CD44hiCD62Llo) in the MLN DC group express LPAM-1 and significantly less E-lig or P-lig (*P < .001, n = 8-15). There is also statistically significantly higher expression of integrin α4 and integrin β7 in the MLN DC group (**P < .05, n = 5-6). Error bars represent SEM. (B) Coculture of PLN and MLN DCs at equivalent concentrations shows that MLN DCs induce LPAM-1 irrespectively of PLN DCs, and negatively affect the induction of E-lig and P-lig (1 representative of 3 independent experiments).
Microarray analysis of allogeneic T cells stimulated with different organ-derived dendritic cells reveals differentially induced genes
To apply this culture system to discover new genes that are differentially induced by organ-derived dendritic cells, we performed microarray analyses of these stimulated T cells using the GeneChip Mouse Genome 430A 2.0 Array (Affymetrix). By directly comparing 2 groups we eliminated all genes that were equally up-regulated in both groups (eg, reflecting an activated state). In addition, comparisons were always done between stimulated T cells from one experiment that consisted of the same initially naive T cells with different organ-derived dendritic cells from the same (pooled) animal. This eliminated interexperiment variability and possible genetic heterogeneity. A first comparison between liver and spleen was performed to identify putative liver-homing molecules (Table 1). In the liver group, 22 genes were up-regulated and 16 genes were down-regulated compared with the spleen group. Two genes that have previously been implicated in liver homing (Cxcr6 and S100a4) were found to be up-regulated. A second comparison between MLNs with PLNs (Table 2) identified 15 and 22 different genes to be up- and down-regulated, respectively. Two genes that are induced by retinoic acid (Stra6 and Tgm2) were up-regulated in the MLN group. Overall, there was a major overlap in each comparison with not more than 22 of approximately 14 000 genes being significantly up- or down-regulated, indicating considerable similarities between the different organ-derived dendritic cells and identifying potential differentially induced homing molecules.
T-cell genes differentially induced by liver-derived dendritic cells
| Fold change . | Gene title . | Gene symbol . | Genebank40 . |
|---|---|---|---|
| Up-regulated | |||
| 2,10 | Regulator of G-protein signaling 16 | Rgs16 | U94828.1 |
| 2,09 | Granzyme G | Gzmg | NM_010375.1 |
| 1,92 | Secreted phosphoprotein 1 | Spp1 | NM_009263.1 |
| 1,89 | Granzyme E* (n=2) | Gzme | NM_010373.1 |
| 1,77 | Chemokine (C-X-C motif) receptor 6 | Cxcr6 | AF301018.1 |
| 1,64 | S100 calcium binding protein A4 | S100a4 | D00208.1 |
| 1,49 | Granzyme C | Gzmc | NM_010371.1 |
| 1,36 | Thioredoxin interacting protein | Txnip | NM_023719.1 |
| 1,32 | S100 calcium binding protein A10 (calpactin) | S100a10 | NM_009112.1 |
| 1,32 | Poly A binding protein, cytoplasmic 1 | Pabpc1 | NM_008774.1 |
| 1,29 | Semaphorin 4A | Sema4a | NM_013658.1 |
| 1,29 | Ribosomal protein S2* (n=2) | Rps2 | AK004568.1 |
| 1,26 | Similar to glyceraldehyde-3-phosphate dehydrogenase | LOC14433 | NM_008084.1 |
| 1,26 | Rho, GDP dissociation inhibitor (GDI) beta | Arhgdib | AK002516.1 |
| 1,26 | Ribosomal protein, large, P1 | Rplp1 | NM_018853.1 |
| 1,26 | Lectin, galactose binding, soluble 1 | Lgals1 | NM_008495.1 |
| 1,26 | Ribosomal protein L5 | Rpl5 | BC026934.1 |
| 1,26 | Ribosomal protein L26 | Rpl26 | NM_009080.1 |
| 1,26 | Ribosomal protein L13 | Rpl13 | NM_016738.1 |
| 1,26 | Actin, beta, cytoplasmic* (n=2) | Actb | M12481.1 |
| 1,26 | Adenylosuccinate lyase | Adsl | K00131.1 |
| 1,26 | Caspase 3, apoptosis related cysteine protease | Casp3 | D86352.1 |
| Down-regulated | |||
| −1,45 | Calponin 3, acidic* (n=3) | Cnn3 | BB724741 |
| −1,49 | Suppressor of cytokine signaling 3* (n=2) | Socs3 | BB241535 |
| −1,49 | RIKEN cDNA 1110035L05 gene | 1110035L05Rik | NM_026125.1 |
| −1,52 | Calponin 3, acidic | Cnn3 | AV172168 |
| −1,55 | Tumor necrosis factor receptor superfamily, member 8 | Tnfrsf8 | NM_009401.1 |
| −1,59 | Interleukin 1 receptor, type II | Il1r2 | NM_010555.1 |
| −1,70 | T-cell receptor gamma chain /// T cell receptor gamma chain | Tcrg | NM_011558.2 |
| −1,74 | Histocompatibility 2, Q region locus 10 | H2-Q10 | BC011215.1 |
| −1,76 | T cell receptor associated transmembrane adaptor 1 | Trat1 | BB622792 |
| −1,78 | FERM domain containing 4B | Frmd4b | BB009122 |
| −1,95 | Aryl-hydrocarbon receptor | Ahr | NM_013464.1 |
| −2,00 | Insulin-like growth factor binding protein 4* (n=4) | Igfbp4 | BC019836.1 |
| −2,45 | Extracellular matrix protein 1 | Ecm1 | NM_007899.1 |
| −3,32 | Lumican | Lum | AK014312.1 |
| −3,35 | Interleukin 17 | Il17 | NM_010552.1 |
| −4,61 | Transglutaminase 2, C polypeptide* (n=4) | Tgm2 | BC016492.1 |
| Fold change . | Gene title . | Gene symbol . | Genebank40 . |
|---|---|---|---|
| Up-regulated | |||
| 2,10 | Regulator of G-protein signaling 16 | Rgs16 | U94828.1 |
| 2,09 | Granzyme G | Gzmg | NM_010375.1 |
| 1,92 | Secreted phosphoprotein 1 | Spp1 | NM_009263.1 |
| 1,89 | Granzyme E* (n=2) | Gzme | NM_010373.1 |
| 1,77 | Chemokine (C-X-C motif) receptor 6 | Cxcr6 | AF301018.1 |
| 1,64 | S100 calcium binding protein A4 | S100a4 | D00208.1 |
| 1,49 | Granzyme C | Gzmc | NM_010371.1 |
| 1,36 | Thioredoxin interacting protein | Txnip | NM_023719.1 |
| 1,32 | S100 calcium binding protein A10 (calpactin) | S100a10 | NM_009112.1 |
| 1,32 | Poly A binding protein, cytoplasmic 1 | Pabpc1 | NM_008774.1 |
| 1,29 | Semaphorin 4A | Sema4a | NM_013658.1 |
| 1,29 | Ribosomal protein S2* (n=2) | Rps2 | AK004568.1 |
| 1,26 | Similar to glyceraldehyde-3-phosphate dehydrogenase | LOC14433 | NM_008084.1 |
| 1,26 | Rho, GDP dissociation inhibitor (GDI) beta | Arhgdib | AK002516.1 |
| 1,26 | Ribosomal protein, large, P1 | Rplp1 | NM_018853.1 |
| 1,26 | Lectin, galactose binding, soluble 1 | Lgals1 | NM_008495.1 |
| 1,26 | Ribosomal protein L5 | Rpl5 | BC026934.1 |
| 1,26 | Ribosomal protein L26 | Rpl26 | NM_009080.1 |
| 1,26 | Ribosomal protein L13 | Rpl13 | NM_016738.1 |
| 1,26 | Actin, beta, cytoplasmic* (n=2) | Actb | M12481.1 |
| 1,26 | Adenylosuccinate lyase | Adsl | K00131.1 |
| 1,26 | Caspase 3, apoptosis related cysteine protease | Casp3 | D86352.1 |
| Down-regulated | |||
| −1,45 | Calponin 3, acidic* (n=3) | Cnn3 | BB724741 |
| −1,49 | Suppressor of cytokine signaling 3* (n=2) | Socs3 | BB241535 |
| −1,49 | RIKEN cDNA 1110035L05 gene | 1110035L05Rik | NM_026125.1 |
| −1,52 | Calponin 3, acidic | Cnn3 | AV172168 |
| −1,55 | Tumor necrosis factor receptor superfamily, member 8 | Tnfrsf8 | NM_009401.1 |
| −1,59 | Interleukin 1 receptor, type II | Il1r2 | NM_010555.1 |
| −1,70 | T-cell receptor gamma chain /// T cell receptor gamma chain | Tcrg | NM_011558.2 |
| −1,74 | Histocompatibility 2, Q region locus 10 | H2-Q10 | BC011215.1 |
| −1,76 | T cell receptor associated transmembrane adaptor 1 | Trat1 | BB622792 |
| −1,78 | FERM domain containing 4B | Frmd4b | BB009122 |
| −1,95 | Aryl-hydrocarbon receptor | Ahr | NM_013464.1 |
| −2,00 | Insulin-like growth factor binding protein 4* (n=4) | Igfbp4 | BC019836.1 |
| −2,45 | Extracellular matrix protein 1 | Ecm1 | NM_007899.1 |
| −3,32 | Lumican | Lum | AK014312.1 |
| −3,35 | Interleukin 17 | Il17 | NM_010552.1 |
| −4,61 | Transglutaminase 2, C polypeptide* (n=4) | Tgm2 | BC016492.1 |
Naive donor-type allogeneic T cells were cocultured for 4 days with liver-derived host-type dendritic cells, total RNA was extracted, and microarray analysis was performed using the GeneChip Mouse Genome 430A 2.0 Array (Affymetrix). Gene expression levels were compared with T cells cultured with spleen-derived dendritic cells. Greater than 1.25-fold changes with P<.001 were chosen.
T-cell genes differentially induced by MLN-derived dendritic cells
| Fold change . | Gene title . | Gene symbol . | Genebank40 . |
|---|---|---|---|
| Up-regulated | |||
| 4.14 | Stimulated by retinoic acid gene 6 | Stra6 | NM_009291.1 |
| 2.37 | Leucine zipper transcription factor-like 1 | Lztfl1 | NM_033322.1 |
| 2.00 | Hypermethylated in cancer 1 | Hic1 | NM_010430.1 |
| 1.73 | Xanthine dehydrogenase | Xdh | AV286265 |
| 1.63 | Granzyme G | Gzmg | NM_010375.1 |
| 1.63 | Purinergic receptor P2X, ligand-gated ion channel 4 | P2rx4 | AJ251462.1 |
| 1.49 | Serine (or cysteine) proteinase inhibitor, clade E, member 2 | Serpine2 | NM_009255.1 |
| 1.49 | Serine (or cysteine) proteinase inhibitor, clade A, member 3G | Serpina3g | BC002065.1 |
| 1.45 | Growth factor independent 1 | Gfi1 | NM_010278.1 |
| 1.45 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | Gcnt2 | BM236768 |
| 1.44 | Transglutaminase 2, C polypeptide* (n=2) | Tgm2 | BB550124 |
| 1.42 | Tribbles homolog 2 (Drosophila) | Trib2 | BC027159.1 |
| 1.38 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | Lilrb4 | U05264.1 |
| 1.35 | Sprouty-related, EVH1 domain containing 2 | Spred2 | AV229054 |
| 1.35 | GTPase, IMAP family member 3 | Gimap3 | NM_031247.1 |
| Down-regulated | |||
| −1.26 | CD44 antigen | Cd44 | BC005676.1 |
| −1.26 | Interleukin 7 receptor | Il7r | NM_008372.2 |
| −1.31 | Asparagine synthetase* (n=2) | Asns | BC005552.1 |
| −1.32 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | St8sia4 | NM_009183.1 |
| −1.39 | Beta galactoside alpha 2,6 sialyltransferase 1 | St6gal1 | BG075800 |
| −1.42 | Inducible T-cell co-stimulator* (n=2) | Icos | NM_017480.1 |
| −1.42 | Membrane-spanning 4-domains, subfamily A, member 4C | Ms4a4c | NM_022429.1 |
| −1.45 | Selectin, lymphocyte* (n=2) | Sell | M36005.1 |
| −1.49 | Prion protein* (n=2) | Prnp | NM_011170.1 |
| −1.50 | RIKEN cDNA 1190002H23 gene* (n=2) | 1190002H23Rik | BB408123 |
| −1.56 | Syndecan binding protein (syntenin) 2 | Sdcbp2 | BC005556.1 |
| −1.61 | Glutamic pyruvate transaminase (alanine aminotransferase) 2 | Gpt2 | BI648645 |
| −1.63 | Interleukin 10 | Il10 | NM_010548.1 |
| −1.64 | Palmitoyl-protein thioesterase precursor | ppt | AF087568.1 |
| −1.69 | Cystathionase (cystathionine gamma-lyase) | Cth | BC019483.1 |
| −1.81 | Interleukin 18 receptor 1 | Il18r1 | NM_008365.1 |
| −1.95 | Plexin C1* (n=2) | Plxnc1 | BB765457 |
| −2.02 | Interleukin 24 | Il24 | AF333251.1 |
| −2.11 | Hemogen | Hemgn | NM_053149.1 |
| −2.23 | Killer cell lectin-like receptor subfamily C, member 1 /// member 2 | Klrc1 /// Klrc2 | AF106008.1 |
| −2.25 | Semaphorin 4A* (n=2) | Sema4a | NM_013658.1 |
| −4.23 | Lamin A | Lmna | NM_019390.1 |
| Fold change . | Gene title . | Gene symbol . | Genebank40 . |
|---|---|---|---|
| Up-regulated | |||
| 4.14 | Stimulated by retinoic acid gene 6 | Stra6 | NM_009291.1 |
| 2.37 | Leucine zipper transcription factor-like 1 | Lztfl1 | NM_033322.1 |
| 2.00 | Hypermethylated in cancer 1 | Hic1 | NM_010430.1 |
| 1.73 | Xanthine dehydrogenase | Xdh | AV286265 |
| 1.63 | Granzyme G | Gzmg | NM_010375.1 |
| 1.63 | Purinergic receptor P2X, ligand-gated ion channel 4 | P2rx4 | AJ251462.1 |
| 1.49 | Serine (or cysteine) proteinase inhibitor, clade E, member 2 | Serpine2 | NM_009255.1 |
| 1.49 | Serine (or cysteine) proteinase inhibitor, clade A, member 3G | Serpina3g | BC002065.1 |
| 1.45 | Growth factor independent 1 | Gfi1 | NM_010278.1 |
| 1.45 | Glucosaminyl (N-acetyl) transferase 2, I-branching enzyme | Gcnt2 | BM236768 |
| 1.44 | Transglutaminase 2, C polypeptide* (n=2) | Tgm2 | BB550124 |
| 1.42 | Tribbles homolog 2 (Drosophila) | Trib2 | BC027159.1 |
| 1.38 | Leukocyte immunoglobulin-like receptor, subfamily B, member 4 | Lilrb4 | U05264.1 |
| 1.35 | Sprouty-related, EVH1 domain containing 2 | Spred2 | AV229054 |
| 1.35 | GTPase, IMAP family member 3 | Gimap3 | NM_031247.1 |
| Down-regulated | |||
| −1.26 | CD44 antigen | Cd44 | BC005676.1 |
| −1.26 | Interleukin 7 receptor | Il7r | NM_008372.2 |
| −1.31 | Asparagine synthetase* (n=2) | Asns | BC005552.1 |
| −1.32 | ST8 alpha-N-acetyl-neuraminide alpha-2,8-sialyltransferase 4 | St8sia4 | NM_009183.1 |
| −1.39 | Beta galactoside alpha 2,6 sialyltransferase 1 | St6gal1 | BG075800 |
| −1.42 | Inducible T-cell co-stimulator* (n=2) | Icos | NM_017480.1 |
| −1.42 | Membrane-spanning 4-domains, subfamily A, member 4C | Ms4a4c | NM_022429.1 |
| −1.45 | Selectin, lymphocyte* (n=2) | Sell | M36005.1 |
| −1.49 | Prion protein* (n=2) | Prnp | NM_011170.1 |
| −1.50 | RIKEN cDNA 1190002H23 gene* (n=2) | 1190002H23Rik | BB408123 |
| −1.56 | Syndecan binding protein (syntenin) 2 | Sdcbp2 | BC005556.1 |
| −1.61 | Glutamic pyruvate transaminase (alanine aminotransferase) 2 | Gpt2 | BI648645 |
| −1.63 | Interleukin 10 | Il10 | NM_010548.1 |
| −1.64 | Palmitoyl-protein thioesterase precursor | ppt | AF087568.1 |
| −1.69 | Cystathionase (cystathionine gamma-lyase) | Cth | BC019483.1 |
| −1.81 | Interleukin 18 receptor 1 | Il18r1 | NM_008365.1 |
| −1.95 | Plexin C1* (n=2) | Plxnc1 | BB765457 |
| −2.02 | Interleukin 24 | Il24 | AF333251.1 |
| −2.11 | Hemogen | Hemgn | NM_053149.1 |
| −2.23 | Killer cell lectin-like receptor subfamily C, member 1 /// member 2 | Klrc1 /// Klrc2 | AF106008.1 |
| −2.25 | Semaphorin 4A* (n=2) | Sema4a | NM_013658.1 |
| −4.23 | Lamin A | Lmna | NM_019390.1 |
Naive donor-type allogeneic T cells were cocultured for 4 days with MLN-derived host-type dendritic cells, total RNA was extracted, and microarray analysis was performed using the GeneChip Mouse Genome 430A 2.0 Array (Affymetrix). Gene expression levels were compared with T cells cultured with PLN-derived dendritic cells. Greater than 1.25-fold changes with P < .001 were chosen.
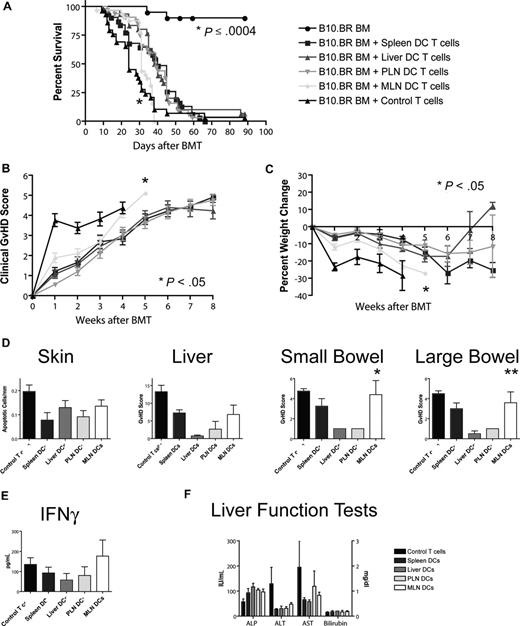
Allogeneic T cells stimulated with MLN DCs induce the highest GVHD mortality and morbidity
To assess the impact of different homing phenotypes on the capacity to induce GVHD, we transplanted T cell–depleted bone marrow and 106 stimulated T cells (B10.BR) into lethally irradiated hosts (C57BL/6). Recipients of T cells from the MLN DC group had the shortest overall survival (Figure 5A), with a median survival time of 31 days as opposed to approximately 40 days for the other groups. This difference was highly statistically significant (MLN vs liver/PLN: P < .001, MLN vs spleen: P = .001) and was also associated with higher overall clinical GVHD scores (MLN vs spleen/PLN: P < .001, MLN vs liver: P = .024, Figure 5B) and more gut GVHD-induced weight loss (MLN vs spleen/PLN: P < .001, MLN vs liver: P = .013, Figure 5C), but not higher clinical skin GVHD scores (data not shown) compared with the other 3 groups. This higher mortality and morbidity correlated with increased GVHD target organ pathology in the gut (MLN vs liver/PLN: P < .05, Figure 5D), but not in liver or skin. At day 14, levels of IFN-γ, IL-12p70, TNF-α, MCP-1, and IL-10 (Figure 5E; Figure S4) and liver function tests (Figure 5F) in the sera of mice that underwent transplantation were similar. We did not find evidence for more skin GVHD in the PLN DC group or more liver GVHD in the liver DC group. T cells stimulated with MLN DCs thus induce the highest GVHD mortality and morbidity after BMT and specifically more gut GVHD, but the absolute difference from the other groups is rather small.
Allogeneic T cells stimulated with host-type MLN DCs in vitro induced the highest GVHD mortality and morbidity after bone marrow transplantation in vivo. (A) Naive allogeneic donor T cells stimulated with organ-derived host-type dendritic cells in vitro were used as donor T cells in an MHC-mismatched B10.BR into C57BL/6 murine BMT model with T cell–depleted 5 to 10 × 106 BM cells and 106 T cells (n = 28-32, 4 combined experiments). Survival was significantly shorter for the MLN DC group (*MLN vs liver/PLN: P < .001, MLN vs spleen: P = .001) with 40 days median survival for spleen-derived dendritic cells, 39 days for liver-derived dendritic cells, 39 days for PLN DCs, and 31 days for MLN DCs. (B) Clinical GVHD score (*MLN vs spleen/PLN: P < .001, MLN vs liver: P = .024) and (C) weight curves (*MLN vs spleen/PLN: P < .001, MLN vs liver: P = .013) show higher GVHD morbidity for the MLN DC group. (D) Histopathologic analysis of the GVHD target organs skin, liver, and small and large bowel at day 14. There is a trend toward higher GVHD scores in the small and large bowel of the MLN DC group (*MLN vs liver/PLN: P < .05, **MLN vs liver: P < .05). (E) Levels of IFN-γ and (F) liver function tests at day 14 on sera of mice that underwent transplantation are comparable among all groups. Error bars represent SEM.
Allogeneic T cells stimulated with host-type MLN DCs in vitro induced the highest GVHD mortality and morbidity after bone marrow transplantation in vivo. (A) Naive allogeneic donor T cells stimulated with organ-derived host-type dendritic cells in vitro were used as donor T cells in an MHC-mismatched B10.BR into C57BL/6 murine BMT model with T cell–depleted 5 to 10 × 106 BM cells and 106 T cells (n = 28-32, 4 combined experiments). Survival was significantly shorter for the MLN DC group (*MLN vs liver/PLN: P < .001, MLN vs spleen: P = .001) with 40 days median survival for spleen-derived dendritic cells, 39 days for liver-derived dendritic cells, 39 days for PLN DCs, and 31 days for MLN DCs. (B) Clinical GVHD score (*MLN vs spleen/PLN: P < .001, MLN vs liver: P = .024) and (C) weight curves (*MLN vs spleen/PLN: P < .001, MLN vs liver: P = .013) show higher GVHD morbidity for the MLN DC group. (D) Histopathologic analysis of the GVHD target organs skin, liver, and small and large bowel at day 14. There is a trend toward higher GVHD scores in the small and large bowel of the MLN DC group (*MLN vs liver/PLN: P < .05, **MLN vs liver: P < .05). (E) Levels of IFN-γ and (F) liver function tests at day 14 on sera of mice that underwent transplantation are comparable among all groups. Error bars represent SEM.
Imprinted allogeneic T cells show differential in vivo homing
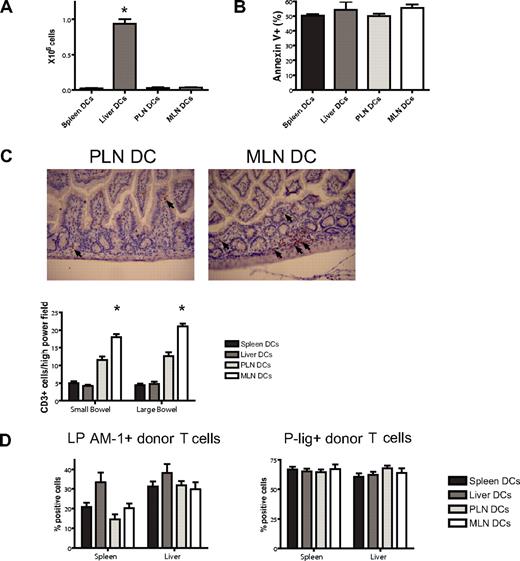
To assess a potential liver-homing phenotype, we quantified donor T-cell numbers in the liver at day 7 after BMT and found reproducibly significantly higher numbers in the liver DC group (Figure 6A). This was not due to less apoptosis as evidenced by staining for annexin V (Figure 6B). To analyze the in vivo homing of the PLN DC and the MLN DC group, we quantified cells in the skin and the gut of mice that underwent transplantation. Although not statistically significant, we did observe more MLN DC T cells in the gut and PLN DC T cells in the skin on days 7 and 23 (data not shown). However, we felt that the isolation of lymphocytes from the skin and the gut was prone to high variability. We therefore opted for additional in situ analyses of gut-infiltrating T cells by immunohistochemical staining and quantification of the number of CD3+ cells per high-power field. On day 7, significantly higher numbers of T cells were found in the small and large bowel of mice that received a transplant of T cells from the MLN DC group compared with the PLN DC group (Figure 6C). We conclude that imprinted allogeneic T cells show differential in vivo migratory behavior leading to preferential infiltration of the corresponding organ of the dendritic cells that were used for stimulation.
Imprinted allogeneic T cells show differential in vivo homing. (A) Donor T-cell numbers in the liver at day 7 after BMT are highest in the liver DC group (*P < .001, n = 4-5). (B) Apoptosis as analyzed by staining for annexin V is comparable among the groups. (C) On day 7, immunohistochemical staining and quantification of the number of CD3+ cells per high-power field shows significantly higher numbers of allogeneic T cells in the small and large bowel of mice in the MLN DC group compared with the PLN DC group (*small bowel: P < .001, large bowel: P < .001, n = 5). Corresponding representative photomicrographs (original magnification: × 200). Images were visualized with an Olympus BX40 microscope (Olympus, Melville, NY) equipped with a 10×/0.65 aperture objective lens. Images were acquired with a JVC digital camera GC-Qx 5HDU (JVC, Wayne, NJ). (D) The expression of the homing molecules LPAM-1 and P-lig on donor T cells in vivo was analyzed at day 6 after pre-vious stimulation with different organ-derived dendritic cells in vitro (n = 5). Both molecules are up-regulated irrespective of previous stimulation in vitro. Error bars represent SEM.
Imprinted allogeneic T cells show differential in vivo homing. (A) Donor T-cell numbers in the liver at day 7 after BMT are highest in the liver DC group (*P < .001, n = 4-5). (B) Apoptosis as analyzed by staining for annexin V is comparable among the groups. (C) On day 7, immunohistochemical staining and quantification of the number of CD3+ cells per high-power field shows significantly higher numbers of allogeneic T cells in the small and large bowel of mice in the MLN DC group compared with the PLN DC group (*small bowel: P < .001, large bowel: P < .001, n = 5). Corresponding representative photomicrographs (original magnification: × 200). Images were visualized with an Olympus BX40 microscope (Olympus, Melville, NY) equipped with a 10×/0.65 aperture objective lens. Images were acquired with a JVC digital camera GC-Qx 5HDU (JVC, Wayne, NJ). (D) The expression of the homing molecules LPAM-1 and P-lig on donor T cells in vivo was analyzed at day 6 after pre-vious stimulation with different organ-derived dendritic cells in vitro (n = 5). Both molecules are up-regulated irrespective of previous stimulation in vitro. Error bars represent SEM.
Stimulated donor T cells express homing molecules in vivo irrespective of previous stimulation in vitro
One possible explanation for this moderate shift in aggressiveness is the reprogramming of T cells that has been described previously for in vitro and in vivo imprinted T cells23,24 and sorted unstimulated T cells.41 We therefore analyzed expression of homing molecules on stimulated donor T cells 6 days after transplantation in vivo (Figure 6D). Analysis of LPAM-1 and P-lig expression showed comparable expression between the groups despite previous stimulation with different dendritic cells. The induction of a defined homing phenotype in vitro thus appears to be transient after adoptive transfer to an allogeneic bone marrow transplant recipient.
Discussion
Host APCs are crucial for the initiation of GVHD, but their possible role in imprinting homing molecules on alloreactive donor T cells has not been addressed previously. Here, we took a combined in vitro and in vivo approach to study the impact of different organ-derived dendritic cells on naive allogeneic T cells in vitro and their capacity to induce GVHD after transplantation in vivo.
We chose to focus on the secondary lymphoid and GVHD target organ spleen, the liver as a GVHD target organ, and the peripheral and mesenteric lymph nodes as secondary lymphoid organs for the GVHD target organs skin and gut. We also performed experiments with dendritic cells from Peyer patches, which were limited by the lower number of dendritic cells that could be obtained. However, the main conclusions regarding MLN DCs, specifically the induction of a gut-homing phenotype and the higher mortality and morbidity after transplantation, also apply to dendritic cells from Peyer patches (data not shown).
A number of recent studies have shown that different organ-derived dendritic cells imprint a homing phenotype on naive T cells.20,,,–24,42 We extend these studies from transgenic autologous model systems to an allogeneic MHC-mismatched BMT model. Whereas only T cells stimulated with MLN DCs became positive for LPAM-1, all T cells up-regulated their expression of E-lig and P-lig. However, the level of E-lig and P-lig up-regulation was lower after stimulation with dendritic cells from MLNs than after stimulation with other dendritic cells. As previously shown for Peyer patch–derived dendritic cells,23 MLN DCs dominantly induced LPAM-1 when cocultured at equivalent ratios with other dendritic cells and this resulted in lower expression of E-lig and P-lig (Figure 4B).
One potential source of bias of our results may be the use of FLT3L to expand dendritic cells. However, the paucity in normal organs precludes the yield of sufficient numbers of dendritic cells for the experiments presented here and most of the previous imprinting studies have made use of this tool. Previous reports examining the effect of FLT3L on alloresponses have compared FLT3L treatment with no treatment. Here, we always compared dendritic cells that were equally exposed to FLT3L and have therefore presumably undergone the same changes.
The liver has long been known for its unique immunologic properties as a tolerogenic organ.43 This can be attributed in part to its dendritic cells,44,45 and possibly to the CD8−CD11b− dendritic cell subset.46 We here extend the latter report, which described less maturity and allostimulatory capacity of liver-derived dendritic cells in a Th2-driven BALB/c into C57BL/6 model, now to a Th1-driven B10.BR into C57BL/6 model. These results may have been biased due to a lack of sensitivity of hepatic dendritic cells to LPS.47 However, this was shown for a much lower concentration than used here, and in some cases we additionally substituted LPS with recombinant CD40L, obtaining the same results (data not shown). Foxp3+CD4+ T cells were not increased (Figure 3F). However, CD4+CD25+ regulatory T cells generated with liver-derived dendritic cells showed a trend toward more effective suppression in a suppression assay (Figure 3G). A number of different molecules has been implicated in liver homing,48 such as LFA-1,39 but we did not find significantly higher levels of LFA-1 in the liver group. However, our microarray analysis showed induction of CXCR6 (Table 1), which has been implicated in liver homing and GVHD.49
In contrast to previous reports using microarray analysis in GVHD,50,51 our culture system allowed us to strictly limit our analysis to genes directly induced in T cells after stimulation with dendritic cells without potentially confounding parenchymal cells or migrating T cells that have been activated in a different organ. Most remarkable was the fact that very few genes were differentially induced. Fewer than 22 of approximately 14 000 genes differed in each comparison (liver vs spleen, and MLN vs PLN). Although there are significant differences between organ-derived dendritic cells, they have many similarities with respect to their effect on T cells. We found 2 genes previously implicated in liver homing, Cxcr649 and S100a4,52 to be induced by liver-derived dendritic cells. Surprisingly, many granzymes were also up-regulated in the liver compared with spleen. Although described in the setting of antitumor activity, the full expression of an effector phenotype in vitro can lead to a paradoxical impairment of function in vivo.53 A similar process with respect to GVHD may apply here. The identification of 2 retinoic acid–dependent genes induced by MLN DCs further underlines the involvement of retinoic acid25 and identifies potential novel downstream candidate genes in gut homing.
Several groups have shown the importance of homing and the expression of homing molecules including CXCR3,54 CD103,55 CCR2,31 L-selectin, and β7 integrin56 for alloreactive donor T cells using gene-deficient mice, blocking antibodies, or inhibitory molecules separately or in combination.29,30 We have previously shown that specifically targeting gut homing by using either cells deficient for the β7 integrin57 or sorted LPAM-1− cells41 can lead to significant improvement of GVHD mortality and morbidity. In this study, we confirm the importance of trafficking by alloreactive T cells to the gut: donor T cells stimulated with MLN DCs induced more severe intestinal GVHD than T cells stimulated with other dendritic cells and consequently higher GVHD mortality. Our data are also in agreement with previous studies regarding the dominant role of the gut in the pathophysiology of GVHD.5
Analysis of the in vivo homing properties confirmed the preferential trafficking of imprinted T cells to their corresponding organs. T cells imprinted with dendritic cells from the liver were found in significantly higher numbers in the liver. This was most likely due to increased homing since decreased apoptosis was not detected. T cells imprinted with dendritic cells from the secondary lymphoid organs PLN and MLN showed remarkable similarities in all aspects with the exception of homing molecules that translated into a higher degree of T-cell infiltration into the corresponding target organs skin and gut, respectively. However, various numbers of T cells from any group were still observed in every organ analyzed.
We observed only a delay in mortality, indicating that a “catch-up” phenomenon is occurring. We attribute this to the dynamic nature of imprinting as previously described in vitro.23,24 Our data suggest that this reprogramming also affects alloreactive T cells in vivo (Figure 6D), which has previously been described to occur within a few days in vitro.23 The use of genetically deficient mice in previous studies of homing molecules obviously prevented this phenomenon. Further contributing to this shift is the fact that, although uniformly stimulated, the donor T cells were heterogeneous with a subset still having a naive phenotype (CD44loCD62Lhi).
We found no evidence for selective skin GVHD in the PLN group. This was not completely unexpected since expression of E-lig and P-lig was comparable in the spleen, liver, and PLN groups. There was also no elevation of transaminases in the liver group (Figure 5F) despite evidence for liver homing (Figure 6A), which was most likely due to the less activated/tolerogenic state of these T cells. Imprinting of homing molecules other than those associated with the gut may therefore only minimally contribute to the strikingly selective clinical involvement of GVHD target organs.10 This is not surprising considering that the pathophysiology of GVHD includes a “cytokine storm” and a high degree of inflammation. Highly expressed inflammatory cytokines and chemokines could be expected to “override” any effect of tissue DC imprinting and regulate GVHD target organ damage.58 As has been shown in numerous previous studies, T-cell polarization and costimulation clearly also influence the degree of GVHD. As a matter of fact, it is therefore surprising that we demonstrated any effect of tissue DC imprinting on target organ GVHD at all.
In vivo imaging of GVHD in mice showed initial trafficking of donor T cells to secondary lymphoid organs where the up-regulation of homing molecules takes place36 followed by infiltration of target organs.37,59 We chose our experimental approach to at least partially recapitulate this initial priming and imprinting phase circumventing the need for trafficking to secondary lymphoid organs. Despite an earlier report that Peyer patches are crucial for the initiation of a graft-versus-host reaction (GVHR),60 there is evidence that this may not apply to GVHD and that secondary lymphoid organs may compensate for each other.61,62 Use of these stimulated T cells in mice without secondary lymphoid organs may further emphasize the differences in GVHD mortality.
In conclusion, by systemically analyzing the effect of different organ-derived dendritic cells on allogeneic T cells in vitro, we found an impact on at least 2 different levels: the degree of activation, as exemplified by less activation by liver dendritic cells, and the imprinting of a homing phenotype, as exemplified by the induction of LPAM-1 by mesenteric lymph nodes. Donor T cells stimulated with dendritic cells from the gut-associated lymphatic tissue MLNs induce significantly higher GVHD mortality and morbidity than donor T cells stimulated with dendritic cells from all other GVHD target organs. However, the absence of significant target organ GVHD induced by these other dendritic cell types indicates that previous imprinting in vitro may be overridden in vivo. Successful targeting of homing to treat or prevent GVHD may therefore need to be continuous and interfere with several pathways.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants HL69929, CA33049, CA023766, CA107096, and P20-CA103694 from the National Institutes of Health; by awards from the Leukemia and Lymphoma Society, the Ryan Gibson Foundation, the Emerald Foundation, the Byrne Fund, and The Experimental Therapeutics Center of the Memorial Sloan-Kettering Cancer Center, funded by Mr William H. Goodwin and Mrs Alice Goodwin and the Commonwealth Foundation for Cancer Research (M.R.M.B.); a Mildred Scheel-Stipendium of the Deutsche Krebshilfe (T.D.K.); Deutscher Akademischer Austausch Dienst (T.H.T.); Boehringer Ingelheim Fonds (T.H.T.); Dr Werner Jackstaedt Stiftung (J.L.Z.); and Deutsche Forschungsgemeinschaft (J.L.Z.). J.L.Z. is the recipient of a fellowship grant from the Lymphoma Research Foundation. O.A. is the recipient of an Amy Strelzer Manasevit Scholar award from the National Marrow Donor Program (NMDP) and The Marrow Foundation.
The authors thank Ulrich H. von Andrian (The CBR Institute for Biomedical Research) for generously providing the B16-FLT3L cell line; Agnes Viale (MSKCC genomics core facility) for support with the microarray analysis; Amgen for generously providing recombinant human FLT3L; and the staff of the Laboratory of Comparative Pathology (Cornell University) and the staff of the Research Animal Resource Center (MSKCC) for excellent assistance and excellent animal care.
National Institutes of Health
Authorship
Contribution: T.D.K. designed and performed research, analyzed data, and wrote the paper; T.H.T., J.L.Z., D.S., A.A.K., M.E.C., C.G.K., C.B., J.G., O.M.S., C.L., and G.F.M. performed research; G.H. performed statistical analysis; O.A. designed research and analyzed data; M.R.M.B. designed research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Marcel R. M. van den Brink, Laboratory of Allogeneic Bone Marrow Transplantation, Memorial Sloan-Kettering Cancer Center, Z1404, Box 111, 1275 York Ave, New York, NY 10021; e-mail: vandenbm@mskcc.org.