Abstract
Regulatory T cells (Tregs) have been shown to play a crucial role in maintaining self-tolerance and suppressing autoimmunity. The forkhead transcription factor, FoxP3, is a key molecule necessary and sufficient for Tregs development and function. However, the molecular mechanisms by which FoxP3 regulates the phenotypic (anergic) and the functional (suppressive) characteristics of Tregs are not well defined. Here we found that the promoter DNA-binding activity of AP-1 transcription factors is selectively inhibited in the naturally occurring CD4+ CD25+ Tregs from mice. The impaired AP-1 DNA binding is not the result of the decreased nuclear translocation of AP-1 family transcription factors, including c-Jun, JunB, and c-Fos. FoxP3 significantly suppresses both the transcriptional activity and promoter DNA-binding of AP-1 by interacting with c-Jun. The N-terminus of FoxP3, but not its C-terminus forkhead domain, specifically interacts with phosphorylated c-Jun and alters c-Jun subnuclear distribution. This N-terminus of FoxP3 with nuclear localization signals (FoxP3N/NLS) is able to suppress AP-1 transcriptional activity. Ectopic expression of FoxP3N/NLS sufficiently induces the unresponsiveness of mouse primary CD4+ CD25− T cells, whereas the full-length FoxP3 is required for the suppressive functions of Tregs. These findings uncover one of the mechanisms underlying how FoxP3 maintains the unresponsiveness of Tregs.
Introduction
Mutation of FoxP3 gene in mice, scurfy mice, causes wasting, exfoliative dermatitis, lymphadenopathy, hepatosplenomegaly, multiorgan lymphocytic infiltrates, and the presence of autoantibodies in males.1-3 FoxP3 was subsequently found to be the gene mutated in the human immune dysregulation, polyendocrinopathy, enteropathy, X-linked syndrome, which is phenotypically similar to Scurfy with affected males displaying systemic autoimmune disease affecting bowel, skin, endocrine organs, and blood.4-7 Recent studies have demonstrated that FoxP3 is a lineage-specific factor of the CD4+ CD25+ regulatory T cells (Tregs), which participate in suppressing autoreactive T cells, thus facilitating self-tolerance.8,9 These CD4+ CD25+ cells appear to play a major role in the suppression of both autoimmune and allergic disease.10 Foxp3 regulates Tregs cell development and function in a dose-dependent, nonbinary manner, and that decreased Foxp3 expression can cause immune disease.11 In addition, Tregs have been implicated in transplantation tolerance in rodent model systems in mice, and ex vivo studies indicate that similar cells exist in humans.4 The significant features of Tregs are: (1) Tregs are anergic; in other words, Tregs do not produce interleukin 2 (IL-2) and do not proliferate in response to activation signals; and (2) Tregs suppress the immune response of the effector T cells by producing inhibitory factors, such as IL-10, TGF-β, and CTLA-4
FoxP3 is a member of the forkhead/winged-helix family of transcriptional regulators.6 In addition to the forkhead (FHK) domain, FoxP3 also contains a single C2H2 Zinc finger and a coiled-coil motif. In vitro assays have indicated that FoxP3 can act as a transcriptional repressor using an IL-2 promoter-based reporter assay.12 Accumulated evidences indicate that FoxP3 controls Tregs functions by interacting with multiple transcription factors.13-15 A recent study demonstrated that FoxP3 may inhibit IL-2 transcription by directly binding to and suppressing the activities of the transcription factors, NF-AT and NF-κB,13,15 which provided the first evidence that FoxP3 may function as a transcription repressor during T-cell activation. FoxP3 can also form a cooperative complex with NF-AT to up-regulate expression of the Treg markers CTLA4 and CD25, and confers suppressor function in a murine model of autoimmune diabetes.15 Ono et al found that a direct cross-talk between FoxP3 and the Runt-related transcription factor 1 (Runx1) plays a crucial role in suppressing the production of IL-2 by Tregs.14 In addition, Greene's group recently found that the N-terminus of FoxP3 recruits both the histone acetyltransferase and the histone deacetylase for Tregs functions.16
c-Jun, a member of the AP-1 transcription factors, participates in the control of cellular responses to stimuli that regulate cell proliferation, differentiation, cell death, and cell transformation.17 In T lymphocytes, activation of c-Jun requires TCR and CD28 stimulation, and its activation is critical for T-cell development, differentiation, and activation.18-21 c-Jun forms either homodimers or heterodimers with other members of Fos-Jun family of transcription factors. It has been shown that the transcriptional activation of c-Jun is inhibited in anergic T cells.22-27 Stimulation of anergic T cells failed to activate the binding of IL-2 promoter by Jun-Jun homodimer and the formation of Jun-Jun/Oct complex.25,27 However, whether and how c-Jun transcription activity is suppressed in Tregs remain unknown.
Here we found that the promoter DNA-binding activity of AP-1 is selectively impaired in the naturally occurring Tregs when stimulated with anti-CD3 plus anti-CD28. FoxP3 inhibits c-Jun–based AP-1 transcriptional activity by interacting with c-Jun in Tregs. More interestingly, FoxP3 specifically recognizes the phosphorylated (which is the activated) form of c-Jun. This interaction inhibits the promoter DNA-binding activity of c-Jun and alters c-Jun subnuclear localization. The N-terminus of FoxP3 mediates its interaction with c-Jun, and expression of this N-terminus is sufficient to induce T-cell anergy. These findings uncover a new mechanism of FoxP3 in regulating Tregs functions.
Methods
The use of mice in this study has been approved by the Institutional Animal Care and Use Committee of the University of Missouri.
Cell line
HEK 293 T cells were maintained in DMEM (Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 200 μg/mL streptomycin, and 0.25 μg/mL amphotericin B.
Antibodies and reagents
Polyclonal antibody against the epitope-tags of Xpress was purchased from Invitrogen; polyclonal antibodies against c-Jun, JunB, c-Fos, and Myc were obtained from Santa Cruz Biotechnology (Santa Cruz, CA). Antiphosphorylated c-Jun, anti-FoxP3, anti-CD3, and anti-CD28 were from eBioscience (San Diego, CA). The antiactin antibody was from Sigma-Aldrich (St Louis, MO). Anti-c-Jun mouse antibody was obtained from BD Biosciences Transduction Laboratories (Lexington, KY); JNK-specific inhibitor (SP600125) and the recombinant human TNF-α were purchased from Calbiochem (San Diego, CA).
Plasmids
Full-length mouse FOXP3 expression plasmid was kind gift from Oukka (Harvard University).13 To generate truncated mutants, fragments of the FOXP3 cDNA were amplified by polymerase chain reaction (PCR) using linker primers that introduced EcoRI site at the 5′end and XbaI site at the 3′ end. PCR products were digested with EcoRI and XbaI and ligated into the pEFmyc/hisB expression vector (Invitrogen). The fusion protein of the N-terminus of YFP (YN) with FoxP3 was generated by ligation of the YN fragment that was amplified by PCR using specific primers with KpnI sites at 5′ end and EcoRI sites at 3′ end. Xpress-tagged c-Jun and c-Jun fusion with the C-terminus CFP (CC-c-Jun) expression plasmids have been described previously.28 To generate FoxP3 FN/NLS mutant, nuclear localization signal (NLS) of the SV40 large T-antigen (PKKKRKV) was introduced at C-terminal end of FoxP3 1-coil DNA. Recombinant retroviral vectors were constructed by inserting Full-length FoxP3 or FoxP3 FN/NLS into pMig plasmid using BglII and EcoRI sites. All the newly generated plasmids in this study were verified by DNA sequencing.
Isolating mouse primary T cells and CD4+ CD25+ Tregs
T cells were isolated from the lymph nodes and spleens of 4- to 6-week-old C57BL6 mice. CD4+ T cells were purified by negative selection with the CD4+ T-cell isolation kit (Miltenyi Biotec, Auburn, CA). CD4+ T cells then underwent positive selection for CD25 expression (Miltenyi Biotec). These cells were maintained in RPMI media (Invitrogen) supplemented with 10% fetal bovine serum, 100 U/mL penicillin, 200 μg/mL streptomycin, and 0.25 μg/mL amphotericin, CD4+ CD25+ or CD4+ CD25− T cells were left unstimulated or stimulated with anti-CD3, or anti-CD3 plus anti-CD28 for various time periods for the study.
Dual luciferase assay
HEK 293 cells in 12-well plates were transfected with pRL-TK (Promega, Madison, WI) and pAP-1 luciferase, along with various expression plasmids, using the Lipofectamine transfection reagent (Invitrogen). The pRL-TK plasmid contains the Renilla reniformis (sea pansy) luciferase gene under the transcriptional control of the herpesvirus thymidine kinase promoter and constitutively expresses low levels of renillar luciferase. Transfected cells were lysed, and the luciferase activities in the cell lysates were analyzed using a Dual Luciferase Reporter assay kit (Promega). Luciferase activity was measured as relative light units using a luminometer (Turner Designs, Sunnyvale, CA).
Transfection, immunoprecipitation, and Western blotting
HEK 293 cells grown in 6-cm2 dishes were transfected with 1 to 2 μg of plasmid DNA. Transfected cells were collected and lysed with Nonidet P-40 (NP-40) lysis buffer (1% NP-40, 20 mM Tris-HCl, pH 7.5, 150 mM NaCl, 5 mM ethylenediaminetetraacetic acid, 5 mM NaPiP, 2 mM Na3VO4, and 10 μg/mL each of aprotinin and leupeptin). Insoluble materials were removed by centrifugation at 13 000g. For immunoprecipitation, lysates were mixed with antibodies (1 μg) for 1 hour, followed by the addition of 30 μL of protein G-Sepharose beads (GE Healthcare, Little Chalfont, United Kingdom) for an additional 2 hours at 4°C. Immunoprecipitates were washed 4 times with NP-40 lysis buffer and boiled in 15 μL of 4 × Laemmli's buffer. Samples were subjected to 8% or 10% sodium dodecyl sulfate (SDS)–polyacrylamide gel electrophoresis analysis and electrotransferred onto nitrocellulose membrane (Hybond ECL, GE Healthcare). After blocking with 4% nonfat dry milk in TBST for 2 hours, membranes were probed with the indicated primary antibodies (usually 1 μg/mL), followed by horseradish peroxidase-conjugated secondary antibodies. Membranes were then washed and visualized with an enhanced chemiluminescence detection system (GE Healthcare). When necessary, membranes were stripped by incubation in stripping buffer (Pierce Chemical, Rockford, IL) and then reprobed with other antibodies as indicated.
Retrovirus production and transduction
Recombinant retrovirus was produced by transient transfection of the ecotropic packaging cell line, PlatE, using Lipofectamine 2000 transfection reagent (Invitrogen). Viral supernatants were harvested 48 and 72 hours after transfection. Primary CD4+ CD25− T cells were cultured with anti-CD3 (5 μg/mL) plus anti-CD28 (1 μg/mL) for 24 hours and 106/well in 6-well plates and centrifuged with 2 mL of the viral supernatants at 1200g at 33°C for 60 minutes. After incubation at 33°C for 6 hours, cells were cultured with complete RPMI 1640 for the indicated periods before experimentation.
Electrophoretic mobility shift assay
Nuclear protein extracts from HEK 293 cells or primary T cells were prepared with the nuclear extraction kit (Active Motif, Carlsbad, CA) according to the manufacturer's protocol. Protein concentration was determined by the Bradford protein assay (Bio-Rad, Hercules, CA). Oligonucleotides for AP-1, NF-κB, and NF-AT (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) were labeled with biotin-11 UTP using a biotin 3′ end DNA labeling kit (Pierce Chemical). Binding reactions were performed in a 25 μL volume. Each reaction contained 2 μg nuclear extract, 2 μg polydeoxyinosinic-deoxycytidylic acid (Sigma-Aldrich), 50 mM NaCl, 10 mM Tris-HCl, 4% (v/v) glycerol, 0.5 mM DTT, 0.5 mM ethylenediaminetetraacetic acid, 5 mM MgCl2, and 20 fmol of biotin-labeled oligonucleotides. Reactions were incubated for 20 minutes at room temperature and then electrophoresed through a 5% or 7% polyacrylamide gel with 0.5× TBE running buffer. Gels were transferred onto Hybond N+ membrane, followed by cross-linking at 120 mJ/cm2 using a commercial UV-light cross-linker Spectrolinker XL-1500UV crosslinker (Spectronics, Lincoln, NE). Biotin-labeled AP-1 was detected by chemiluminescence using Phototope-Star detection kit (New England Biolabs, Ipswich, MA). The specificity of the binding reaction was confirmed using mutants of AP-1, NF-AT, and NF-κB oligos as shown in Table S1. No DNA-binding activity was detected when mutant oligos were used (data not shown).
Immunofluorescence assay and fluorescence complementation assay
For immunofluorescence assay, cells transfected with plasmids encoding c-Jun-CFP and Myc-tagged FoxP3 were fixed with 4% paraformaldehyde. Fixed cells were permeabilized with 0.1% Triton X-100 in phosphate-buffered saline and incubated with anti-Myc antibody followed by secondary antibody labeled with Texas-Red. Nuclei were identified with 4,6-diamidino-2-phenylindole (DAPI) staining. Fluorescence was observed under fluorescence microscope.
For florescence complementation assay, cells transfected with plasmids encoding YN-FoxP3 and CC-c-Jun were incubated at 37°C for 24 hours and then transferred to 30°C for 12 to 16 hours to promote fluorophore maturation. Nuclei were identified with DAPI staining. The fluorescence emissions of the cells were observed under fluorescence microscope.
T-cell proliferation assay
Mouse primary T cells (1 × 105 cells/well) were stimulated with anti-CD3 plus anti-CD28 for 24 hours and then pulsed with 1 μCi/mL of [3H]-thymidine and incubated for additional 12 hours. [3H]Thymidine incorporation was measured with a scintillation counter.
Chromatin immunoprecipitation assay
Chromatin immunoprecipitation assay was performed according to the method recommended by Upstate Biotechnology (Charlottesville, VA); 1 × 106 cells were cross-linked with 1% formaldehyde and lysed with SDS lysis buffer. Cell lysates were sonicated, and 10% of cell lysate was removed and used to determine the total amount of target DNA in input. Remaining cell lysates were diluted in chromatin immunoprecipitation dilution buffer. To reduce nonspecific background, Salmon sperm DNA/protein agarose-50% slurry was added to the diluted cell supernatant. Immunoprecipitation was performed with 4 μg of polyclonal anti-c-Jun antibodies (Santa Cruz Biotechnology) at 4°C overnight. Immune complexes were then mixed with Salmon sperm DNA/protein agarose-50% slurry 4°C for 1 hour. After immunoprecipitates were washed sequentially with low salt buffer, high salt buffer, LiCl wash buffer, and TE buffer, DNA-protein complexes were eluted with elution buffer and cross-linking was reversed. Genomic DNA was extracted using phenol/chloroform and ethanol-precipitated DNA was resuspended in TE. PCR was performed with specific primers for mouse IL-2 promoter. The forward primer is 5′-CATACAGAAGGCGTTCATTG-3′ and reverse primer is 5′-AGCTCTTCAGCATGGGAGGC-3′.
Results
AP-1 promoter DNA-binding is selectively impaired in naturally occurring Tregs in mice
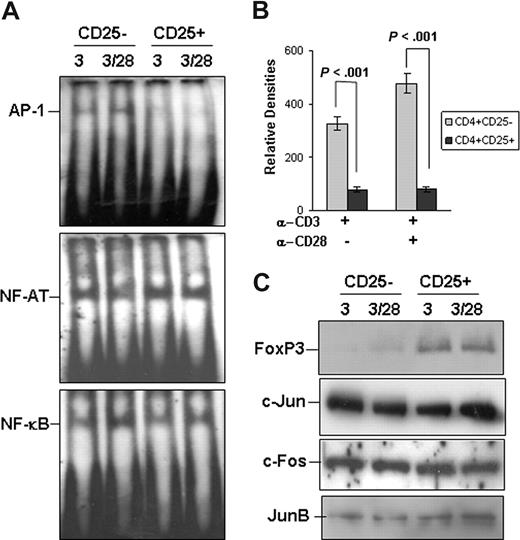
We compared the AP-1 promoter DNA-binding activity of the nuclear extracts from naturally developed Tregs (CD4+ CD25+) with that from CD4+ CD25− T cells after stimulation with anti-CD3 or anti-CD3 plus anti-CD28. The DNA-binding activity of AP-1 was significantly reduced in CD4+ CD25+ regulatory T cells (Figure 1A). Results from quantifications of band densities of 5 independent electrophoretic mobility shift assay experiments revealed there was an approximately 90% reduction of AP-1 promoter-binding activity in Tregs compared with that of naive CD4+ CD25− T cells on stimulation (Figure 1B). The NF-κB and NF-AT DNA-binding activities were comparable between these 2 types of T cells after stimulation (Figure 1A). As controls, the DNA binding activities of AP-1, NF-κB, and NF-AT were undetectable both in CD4+ CD25+ and in CD4+ CD25− T cells without stimulation (data not shown). The reduced AP-1 DNA-binding activity in Tregs was not the result of the decreased nuclear translocation of AP-1 family transcription factors, including c-Jun, c-Fos, and JunB because Western blotting analysis revealed that the protein levels of these proteins in the nuclear extracts were comparable between those 2 types of CD4+ T cells (Figure 1C). These results indicate that the AP-1 transcriptional activities are selectively inhibited in naturally developed Tregs in mice.
Impaired AP-1 promoter DNA-binding in naturally developed Tregs. (A) Purified CD4+ CD25+ regulatory T cells, with CD4+ CD25− naive T cells as controls, were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 36 hours. Nuclear extracts were isolated from these stimulated cells and used for electrophoretic mobility shift assay assay using biotin-labeled DNA fragments corresponding to the promoter region of AP-1 (top panel), NF-AT (middle panel), or NF-κB (bottom panel). (B) The bands' density of AP-1 DNA-binding was measured using National Institutes of Health 1.63 software. Error bars represent data from 5 independent experiments. Student t test was used for the statistical analysis. (C) The protein expression of FoxP3, c-Jun, c-Fos, and JunB from the nuclear extracts used in panel A was analyzed in parallel by Western blotting.
Impaired AP-1 promoter DNA-binding in naturally developed Tregs. (A) Purified CD4+ CD25+ regulatory T cells, with CD4+ CD25− naive T cells as controls, were stimulated with anti-CD3 or anti-CD3 plus anti-CD28 for 36 hours. Nuclear extracts were isolated from these stimulated cells and used for electrophoretic mobility shift assay assay using biotin-labeled DNA fragments corresponding to the promoter region of AP-1 (top panel), NF-AT (middle panel), or NF-κB (bottom panel). (B) The bands' density of AP-1 DNA-binding was measured using National Institutes of Health 1.63 software. Error bars represent data from 5 independent experiments. Student t test was used for the statistical analysis. (C) The protein expression of FoxP3, c-Jun, c-Fos, and JunB from the nuclear extracts used in panel A was analyzed in parallel by Western blotting.
FoxP3 inhibits AP-1 transcription activity
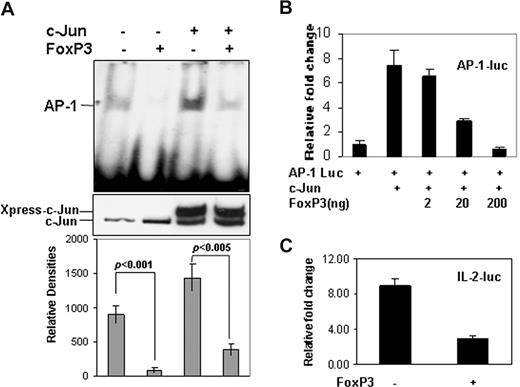
FoxP3 has been identified as a lineage marker of Tregs.8 Previous studies have demonstrated that FoxP3 inhibits NF-κB, NF-AT, and Runx1 transcriptional activity.13 Here, we investigated whether FoxP3 is responsible for the reduced AP-1 DNA-binding activity. Forced expression of FoxP3 in HEK 293 cells significantly suppressed AP-1 DNA binding. The AP-1 binding activity was inhibited by FoxP3 even when c-Jun was coexpressed, whereas the expression of c-Jun, both its endogenous forms and the overexpression ones, was not affected by FoxP3 expression (Figure 2A). These results indicate that FoxP3 inhibits AP-1 DNA-binding activity but not c-Jun protein expression. Cotransfection of FoxP3 plasmid inhibited c-Jun–mediated AP-1 transcriptional activity in a dose-dependent manner, and more than 90% of AP-1 dependent luciferase activity was inhibited when 200 ng of FoxP3 plasmid was cotransfected (Figure 2B). The IL-2 promoter region contains multiple AP-1 binding sites; we therefore tested the effects of FoxP3 on IL-2 promoter activation. As shown in Figure 2C, cotransfection of FoxP3 expression plasmid DNA caused approximately 80% reduction of IL-2 promoter dependent luciferase activity (Figure 2C). These results indicate that FoxP3 is a suppressor of c-Jun–based AP-1 transcription activity.
FoxP3 inhibits the DNA-binding and transcriptional activity of AP-1. (A) HEK 293 cells were transfected without or with FoxP3 and c-Jun expression plasmids as indicated. Nuclear extracts were isolated. Equal amounts of nuclear proteins were incubated with biotin-labeled AP-1 promoter fragments, and gel shift assay was performed (top panel). The protein expression levels of c-Jun in the nuclear extracts were detected by Western blotting using anti-Jun antibody (middle panel). The densities of these AP-1 DNA-binding bands were measured using NIH 1.63 software (bottom panel). Error bars represent data from 3 independent experiments. Data were statistically analyzed with Student t test. (B) AP-1 luciferase, c-Jun, and FoxP3 expression plasmids were cotransfected into HEK 293 cells. The luciferase activities were determined. Error bars represent data from 3 independent experiments. (C) IL-2 luciferase reporter plasmid was cotransfected without or with FoxP3 plasmid into HEK 293 cells. The luciferase activities were determined. Error bars represent data from 3 independent experiments.
FoxP3 inhibits the DNA-binding and transcriptional activity of AP-1. (A) HEK 293 cells were transfected without or with FoxP3 and c-Jun expression plasmids as indicated. Nuclear extracts were isolated. Equal amounts of nuclear proteins were incubated with biotin-labeled AP-1 promoter fragments, and gel shift assay was performed (top panel). The protein expression levels of c-Jun in the nuclear extracts were detected by Western blotting using anti-Jun antibody (middle panel). The densities of these AP-1 DNA-binding bands were measured using NIH 1.63 software (bottom panel). Error bars represent data from 3 independent experiments. Data were statistically analyzed with Student t test. (B) AP-1 luciferase, c-Jun, and FoxP3 expression plasmids were cotransfected into HEK 293 cells. The luciferase activities were determined. Error bars represent data from 3 independent experiments. (C) IL-2 luciferase reporter plasmid was cotransfected without or with FoxP3 plasmid into HEK 293 cells. The luciferase activities were determined. Error bars represent data from 3 independent experiments.
FoxP3 interacts with c-Jun in vitro and in Tregs
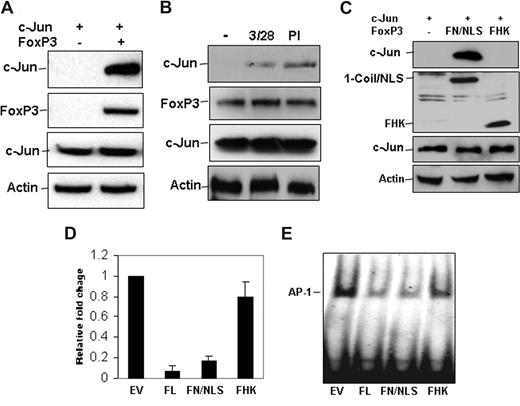
Next, we investigated whether FoxP3 inhibits AP-1 transcriptional activity by interacting with c-Jun. Coimmunoprecipitation and Western blotting experiments demonstrated that FoxP3 interacted with c-Jun in HEK 293 cells that have been transiently transfected with FoxP3 and c-Jun expression plasmids (Figure 3A), suggesting that FoxP3 suppresses AP-1 activity by forming a complex with c-Jun. To support this, we further demonstrated that the endogenous FoxP3 was physically associated with c-Jun in Tregs. Interestingly, their interaction can be detected in Tregs only when cells were stimulated with anti-CD3 plus anti-CD28 or PMA plus ionomycin (Figure 3B). These results indicate that FoxP3 interacts with c-Jun, and this interaction is regulated in Tregs. In addition, FoxP3 also interacts with c-Fos, but not JunB and ATF-2 (Figure S1), suggesting that FoxP3 selectively interacts with AP-1 family transcription factors, particularly with c-Jun/c-Fos. In this study, we focus our effort on the analysis of c-Jun/FoxP3 cross-talk because FoxP3 significantly suppresses c-Jun–based AP-1 transcriptional activity.
FoxP3 interacts with c-Jun. (A) Xpress-tagged c-Jun Myc-FoxP3 expression plasmids were cotransfected into HEK 293 cells. Transfected cells were collected and lysed 48 hours after transfection. FoxP3 in the cell lysates was immunoprecipitated with anti-Myc antibody. The bound c-Jun was detected by Western blotting using anti-Xpress antibody (top panel). The same membrane was reprobed with anti-FoxP3 antibody (second panel from top). As controls, the levels of c-Jun and Actin in the whole cell lysates were determined with Western blotting using anti-Xpress and Actin, respectively (bottom 2 panels). (B) CD4+ CD25+ T cells were isolated and stimulated with anti-CD3 plus anti-CD28 or PMA plus ionomycin for 30 minutes. Stimulated cells were lysed, and FoxP3 in the cell lysates was immunoprecipitated with anti-FoxP3 antibody; the bound c-Jun in the immunoprecipitates was detected with anti-c-Jun antibody (top panel). The same membrane was reprobed with anti-FoxP3 antibody (second panel from top). As controls, the protein levels of c-Jun and Actin in the whole cell lysates were determined with Western blotting using anti-c-Jun and anti-Actin antibodies, respectively (bottom 2 panels). (C) Xpress-tagged c-Jun and Myc-tagged FoxP3 mutants, FN/NLS or FHK, expression plasmids, were cotransfected into HEK 293 cells. FoxP3 mutant proteins in these lysates from transfected cells were immunoprecipitated with anti-Myc antibody, and the bound c-Jun was detected with anti-Xpress antibody (top panel). The same membrane was reprobed with anti-Myc antibody for detecting the expression of FoxP3 mutants (second panel from top). As controls, the protein levels of c-Jun and Actin were determined by Western blotting with anti–c-Jun and anti-Actin, respectively (bottom 2 panels). (D) AP-1-luciferase and c-Jun expression plasmids were cotransfected without or with FoxP3 or with each of FoxP3 mutants. The luciferase activities in the cell lysates were detected. Error bars represent data from 3 independent experiments. (E) HEK293 cells were transfected with empty plasmid or with plasmid encoding full-length FoxP3 (FoxP3 FL) or each of FoxP3 truncated mutants (FN/NLS and FHK) as indicated. Nuclear extracts were isolated 48 hours after transfection. Equal amounts of nuclear proteins were incubated with biotin-labeled AP-1 promoter fragments and gel shift assay was performed.
FoxP3 interacts with c-Jun. (A) Xpress-tagged c-Jun Myc-FoxP3 expression plasmids were cotransfected into HEK 293 cells. Transfected cells were collected and lysed 48 hours after transfection. FoxP3 in the cell lysates was immunoprecipitated with anti-Myc antibody. The bound c-Jun was detected by Western blotting using anti-Xpress antibody (top panel). The same membrane was reprobed with anti-FoxP3 antibody (second panel from top). As controls, the levels of c-Jun and Actin in the whole cell lysates were determined with Western blotting using anti-Xpress and Actin, respectively (bottom 2 panels). (B) CD4+ CD25+ T cells were isolated and stimulated with anti-CD3 plus anti-CD28 or PMA plus ionomycin for 30 minutes. Stimulated cells were lysed, and FoxP3 in the cell lysates was immunoprecipitated with anti-FoxP3 antibody; the bound c-Jun in the immunoprecipitates was detected with anti-c-Jun antibody (top panel). The same membrane was reprobed with anti-FoxP3 antibody (second panel from top). As controls, the protein levels of c-Jun and Actin in the whole cell lysates were determined with Western blotting using anti-c-Jun and anti-Actin antibodies, respectively (bottom 2 panels). (C) Xpress-tagged c-Jun and Myc-tagged FoxP3 mutants, FN/NLS or FHK, expression plasmids, were cotransfected into HEK 293 cells. FoxP3 mutant proteins in these lysates from transfected cells were immunoprecipitated with anti-Myc antibody, and the bound c-Jun was detected with anti-Xpress antibody (top panel). The same membrane was reprobed with anti-Myc antibody for detecting the expression of FoxP3 mutants (second panel from top). As controls, the protein levels of c-Jun and Actin were determined by Western blotting with anti–c-Jun and anti-Actin, respectively (bottom 2 panels). (D) AP-1-luciferase and c-Jun expression plasmids were cotransfected without or with FoxP3 or with each of FoxP3 mutants. The luciferase activities in the cell lysates were detected. Error bars represent data from 3 independent experiments. (E) HEK293 cells were transfected with empty plasmid or with plasmid encoding full-length FoxP3 (FoxP3 FL) or each of FoxP3 truncated mutants (FN/NLS and FHK) as indicated. Nuclear extracts were isolated 48 hours after transfection. Equal amounts of nuclear proteins were incubated with biotin-labeled AP-1 promoter fragments and gel shift assay was performed.
The DNA binding domain of FoxP3, FHK, which has been found to interact with NF-AT,15 was not involved in the interaction with c-Jun (Figure 3C). A previous study has found that the N-terminus of FoxP3 distributes in the cytoplasm. To test the interaction of FoxP3 N-terminus with the nuclear protein, c-Jun, we fused the N-terminus of FoxP3 with a SV40 nuclear localization sequence (FN/NLS). This SV40 nuclear localization sequence relocated FoxP3 N-terminus into the nucleus (Figure S2). Co-IP and Western blotting revealed that FN/NLS interacted with c-Jun (Figure 3C). Expression of FN/NLS sufficiently inhibited both AP-1 luciferase activity (Figure 3E) and DNA binding (Figure 3F). Expression of FHK domain only, which is predominantly expressed in the nucleus (Figure S2), had little effects on AP-1 transcriptional activity (Figure 3E,F). These findings suggest that the N-terminal region of FoxP3 that mediates its interaction with c-Jun is sufficient to inhibit c-Jun–based AP-1 transcriptional activity.
JNK-mediated phosphorylation of c-Jun is required for c-Jun–FoxP3 interaction
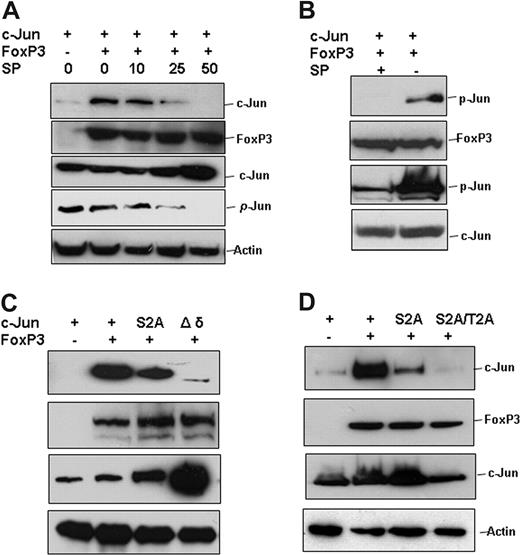
Our findings that the interaction of FoxP3 with c-Jun in Tregs requires both TCR and CD28 signaling suggest that their interaction is regulated. JNK-mediated phosphorylation of c-Jun at its transcriptional activation domain is triggered by TCR and CD28 stimulation and is required for c-Jun transcription activity.19,29 We therefore determined whether this phosphorylation event affects c-Jun interaction with FoxP3. In the presence of JNK specific inhibitor, the interaction of c-Jun with FoxP3, as well as c-Jun phosphorylation, was inhibited in a dose-dependent manner and their interaction was completely blocked when cells were treated with 50 nM JNK inhibitor, suggesting that JNK-mediated phosphorylation is required for c-Jun and FoxP3 interaction (Figure 4A). We further demonstrated that the c-Jun bound to FoxP3 is phosphorylated (Figure 4B). Mutations of the serine 63 and 73, which have been identified as predominant modification sites by JNK, only partially suppressed c-Jun interaction, suggesting that other phosphorylation sites are also involved in mediating c-Jun/FoxP3 interaction. Indeed, deletion of the δ domain, the JNK docking site of c-Jun, almost completely blocked the interaction of c-Jun with FoxP3 (Figure 4C). Further mutation of the 91 and 93 threonine site on the S63,73A mutant, abolished c-Jun interaction with FoxP3 (Figure 4D). Based on these results, we collectively conclude that phosphorylation of c-Jun by JNK is required for its interaction with FoxP3, suggesting that FoxP3 inhibits AP-1 activation by selectively binding to the transcriptionally active form of c-Jun.
c-Jun phosphorylation is required for its interaction with FoxP3. (A) Xpress-tagged c-Jun expression plasmids were cotransfected without or with Myc-tagged FoxP3 into HEK 293 cells. Cells were treated with JNK inhibitor SP600125 at the indicated doses for 6 hours and then collected. FoxP3 protein was immunoprecipited with anti-Myc antibody, and the interaction of c-Jun was detected with anti-Xpress antibody (top panel). The same membrane was reprobed with anti-FoxP3 antibody (second panel from top). The protein levels of c-Jun, phosphorylated c-Jun, and actin in the whole cell lysates were detected by Western blotting (bottom 3 panels). (B) c-Jun and FoxP3 expression plasmids were cotransfected into HEK 293 cells and cultured for 36 hours. Transfected cells were then treated with or without 50 nM of JNK inhibitor SP600125 for 6 hours. The protein of FoxP3 in the lysates of transfected cells was immunoprecipitated with anti-Myc antibody. The immunoprecipitates were analyzed by SDS-PAGE and then detected with anti-phospho–c-Jun antibody (top panel). (C,D) Xpress-tagged c-Jun or its mutants, including mutation of the 63, 73 serine to alanine (S2A), deletion of the δdomain (Δδ), and mutation of the 63, 73 serine and 91, 94 threonine to alanine (S2A/T2A), were cotransfected without or with Myc-tagged FoxP3 into 293 cells. FoxP3 protein in the lysates from transfected cells was immunoprecipitated with anti-Myc antibody; the bound c-Jun or its mutants were detected with anti-Xpress antibody (top panels). The same membranes were reprobed with anti-FoxP3 antibody (second panel from top). The expression of c-Jun or its mutants and actin in the whole cell lysates was determined with Western blotting as controls (bottom 2 panels).
c-Jun phosphorylation is required for its interaction with FoxP3. (A) Xpress-tagged c-Jun expression plasmids were cotransfected without or with Myc-tagged FoxP3 into HEK 293 cells. Cells were treated with JNK inhibitor SP600125 at the indicated doses for 6 hours and then collected. FoxP3 protein was immunoprecipited with anti-Myc antibody, and the interaction of c-Jun was detected with anti-Xpress antibody (top panel). The same membrane was reprobed with anti-FoxP3 antibody (second panel from top). The protein levels of c-Jun, phosphorylated c-Jun, and actin in the whole cell lysates were detected by Western blotting (bottom 3 panels). (B) c-Jun and FoxP3 expression plasmids were cotransfected into HEK 293 cells and cultured for 36 hours. Transfected cells were then treated with or without 50 nM of JNK inhibitor SP600125 for 6 hours. The protein of FoxP3 in the lysates of transfected cells was immunoprecipitated with anti-Myc antibody. The immunoprecipitates were analyzed by SDS-PAGE and then detected with anti-phospho–c-Jun antibody (top panel). (C,D) Xpress-tagged c-Jun or its mutants, including mutation of the 63, 73 serine to alanine (S2A), deletion of the δdomain (Δδ), and mutation of the 63, 73 serine and 91, 94 threonine to alanine (S2A/T2A), were cotransfected without or with Myc-tagged FoxP3 into 293 cells. FoxP3 protein in the lysates from transfected cells was immunoprecipitated with anti-Myc antibody; the bound c-Jun or its mutants were detected with anti-Xpress antibody (top panels). The same membranes were reprobed with anti-FoxP3 antibody (second panel from top). The expression of c-Jun or its mutants and actin in the whole cell lysates was determined with Western blotting as controls (bottom 2 panels).
FoxP3 colocalizes with c-Jun and alters the nuclear localization of c-Jun
To further investigate the mechanisms underlying the inhibition of AP-1 transcriptional activity by FoxP3, we determined whether FoxP3 alters the nuclear localization of c-Jun because FoxP3 blocked its DNA binding. In HEK 293 cells transfected with c-Jun-CFP, the fluorescence predominantly concentrated in the nucleoli and shown as multiple dots (Figure 5A). Interestingly, when FoxP3 was cotransfected, the subnuclear localization of c-Jun-CFP was altered and shown as a defused distribution in the nucleus (Figure 5B), and FoxP3 sequestered c-Jun in a dose-dependent manner (Figure S3). We then confirmed these findings using the fluorescence complementation assay as we described previously because this assay allows a direct visualization of protein-protein interactions.28,30 We visualized green fluorescence, which indicates the c-Jun/FoxP3 interaction, in the nuclei of 293T cells when YN-FoxP3 and Jun-CC were transfected, but not in the control cells transfected with YN-FoxP3 or Jun-CC individually. The interaction of FoxP3 with c-Jun showed defused nucleus distribution in all the fluorescent cells (Figure S4). Immunostaining of CD4+ CD25+ Tregs revealed that both FoxP3 and c-Jun showed a heterogeneous nuclear distribution, which contains both diffused and sparkled distribution in Tregs. A partial colocalization of FoxP3 with c-Jun was detected essentially in the nuclei of all FoxP3+ cells (Figure 5C). Interestingly, c-Jun distributed in a different pattern in FoxP3− CD4+ T cells, with bigger sparkles and less diffused signals in the nuclei (Figure 5D). Thus, FoxP3 colocalizes with and redistributes c-Jun into different subnuclear com-partments in Tregs.
FoxP3 alters c-Jun subnuclear localization. (A,B) c-Jun-CFP expression plasmids were transfected without (A) or with (B) FoxP3 expression plasmids into HEK 293 cells. Transfected cells were immunostained with anti-FoxP3 antibody followed by a second antibody that has been labeled by Texas-Red. The nuclear DNA was stained with DAPI (blue). Cells were then visualized under the fluorescence microscopy. (C,D) CD4+ CD25+ cells and CD4+ CD25− cells were purified and stimulated with anti-CD3 and CD28 for 24 hours. Cells were cyto-spun onto slides, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and immunostained with anti-FoxP3 (rat) and anti–c-Jun (rabbit), followed by anti–rat-Alexa 594 (red) and anti–rabbit-Alexa 488 (green). The nuclear DNA was stained with DAPI (blue). Cells were then visualized under the fluorescence microscopy and representative images were selected.
FoxP3 alters c-Jun subnuclear localization. (A,B) c-Jun-CFP expression plasmids were transfected without (A) or with (B) FoxP3 expression plasmids into HEK 293 cells. Transfected cells were immunostained with anti-FoxP3 antibody followed by a second antibody that has been labeled by Texas-Red. The nuclear DNA was stained with DAPI (blue). Cells were then visualized under the fluorescence microscopy. (C,D) CD4+ CD25+ cells and CD4+ CD25− cells were purified and stimulated with anti-CD3 and CD28 for 24 hours. Cells were cyto-spun onto slides, fixed with 4% paraformaldehyde, permeabilized with 0.1% Triton X-100, and immunostained with anti-FoxP3 (rat) and anti–c-Jun (rabbit), followed by anti–rat-Alexa 594 (red) and anti–rabbit-Alexa 488 (green). The nuclear DNA was stained with DAPI (blue). Cells were then visualized under the fluorescence microscopy and representative images were selected.
To further determine whether FoxP3 sequesters c-Jun from the chromatin of IL-2 promoter, we carried out a chromatin immunoprecipitation assay using anti-c-Jun antibody. As shown in the Figure S5, c-Jun was recruited to IL-2 promoter in CD4+ CD25− T cells after stimulation with anti-CD3 plus anti-CD28 for 24 hours, but not in Tregs even after stimulation (Figure S5). This result indicates that FoxP3 sequesters c-Jun from the chromatin at the IL-2 promoter region.
The N-terminus of FoxP3 is sufficient to maintain the unresponsiveness of Tregs
AP-1 has been identified as a unique target for T-cell tolerance.31 Thus, FoxP3 may maintain Tregs anergy by suppressing AP-1 transcriptional activity. We thus used small RNA interference (siRNA) to knock down FoxP3 expression in Tregs and tested whether knockdown of FoxP3 rescued AP-1 activity. siRNA significantly inhibited FoxP3 expression (Figure S6), and knockdown of FoxP3 expression in mouse Tregs resulted in partially rescuing the proliferation of Tregs (Figure 6A). This proliferation of Tregs is the result of a restoration of AP-1 transcriptional activities (Figure S7). Because we have found that the N-terminus of FoxP3 with an NLS (FN/NLS) sufficiently inhibited both AP-1 and IL-2 luciferase activities, we reasoned that whether ecotropic expression of this FN/NLS could induce the unresponsiveness of CD4+ T cells. As previously reported, expression of FoxP3 sufficiently changed CD4+ CD25− T cells into Tregs because these cells are both unresponsive to activation signaling and suppressive to the proliferation of FoxP3− T cells (Figure 6C,D). Interestingly, CD4+ T cells that express FN/NLS are anergic because they do not proliferate when stimulated with anti-CD3 plus and CD28. However, these T cells failed in suppressing the proliferation of CD4+ CD25− T cells (Figure 7C,D). These results indicate that the N-terminus of FoxP3 plays a sufficient role in maintaining or inducing Tregs anergy, whereas the full-length FoxP3 is required for the suppressive functions of Tregs.
The N-terminus of FoxP3 induces T-cell anergy. (A) Suppression of FoxP3 expression breaks Treg anergy. Purified mouse CD4+ CD25+ T cells were infected with lentivirus that carry GFP only or GFP with siRNA against FoxP3. GFP+ cells were sorted and restimulated with anti-CD3 plus anti-CD28 for 24 hours. Cell proliferation was examined by [3H]-thymidine incorporation. Error bars represent data from 3 independent experiments. (B) Suppression of FoxP3 restores AP-1 DNA-binding in Tregs. GFP+ cells from panel A were restimulated with anti-CD3 plus anti-CD28. Nuclear extracts were isolated, and AP-1 DNA-binding was determined. (C) Expression of FoxP3 N-terminus inhibits T-cell proliferation. Mouse CD4+ CD25− T cells were infected with retroviruses that carry GFP only, GFP with FoxP3 or with FoxP3 N-terminus. GFP+ T cells were sorted and restimulated with anti-CD3 plus anti-CD28 for 24 hours. Cell proliferation was examined by [3H]-thymidine. Error bars represent data from 3 independent experiments. (D) Lack of suppressive functions of T cells that carry FoxP3 N-terminus. CD4+ CD25− T cells were isolated and plated at 105 cells/well in 96-well plate with anti-CD3 plus anti-CD28. GFP+ cells in panel C were added to the wells at the indicated ratios. Cells were cultured for 24 hours followed by adding 1 μCi/well H-thymidine and cultured for another 16 hours. Error bars represent data from 3 independent experiments.
The N-terminus of FoxP3 induces T-cell anergy. (A) Suppression of FoxP3 expression breaks Treg anergy. Purified mouse CD4+ CD25+ T cells were infected with lentivirus that carry GFP only or GFP with siRNA against FoxP3. GFP+ cells were sorted and restimulated with anti-CD3 plus anti-CD28 for 24 hours. Cell proliferation was examined by [3H]-thymidine incorporation. Error bars represent data from 3 independent experiments. (B) Suppression of FoxP3 restores AP-1 DNA-binding in Tregs. GFP+ cells from panel A were restimulated with anti-CD3 plus anti-CD28. Nuclear extracts were isolated, and AP-1 DNA-binding was determined. (C) Expression of FoxP3 N-terminus inhibits T-cell proliferation. Mouse CD4+ CD25− T cells were infected with retroviruses that carry GFP only, GFP with FoxP3 or with FoxP3 N-terminus. GFP+ T cells were sorted and restimulated with anti-CD3 plus anti-CD28 for 24 hours. Cell proliferation was examined by [3H]-thymidine. Error bars represent data from 3 independent experiments. (D) Lack of suppressive functions of T cells that carry FoxP3 N-terminus. CD4+ CD25− T cells were isolated and plated at 105 cells/well in 96-well plate with anti-CD3 plus anti-CD28. GFP+ cells in panel C were added to the wells at the indicated ratios. Cells were cultured for 24 hours followed by adding 1 μCi/well H-thymidine and cultured for another 16 hours. Error bars represent data from 3 independent experiments.
Studies by Liew's laboratory recently demonstrated that stimulation of Tregs with either IL-2 or the ligand of toll-like receptor 2 (TLR2), BLP (bacterial lipoprotein), breaks Treg tolerance by down-regulating FoxP3 mRNA transcription.32 In an attempt to determine whether TLR2-mediated signaling reverses Tregs anergy by interrupting FoxP3/c-Jun interaction, we analyzed the effects of BLP on the interaction of FoxP3 with c-Jun in Tregs. However, the interaction of FoxP3 with c-Jun was not detectable because stimulation of Tregs with BLP quickly induced FoxP3 degradation. More than 80% of FoxP3 proteins were degraded in Tregs after 1 hour of stimulation with BLP (Figure S7A). Further analysis indicated that FoxP3 is any unstable protein, the half-life of which is less than 30 minutes (Figure S7B,C). These results indicate that BLP reverses Tregs tolerance by down-regulating FoxP3 expression.
Discussion
The regulatory T cells do not proliferate well on the combined stimulations of TCR and costimulatory molecules; in other words, Tregs are anergic. Inhibition of both AP-1 and NF-κB activation has been demonstrated to be responsible for T-cell tolerance induced ex vivo and in mice.23,31 Here we demonstrated that AP-1 promoter-binding activity is selectively impaired in naturally rising Tregs isolated from mice. FoxP3 plays an essential role in suppressing AP-1 DNA-binding activity and consequently inhibits AP-1 transcription activity because expression of FoxP3 significantly blocked AP-1 luciferase activity and promoter DNA-binding. Knockdown FoxP3 in Tregs restored both AP-1 DNA-binding and the proliferation of Tregs. Thus, our findings uncover a novel mechanism underlying how FoxP3 maintains the unresponsiveness of Tregs.
Recent studies suggested that FoxP3 competes with Fos-Jun heterodimers in the NF-AT/AP-1 promoter and this competition allows FoxP3 to cooperate with NF-AT for the transcription of the suppressive genes in Tregs.15 However, c-Jun is an abundant protein in the nucleus of T cells. The simple competition between AP-1 and FoxP3 for cooperative binding with NF-AT at NF-AT/AP-1 site may not be sufficient to suppress IL-2 production because an N-terminal deletion mutant that still interacts with NF-AT is not able to inhibit IL-2 expression.15 Our finding that FoxP3 specifically recruits phosphorylated c-Jun indicates that FoxP3 selectively competes with the activated form of c-Jun in Tregs. Therefore, FoxP3, on one hand, blocks the AP-1 DNA binding to suppress IL-2 transcriptions and to maintain the anergic features of Tregs. On the other hand, this inhibition allows FoxP3 to cooperate with NF-AT for the transcription of suppressive factors and specific surface markers of Tregs, such as IL-10, TGF-β, CTLA-4, and CD25.15 FoxP3 expression is therefore sufficient for maintaining the anergic feature and suppressive functions of Tregs.
FoxP3 governs Tregs functions via multiple intramolecular regions. The forkhead domain of FoxP3 interacts with NF-AT for the transcription of Tregs specific gene.15 The linker region between the coiled-coil and FHK domain of FoxP3 has been found to mediate its interaction with Runx1, and this interaction plays a critical role in suppressing IL-2 production by Tregs.15 More recently, Greene's group demonstrated that the N-terminal transcription activation domain of FoxP3 specifically recruits the histone acetyltransferase, TIP60, and the class II histone deacetylases, HDAC7 and HDAC9 for the suppressive functions of Tregs.16 Thus, both the N-terminus and the FHK domain of FoxP3 are required for the suppressive functions of Tregs. Here we found that expression of the N-terminus of FoxP3 without the DNA-binding FHK domain is sufficient to maintain the unresponsiveness of Tregs. This N-terminus of FoxP3 possibly inhibits IL-2 production and maintains Tregs anergy by recruiting TIP60, HDAC7/HDAC9, and c-Jun proteins. Acetylation of c-Jun may play important roles in AP-1 transcriptional activation.33,34 It will be interesting to further determine whether the TIP60/HDAC complex regulates c-Jun acetylation/deacetylation in Tregs.
The means to break tolerance without losing the suppressive functions of Tregs have great potential to expand Tregs for therapeutic uses. Accumulated evidence indicates that TLR-mediated signaling, TLR8 in human and TLR2 in mouse, reverses Tregs tolerance.32,35 However, treatment of Tregs with agonists of those TLRs also dismisses the suppressive functions of Tregs because BLP treatment down-regulates FoxP3 expression at both the transcriptional and protein levels. Because FoxP3 maintains Tregs tolerance by inhibiting AP-1 transcription activity, reversion of AP-1 activity could potentially break Tregs tolerance for Tregs expansion while maintaining suppressive function.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the unknown reviewers of this manuscript for their constructive comments as well as Mohamed Oukka (Harvard Medical School) for FoxP3 expression plasmids, Jessica Burroughs-Garcia and Danielle Tatar for technical support, and Fang laboratory members for constructive comments.
Authorship
Contribution: S.-M.L. and D.F. designed the study, analyzed data, and wrote the manuscript; S.-M.L. and B.G. performed the research.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Deyu Fang, Departments of Otolaryngology-Head and Neck Surgery, University of Missouri-Columbia School of Medicine, 1 Hospital Drive, Columbia, MO 65212; e-mail: FangD@missouri.edu.


![Figure 6. The N-terminus of FoxP3 induces T-cell anergy. (A) Suppression of FoxP3 expression breaks Treg anergy. Purified mouse CD4+ CD25+ T cells were infected with lentivirus that carry GFP only or GFP with siRNA against FoxP3. GFP+ cells were sorted and restimulated with anti-CD3 plus anti-CD28 for 24 hours. Cell proliferation was examined by [3H]-thymidine incorporation. Error bars represent data from 3 independent experiments. (B) Suppression of FoxP3 restores AP-1 DNA-binding in Tregs. GFP+ cells from panel A were restimulated with anti-CD3 plus anti-CD28. Nuclear extracts were isolated, and AP-1 DNA-binding was determined. (C) Expression of FoxP3 N-terminus inhibits T-cell proliferation. Mouse CD4+ CD25− T cells were infected with retroviruses that carry GFP only, GFP with FoxP3 or with FoxP3 N-terminus. GFP+ T cells were sorted and restimulated with anti-CD3 plus anti-CD28 for 24 hours. Cell proliferation was examined by [3H]-thymidine. Error bars represent data from 3 independent experiments. (D) Lack of suppressive functions of T cells that carry FoxP3 N-terminus. CD4+ CD25− T cells were isolated and plated at 105 cells/well in 96-well plate with anti-CD3 plus anti-CD28. GFP+ cells in panel C were added to the wells at the indicated ratios. Cells were cultured for 24 hours followed by adding 1 μCi/well H-thymidine and cultured for another 16 hours. Error bars represent data from 3 independent experiments.](https://ash.silverchair-cdn.com/ash/content_public/journal/blood/111/7/10.1182_blood-2007-09-115014/5/m_zh80070817580006.jpeg?Expires=1765883511&Signature=U8cAlrqsTOOzVrRVANHun6Qr7qGneCrc0kJgfY9fm3IjJQN9wfr~pbRakBw9Y3bGh9Uj1hVoc03E3jlnsbplNzgdSNKOtg1pGv9aH1Hxg7pX-zebr1hA2hykJDFBa2Ordef447m9S4c10S3cqmAvOYObeQr4jkT0EKNpQJXfQcoq5r1HGhYcgQxcDh5YpwR~4zU3G0m~Bwp13B6SKtMfljvO2tyv7ywFBUfPtjR99SiqK9EZ97ildp0-7g8NgcLdv9TwYBHvKTuK5RcUj~XxH5IsE2UEunTpM23oPrU5wf5~~rt-KvJFvtuqxh-77B~1VnfBEarpNGvou9HOsxyJhg__&Key-Pair-Id=APKAIE5G5CRDK6RD3PGA)
This feature is available to Subscribers Only
Sign In or Create an Account Close Modal