Abstract
We prospectively observed a child exposed to intensive multimodality therapy for metastatic neuroblastoma from emergence of a MLL translocation to disease diagnosis. The t(4;11)(p12;q23) was detected in the marrow 17 months after starting treatment following topoisomerase II poisons, alkylating agents, local radiation, hematopoietic stem cell transplantation, anti-GD2 monoclonal antibody with granulocyte macrophage–colony-stimulating factor, and a high cumulative dose of oral etoposide. Reciprocal genomic breakpoint junctions and fusion transcripts joined MLL with FRYL, the Drosophila melanogaster protein homologue of which regulates cell fate. Etoposide metabolites induced topoisomerase II cleavage complexes that could form both breakpoint junctions. Cells harboring the translocation replaced the marrow without clinical evidence of leukemia and differentiation appeared unaffected for 37 months. Subsequent bilineage dysplasia and increased blasts in addition to the translocation fulfilled criteria for MDS. The MEIS1 target gene of typical MLL fusion oncoproteins was underexpressed before and at MDS diagnosis. These results are consistent with repair of topoisomerase II cleavage from etoposide metabolites as the translocation mechanism, whereas other agents in the regimen may have contributed to progression of the clone with the translocation to MDS. MLL-FRYL did not increase MEIS1 expression, conferred a proliferative advantage without altering differentiation, and had protracted latency to disease.
Introduction
Epipodophyllotoxins, anthracyclines, and other chemotherapeutic topoisomerase II poisons are associated with leukemias with translocations of the MLL gene at chromosome band 11q23. The MLL gene product is a multidomain oncoprotein that functions in a macromolecular protein complex to regulate transcription.1-3 Recently dubbed the “MLL recombinome,” the approximately 50 MLL partner genes encode diverse nuclear transcription factors, transcriptional regulatory proteins, and cytoplasmic or cell membrane proteins.4 Murine experiments have converged upon a model where heterogeneous MLL fusion oncoproteins affect the incidence or phenotype of leukemia by altering Hox expression.5,6 It has been proposed that MLL translocations fall into a group of mutations that impair differentiation7 and that cooperating mutations that confer a proliferative and/or survival advantage to hematopoietic progenitors (eg, mutation or overexpression of FLT38-10 ) are required for leukemia to occur
Many MLL fusion proteins from the der(11) chromosome immortalize murine hematopoietic progenitor cells in serial replating assays and/or cause leukemia in mice.11-17 However, certain MLL translocations with genes encoding some of the cytoplasmic partner proteins (eg, GRAF, FBP17, ABI1, LASP1) are inactive in serial replating assays, which may reflect the requirement for cooperating alterations.18,19 Although nearly all known MLL translocations, including those inactive in serial replating assays,18-21 manifest as leukemia/myelodysplastic syndrome (MDS) in patients; in one patient, the morphologically normal clone harboring an MLL-ARHGEF17 translocation steadily declined over 30 months, and neither leukemia nor MDS occurred.22
We characterized a chemotherapy-associated MLL translocation with the partner gene FRYL (furry homolog-like; originally called MIFL [MLL Insufficient for Leukemia]), and prospectively followed the clinical course of the affected patient. First MLL-FRYL resulted in clonal replacement of the marrow without clinical disease. Although MLL-FRYL ultimately proved a harbinger to secondary MDS, different gene expression patterns, including low MEIS1 expression and the long latency and protracted course without clinical disease, indicate that MLL-FRYL is unlike typical MLL translocations. The same partner gene called AF4p12 was identified in a case of secondary acute lymphoblastic leukemia (ALL),23 during the time when the patient we describe was being observed, indicating that FRYL is a recurrent partner gene of MLL.
Methods
Patient and samples
The patient was treated for stage IV neuroblastoma at Memorial Sloan-Kettering Cancer Center on the Internal Review Board (IRB)–approved N8 protocol. Informed consent was obtained in accordance with the Declaration of Helsinki from parents. IRBs at the Children's Hospital of Philadelphia and Memorial Sloan-Kettering Cancer Center approved the molecular analyses.
MLL genomic translocation breakpoint junction cloning
Two different MLL exon numbering systems have continued to be widely used. Rasio et al24 designated the breakpoint cluster region (bcr) as the 8.3-kb BamHI fragment between exons 5 and 11 and additional exons not appreciated when the gene was first discovered were designated as exons 4b and 4c. Around the same time, Nilson et al25 described an MLL exon numbering system accounting for the 2 additional exons and a new exon 2 present in less than 10% of transcripts26 in which the same bcr is between exons 8 and 14. Exon/intron numbers used in the current report are Rasio et al24 designations followed by Nilson et al25 designations in parentheses.
The B859 cDNA probe was used for Southern blot analysis of the MLL bcr in genomic DNAs digested with BamHI, BglII, or BamHI and BglII.27 BglII reverse-panhandle polymerase chain reaction (PCR)28 was used to clone the unknown 5′-partner gene-MLL-3′ rearrangement on the der(4) chromosome. The products were purified and subcloned into the pCR 2.1-TOPO vector (Invitrogen, Carlsbad, CA).28 Subclones identified by the PCR screen28 containing inserts of the desired size were sequenced. The der(4) genomic breakpoint junction was confirmed by PCR with gene-specific primers 5′-GCT GAA ACC ATG GAA GGA AA-3′ from positions 1149-1168 of FRYL intron 50 (GenBank accession no. NM_015030) and 5′-AAA AAT TCG CAT GGA GGA GA-3′ from MLL bcr positions 5604-5585 in intron 8(11) (GenBank accession no. U04737); the product was directly sequenced.
The der(4) genomic breakpoint junction informed primers 5′-TCT ACA AGT GCC AGG GGT CT-3′ from MLL bcr positions 5315-5334 in intron 8(11) and 5′-AAA GCC AAG CTT CTC CAT CA-3′ from positions 1680-1661 of FRYL intron 50 to amplify the der(11) breakpoint junction.
Characterization of MLL fusion transcripts
Random hexamer-primed first-strand cDNA was reverse-transcribed from 10 μg of total RNA from whole bone marrow using a High-Capacity cDNA Archive Kit (Applied Biosystems, Foster City, CA). One hundred nanograms of first-strand cDNA was amplified in a 50-μl PCR containing 2.6 units of Taq/Tgo DNA polymerase (Expand High Fidelity PCR System; Roche, Indianapolis, IN), each dNTP at a 200 μmol/L final concentration, 1× PCR buffer, and 12.5 pmol of each primer. The der(11) transcripts were identified with the MLL exon 7(10) forward primer 5′-GAA TGC AGG CAC TTT GAA CA-3′ (cDNA positions 4116-4135; GenBank accession no. L04284) and the FRYL exon 53 reverse primer 5′-TCA CCG ACT TCC AAA TCC TC-3′ (cDNA positions 7783-7764). The der(4) transcript was identified with the FRYL exon 50 forward primer 5′-CTG GAA GGA AGC CCT TAA CA-3′ (cDNA positions 7319-7338), and the MLL exon 10(13) reverse primer 5′-TTT GGT GGG GTA GTT TGG TC-3′ (cDNA positions 4551-4532). The products were isolated in an agarose gel, purified with a Geneclean III kit (Qbiogene, Irvine, CA) and directly sequenced or subcloned into the pCR 2.1-TOPO vector (Invitrogen) before sequencing.
Otherwise, 107 bone marrow cells from 29 months after neuroblastoma diagnosis were stained with a phycoerythrin-conjugated anti-human CD34 antibody (Invitrogen), CD34+ and CD34− fractions were sorted on a MoFlo instrument (Dako Colorado, Fort Collins, CO), and cDNAs from these fractions and from whole bone marrow from this time point were examined for the fusion transcripts as described above.
Tracing der(11) genomic breakpoint junction in sequential marrows
Genomic DNAs were isolated from bone marrow smears on glass slides with the Puregene DNA Isolation Kit (Gentra Systems, Minneapolis, MN) or from cryopreserved bone marrow on a GITC-CsCl gradient. One hundred nanograms of DNA per time point was amplified with primers specific for the der(11) genomic breakpoint junction, and 1-μL aliquots of the products were used in hemi-nested PCRs. For the initial PCR, a forward primer corresponding to MLL bcr positions 4997-5016 in intron 8(11) (5′-GCT GCA GTG AGC CAT TAT CA-3′) was used with a reverse primer from positions 1680-1661 of FRYL intron 50. For the hemi-nested PCR, a forward primer from MLL bcr positions 5315-5334 (5′-TCT ACA AGT GCC AGG GGT CT-3′) was used with the same reverse primer.
Sensitivity was determined using serial 1:10 dilutions of bone marrow DNA from 24 months after neuroblastoma diagnosis, which was 100% replaced with cells containing the t(4;11) by karyotype analysis, into peripheral blood lymphocyte DNA from a normal subject. The reactions contained 100 ng of DNA (DNA equivalent of approximately 104 cells).
Topoisomerase II cleavage assays
DNA fragments of MLL bcr positions 5382-5570 in intron 8(11) or positions 1309-1479 of FRYL intron 50 were amplified from DNA of a normal subject and subcloned into the TOPO pCR2.1 vector (Invitrogen). The 5′ to 3′ orientation and exact matches of the insert sequences with the MLL and partner gene sequences on the der(11) and der(4) chromosomes were ascertained by sequencing. Twenty micrograms of plasmid DNA were treated with T4 DNA ligase,29 and the double-stranded inserts were released by BamHI and NotI (both from Invitrogen) digestion. The DNA was dephosphorylated, and the kinase reaction was performed as described with [γ-32P]ATP (GE Healthcare, Chalfont St Giles, United Kingdom) or ATP (Roche),29 followed by digestion with SpeI (Promega, Madison, WI) to generate singly 5′ end-labeled or nonradiolabeled BamHI-SpeI fragments containing the normal homologues of the translocation breakpoints. Topoisomerase II in vitro cleavage assays were performed on 25 ng of substrate DNA containing 30 000 cpm of labeled DNA and the remainder nonradiolabeled DNA29 without drug or with 20 μmol/L etoposide, etoposide catechol, etoposide quinone, or doxorubicin. Select reactions were heated at 75°C for 10 minutes before trapping the cleavage complexes to test heat stability.30 Because doxorubicin has concentration-dependent mixed effects of a topoisomerase II poison and catalytic inhibitor,31 doxorubicin was also examined at concentrations from 0.001 to 20 μmol/L. Deproteinized cleavage complexes were electrophoresed in a denaturing acrylamide gel alongside a DNA sequencing ladder of the plasmid beginning at the same 5′ end as the substrate.
Real-time quantitative PCR analysis of leukemia-associated gene expression
Random hexamer-primed first-strand cDNAs were synthesized from 1 to 10 μg of total RNA from patients' bone marrows using the High Capacity cDNA Archive Kit (Applied Biosystems). Assay-On-Demand kits were used for real-time quantitative-PCR analysis of HOXA4, HOXA5, HOXA7, HOXA9, PBX3, MEIS1, FLT3, and BCL-2 target genes (Applied Biosystems). A Predeveloped TaqMan Assay Reagent kit was used for the 18S rRNA endogenous control (Applied Biosystems). Final 20-μL reactions contained 100 ng of cDNA, Taqman Universal Master Mix, and target gene-specific or 18S rRNA-specific primers. Amplifications were performed in a 384-well thermal cycler using the ABI Prism 7900HT Sequence Detection system (Applied Biosystems). Data were collected and analyzed with Sequence Detection System 2.1 software (Applied Biosystems). Gene expression was quantified using the comparative CT method.32
Results
Case report
A 4-year-old boy with metastatic neuroblastoma was treated on the N8 protocol, which includes 3 cycles of vincristine, doxorubicin, and cyclophosphamide; 2 cycles of cisplatin and etoposide; surgery; local radiation; anti-GD2 monoclonal antibody (3F8); myeloablative therapy with thiotepa, carboplatin, and topotecan; and autologous peripheral blood stem cell rescue with cells harvested after the first cycle of cisplatin and etoposide. The transplant was followed by 7 cycles of 3F8 antibody with granulocyte macrophage–colony-stimulating factor (GM-CSF), 4 cycles of oral etoposide administered at 50 mg/m2 daily for 21 days per cycle, and 5 cycles of oral 13-cis-retinoic acid (isotretinoin [Accutane]). The cumulative dose of doxorubicin was 225 mg/m2. The cumulative dose of etoposide was 5.4 g/m2 (4.2 g/m2 oral; Figure 1A). The protocol specifies routine bone marrow surveillance for neuroblastoma relapse or occurrence of leukemia every 3 to 6 months.
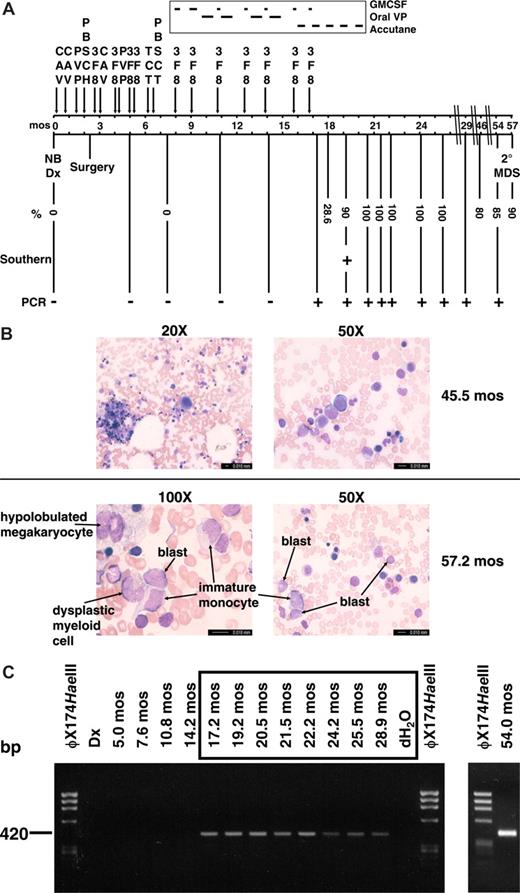
Summary of therapy, clinical course, and cytogenetic and molecular analyses of sequential marrows. (A) Timeline with scale in months (mos) from neuroblastoma diagnosis. Percentages indicate maximum abnormal marrow cells by karyotype, FISH with dual color probe for MLL (Vysis, Downers Grove, IL), or chromosome 4 and 11 painting probes (Vysis). Abbreviations: CAV, cyclophosphamide (2100 mg/m2 per day × 2), doxorubicin (25 mg/m2 per day × 3), vincristine (0.66 mg/m2 per day × 3); PVP, cisplatin (50 mg/m2 per day × 4), etoposide (200 mg/m2 per day × 3); PBSCH, autologous peripheral blood stem cell harvest; 3F8, anti-GD2 antibodies; TCT, thiotepa (300 mg/m2 per day × 3), carboplatin (500 mg/m2 per day × 3), topotecan (2 mg/m2 per day × 5); PBSCT, autologous peripheral blood stem cell transplant; GMCSF, granulocyte macrophage colony stimulating factor; oral VP, oral etoposide (50 mg/m2 per day × 21 days). (B) Representative bone marrow aspirate smear from 45.5 months after neuroblastoma diagnosis at low-power magnification showing normal morphology and trilinear hematopoiesis (top left), and higher power magnification of normal erythroid and myeloid cells in same bone marrow aspirate (top right). Representative bone marrow aspirate smear from 57.2 months after neuroblastoma diagnosis at high-power magnification showing blasts, immature monocytes, a hypolobulated megakaryocyte, and a dysplastic myeloid cell (bottom left and right). Bone marrow aspirates were visualized using a Leica DMLB microscope (20×/0.40 aperature; 50×/0.90 aperature; 100×/1.25 aperature) captured with a Leica DFC420 camera (imaging medium of air for 20×, immersion oil for 50× and 100×), acquired using Leica Application Suite and processed using Microsoft (Redmond, WA) PowerPoint. (C) Tracing of der(11) genomic breakpoint junction in sequential marrows. The clonotypic der(11) genomic breakpoint junction was detectable by PCR in sample from 17 months after neuroblastoma diagnosis and all specimens thereafter. Time represents months from neuroblastoma diagnosis. All positive samples were sequenced to verify the breakpoint junction.
Summary of therapy, clinical course, and cytogenetic and molecular analyses of sequential marrows. (A) Timeline with scale in months (mos) from neuroblastoma diagnosis. Percentages indicate maximum abnormal marrow cells by karyotype, FISH with dual color probe for MLL (Vysis, Downers Grove, IL), or chromosome 4 and 11 painting probes (Vysis). Abbreviations: CAV, cyclophosphamide (2100 mg/m2 per day × 2), doxorubicin (25 mg/m2 per day × 3), vincristine (0.66 mg/m2 per day × 3); PVP, cisplatin (50 mg/m2 per day × 4), etoposide (200 mg/m2 per day × 3); PBSCH, autologous peripheral blood stem cell harvest; 3F8, anti-GD2 antibodies; TCT, thiotepa (300 mg/m2 per day × 3), carboplatin (500 mg/m2 per day × 3), topotecan (2 mg/m2 per day × 5); PBSCT, autologous peripheral blood stem cell transplant; GMCSF, granulocyte macrophage colony stimulating factor; oral VP, oral etoposide (50 mg/m2 per day × 21 days). (B) Representative bone marrow aspirate smear from 45.5 months after neuroblastoma diagnosis at low-power magnification showing normal morphology and trilinear hematopoiesis (top left), and higher power magnification of normal erythroid and myeloid cells in same bone marrow aspirate (top right). Representative bone marrow aspirate smear from 57.2 months after neuroblastoma diagnosis at high-power magnification showing blasts, immature monocytes, a hypolobulated megakaryocyte, and a dysplastic myeloid cell (bottom left and right). Bone marrow aspirates were visualized using a Leica DMLB microscope (20×/0.40 aperature; 50×/0.90 aperature; 100×/1.25 aperature) captured with a Leica DFC420 camera (imaging medium of air for 20×, immersion oil for 50× and 100×), acquired using Leica Application Suite and processed using Microsoft (Redmond, WA) PowerPoint. (C) Tracing of der(11) genomic breakpoint junction in sequential marrows. The clonotypic der(11) genomic breakpoint junction was detectable by PCR in sample from 17 months after neuroblastoma diagnosis and all specimens thereafter. Time represents months from neuroblastoma diagnosis. All positive samples were sequenced to verify the breakpoint junction.
By karyotype analysis, the t(4;11)(p12;q23) first appeared in the bone marrow 18 months after neuroblastoma diagnosis, at which time painting probes showed the t(4;11) in 28.6% of cells. By 19 months after neuroblastoma diagnosis, the t(4;11) was detected in 18 of 20 metaphases, 2 of which acquired t(2;9)(q21;q34). Cells with the t(4;11) replaced all ensuing marrows shown in Figure 1A, with the t(2;9) also present in between 0 and 6 of 20 cells in later specimens. All marrows had normal cellularity, trilinear morphology (Figure 1B top) and normal flow cytometric markers and showed no evidence of leukemia until 54 months after neuroblastoma diagnosis, 37 months after the translocation first appeared. At that time, the karyotype showed t(4;11) in 17 of 20 cells, 4 of which also had −16, add (18q). The patient remained asymptomatic, and the marrow cellularity and peripheral complete blood count remained normal, without cytopenias or increased blasts; occasional neutrophils had the Pelger Huet anomaly, and there were rare micro- and monolobulated megakaryocytes. Three months later, 90% of cells in the marrow showed the t(4;11) by fluorescence in situ hybridization (FISH) analysis. Increased blasts (8% of 200 cells), immature monocytes, dysplastic myeloid cells, and hypolobulated megakaryocytes were present (Figure 1B bottom). The clonal cytogenetic abnormality, bilineage myelodysplasia, and increased blasts fulfilled 3 of 4 WHO criteria for MDS in children,33 indicating that the clone with the MLL-FRYL translocation acquired additional features of secondary MDS.
The patient received cytosine arabinoside and asparaginase for secondary MDS and central nervous system prophylaxis with intrathecal methotrexate and cytosine arabinoside. He underwent human leukocyte antigen-matched sibling hematopoietic stem cell transplantation after a preparative regimen consisting of busulfan and melphalan, followed by post-transplant methotrexate and tacrolimus for graft-versus-host disease prophylaxis. The patient is disease-free 74 months after neuroblastoma diagnosis and 14 months after transplant for secondary MDS.
Characterization of genomic breakpoint junctions of t(4;11)(p12;q23)
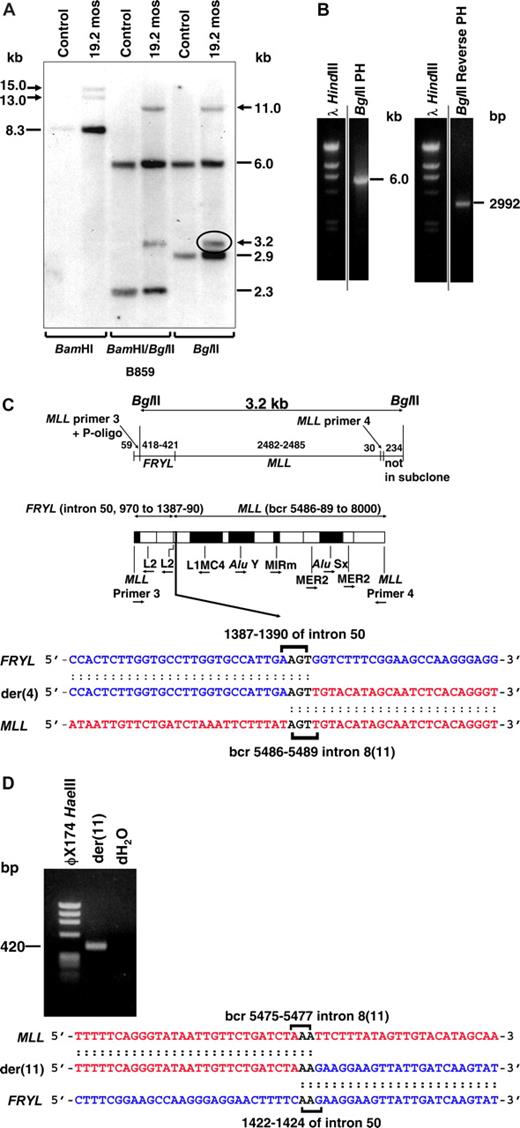
Reciprocal MLL genomic breakpoint junctions and fusion transcripts were characterized in cryopreserved bone marrow cells obtained 19 months after neuroblastoma diagnosis, in which 18 of 20 (90%) of the cells contained this translocation by karyotype analysis (Figure 1A). Southern blot analysis of BamHI or BglII digested DNA revealed 2 MLL bcr rearrangements, consistent with both derivative chromosomes from the translocation (Figure 2A). Because the BamHI rearrangements were large and not reduced by combined BamHI and BglII digestion (Figure 2A), BglII panhandle PCR and BglII reverse-panhandle PCR28 were attempted to clone the breakpoint junctions. A 6.0-kilobase (kb) product consistent with the germline MLL allele, but not the der(11) breakpoint junction, was obtained with BglII panhandle PCR (Figure 2B left).
Molecular characterization of MLL-FRYL translocation. Marrow from 19 months after neuroblastoma diagnosis, which was 82% replaced by cells with t(4;11)(p12;q23) by FISH and 90% replaced by cells with t(4;11)(p12;q23) by karyotype analysis, was used for Southern blot and molecular cloning. (A) Southern blot analysis performed with B859 cDNA probe27 and indicated restriction enzyme digests. Dashes indicate germline bands; arrows show rearrangements. (B) BglII panhandle PCR gave a 6.0-kb BglII germline product from normal MLL allele but no larger or smaller product consistent with a rearrangement (left). BglII reverse-panhandle PCR product, the size of which indicated that 3.2-kb BglII fragment on the Southern blot (panel A, circled) was der(4) rearrangement (right). Vertical lines have been inserted to indicate repositioned gel lanes. (C) Summary of der(4) genomic breakpoint junction in subclones of BglII reverse panhandle PCR product generated by TOPO TA cloning (Invitrogen). One subclone was sequenced in entirety; the breakpoint junction was verified in 2 more subclones and confirmed by PCR with gene-specific primers. Fifty-nine base pairs of 5′ sequence are from MLL primer 3 through ligated oligonucleotide (P-oligo); 418-421 bp of 5′ sequence are the partner gene. The partner DNA sequence corresponded with GenBank accession no. NM_015030 (FRYL, KIAA0826). The 3′ 2512-2515 bp include MLL bcr sequence beginning at position 5486, 5487, 5488, or 5489 (GenBank accession no. U04737), which is in intron 8(11) through MLL primer 4. Brackets show possible FRYL and MLL breakpoint positions and 5′-AGT-3′ in both genes that precluded more precise breakpoint assignments (bottom). Repetitive sequences are shown (middle). MLL intron number is Rasio et al24 designation with parenthetical Nilson et al25 designation alongside. (D) PCR product obtained with gene-specific primers containing der(11) genomic breakpoint junction, which was directly sequenced. MLL and FRYL breakpoints (brackets) could not be assigned precisely because of 5′-AA-3′ in both genes. Short sequence homologies (brackets) and loss of 8-13 bases from MLL and 31-36 bases from FRYL, indicated by comparison with the der(4) genomic breakpoint junction (B), suggested imprecise NHEJ repair.
Molecular characterization of MLL-FRYL translocation. Marrow from 19 months after neuroblastoma diagnosis, which was 82% replaced by cells with t(4;11)(p12;q23) by FISH and 90% replaced by cells with t(4;11)(p12;q23) by karyotype analysis, was used for Southern blot and molecular cloning. (A) Southern blot analysis performed with B859 cDNA probe27 and indicated restriction enzyme digests. Dashes indicate germline bands; arrows show rearrangements. (B) BglII panhandle PCR gave a 6.0-kb BglII germline product from normal MLL allele but no larger or smaller product consistent with a rearrangement (left). BglII reverse-panhandle PCR product, the size of which indicated that 3.2-kb BglII fragment on the Southern blot (panel A, circled) was der(4) rearrangement (right). Vertical lines have been inserted to indicate repositioned gel lanes. (C) Summary of der(4) genomic breakpoint junction in subclones of BglII reverse panhandle PCR product generated by TOPO TA cloning (Invitrogen). One subclone was sequenced in entirety; the breakpoint junction was verified in 2 more subclones and confirmed by PCR with gene-specific primers. Fifty-nine base pairs of 5′ sequence are from MLL primer 3 through ligated oligonucleotide (P-oligo); 418-421 bp of 5′ sequence are the partner gene. The partner DNA sequence corresponded with GenBank accession no. NM_015030 (FRYL, KIAA0826). The 3′ 2512-2515 bp include MLL bcr sequence beginning at position 5486, 5487, 5488, or 5489 (GenBank accession no. U04737), which is in intron 8(11) through MLL primer 4. Brackets show possible FRYL and MLL breakpoint positions and 5′-AGT-3′ in both genes that precluded more precise breakpoint assignments (bottom). Repetitive sequences are shown (middle). MLL intron number is Rasio et al24 designation with parenthetical Nilson et al25 designation alongside. (D) PCR product obtained with gene-specific primers containing der(11) genomic breakpoint junction, which was directly sequenced. MLL and FRYL breakpoints (brackets) could not be assigned precisely because of 5′-AA-3′ in both genes. Short sequence homologies (brackets) and loss of 8-13 bases from MLL and 31-36 bases from FRYL, indicated by comparison with the der(4) genomic breakpoint junction (B), suggested imprecise NHEJ repair.
BglII reverse panhandle PCR gave a 2992-base pair (bp) product that contained the der(4) breakpoint junction (Figure 2B right), indicating that the 3.2-kb BglII fragment on the Southern blot (Figure 2A,C) was from the der(4) rearrangement. BLAT query (http://www.genome.ucsc.edu/) of the partner gene returned a sequence with 100% identity with a gene within chromosome band 4p12, the conforming name of which became FRYL (furry homolog-like; GenBank accession no. NM_015030). The der(4) genomic breakpoint in FRYL was position 1387, 1388, 1389, or 1390 relative to the start of intron 50. The der(4) MLL genomic breakpoint was position 5486, 5487, 5488, or 5489 of the bcr (GenBank accession number U04737), which is in intron 8(11). These breakpoints could not be assigned more precisely because of identical 5′-AGT-3′ sequences in both genes. PCR with gene-specific primers confirmed this breakpoint junction sequence.
PCR with gene-specific primers for the der(11) breakpoint junction gave the 420-bp product in Figure 2D rather than the predicted 464-bp product. The der(11) MLL breakpoint was position 5475, 5476, or 5477 in the bcr, which is in intron 8(11), and the der(11) FRYL breakpoint was position 1422, 1423, or 1424 relative to the start of intron 50, indicating that the translocation resulted in deletion of 8 to 13 bases from MLL and 31-36 bases from FRYL. Homologous 5′-AA-3′ sequences at the breakpoints in both genes also precluded more precise der(11) breakpoint assignments. Similar to other MLL genomic breakpoint junctions in treatment-related leukemias,34,35 the short sequence homologies and small deletions suggested limited processing consistent with nonhomologous end-joining (NHEJ) DNA repair.36-38 Although the FRYL breakpoints on both derivative chromosomes were in an L2 repeat, neither MLL breakpoint was within any repetitive sequence.
BLASTP queries of the FRYL transcript across all 6 reading frames identified a protein with regions of 64%, 33%, 39%, 42%, and 25% amino acid identity with regions 1 through 5 of the Fry protein, respectively (GenBank accession no. AAG41424), which is involved in cell fate and bristle morphogenesis in Drosophila melanogaster.39
t (4;11) (p12;q23) produces reciprocal der(11) and der(4) fusion transcripts
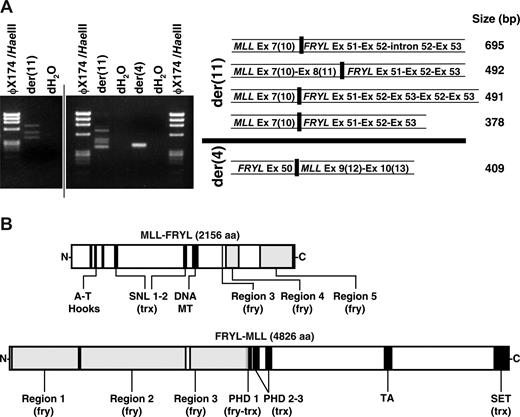
Reverse transcriptase (RT)-PCR demonstrated that the translocation resulted in multiple fusion transcripts (Fig 3A). MLL exon 8(11) or exon 7(10) was alternatively spliced to exon 51 of the 64-exon FRYL gene (GenBank accession no. NM_015030) in 2 in-frame transcripts from the der(11) chromosome. One 5′-MLL-FRYL-3′ transcript was incompletely processed, and another contained a duplication of FRYL exons 52 and exon 53. A reciprocal in-frame transcript from the der(4) chromosome was detected fusing FRYL exon 50 and MLL exon 9(12). Fusion proteins predicted from these transcripts are in Fig 3B.
Reciprocal fusion transcripts and predicted fusion proteins from MLL-FRYL translocation. (A) MLL exon 7(10) sense and FRYL exon 53 antisense primers were used to amplify der(11) transcripts from randomly primed first-strand cDNA. FRYL exon 50 sense and MLL exon 10(13) antisense primers were used for the der(4) transcript. Products are shown in the gels on the left. Sequences of fusion transcripts obtained by direct sequencing or sequencing of subclones were compared with MLL (GenBank accession no. L04284) and FRYL (KIAA0826; GanBank accession no. NM_015030) cDNAs for exon designations (right). MLL exon/intron numbers are Rasio et al designations24 with parenthetical Nilson et al designations25 alongside. A vertical line has been inserted to indicate 2 distinct gels. (B) Schematics of reciprocal fusion proteins predicted from der(11) and der(4) fusion transcripts. Regions of MLL and FRYL are black and gray, respectively. Examples of predicted fusion proteins shown are those from 5′-MLL exon 8(11)-FRYL exon 51-3′ and 5′-FRYL exon 50-MLL exon 9(12)-3′ transcripts, which were not alternatively spliced, incompletely processed and did not contain FRYL exon duplications. Predicted amino acid sequences of the fusion proteins were analyzed using SMART (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/). The amino acids from PHD 1 of MLL retained in the predicted N-MLL-FRYL-C (VCFLCASSGHVE) no longer comprise a PHD domain. However, SMART analysis of the predicted N-FRYL-MLL-C detected a PHD 1 domain (GKTLDFHFDISEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCK) with the first 12 amino acids contributed by FRYL (plain text) and the remaining C terminal amino acids (bold) derived from MLL. Therefore the predicted N-FRYL-MLL-C still contains 3 PHD domains.
Reciprocal fusion transcripts and predicted fusion proteins from MLL-FRYL translocation. (A) MLL exon 7(10) sense and FRYL exon 53 antisense primers were used to amplify der(11) transcripts from randomly primed first-strand cDNA. FRYL exon 50 sense and MLL exon 10(13) antisense primers were used for the der(4) transcript. Products are shown in the gels on the left. Sequences of fusion transcripts obtained by direct sequencing or sequencing of subclones were compared with MLL (GenBank accession no. L04284) and FRYL (KIAA0826; GanBank accession no. NM_015030) cDNAs for exon designations (right). MLL exon/intron numbers are Rasio et al designations24 with parenthetical Nilson et al designations25 alongside. A vertical line has been inserted to indicate 2 distinct gels. (B) Schematics of reciprocal fusion proteins predicted from der(11) and der(4) fusion transcripts. Regions of MLL and FRYL are black and gray, respectively. Examples of predicted fusion proteins shown are those from 5′-MLL exon 8(11)-FRYL exon 51-3′ and 5′-FRYL exon 50-MLL exon 9(12)-3′ transcripts, which were not alternatively spliced, incompletely processed and did not contain FRYL exon duplications. Predicted amino acid sequences of the fusion proteins were analyzed using SMART (Simple Modular Architecture Research Tool; http://smart.embl-heidelberg.de/). The amino acids from PHD 1 of MLL retained in the predicted N-MLL-FRYL-C (VCFLCASSGHVE) no longer comprise a PHD domain. However, SMART analysis of the predicted N-FRYL-MLL-C detected a PHD 1 domain (GKTLDFHFDISEFVYCQVCCEPFHKFCLEENERPLEDQLENWCCRRCK) with the first 12 amino acids contributed by FRYL (plain text) and the remaining C terminal amino acids (bold) derived from MLL. Therefore the predicted N-FRYL-MLL-C still contains 3 PHD domains.
t(4,11) (p12;q23) emerges late during neuroblastoma treatment
The der(11) genomic breakpoint junction was first detectable in the bone marrow by hemi-nested PCR 17 months after neuroblastoma diagnosis. This was after completion of all therapy in the intensive multimodality regimen except isotretinoin, after the 3F8 with GM-CSF, and after oral etoposide when the cumulative etoposide dose reached 5.4 g/m2 (Figure 1A,C). The clonotypic der(11) breakpoint junction remained detectable in all specimens until the diagnosis of secondary MDS was made. The t(4;11) could not be detected in any prior bone marrow samples, including the bone marrow from neuroblastoma diagnosis (Figure 1A,C). Analysis of serial dilutions of the sample from 24 months after neuroblastoma diagnosis, which was completely replaced with cells harboring the t(4;11) (Figure 1A), into peripheral blood lymphocyte DNA from a healthy subject indicated that the sensitivity of the assay for detection of the translocation was between approximately 1 cell in 10 000 and 1 cell in 100 000 cells (data not shown).
Consistent with the presence of the translocation in the bone marrow progenitor cell population, RT-PCR detected the 5′-MLL-FRYL-3′ transcript fusing MLL exon 8(11) to FRYL exon 51 and the reciprocal 5′-FRYL-MLL-3′ transcript fusing FRYL exon 50 to MLL exon 9(12) in the CD34+, CD34−, and whole bone marrow fractions obtained 29 months after neuroblastoma diagnosis (data not shown).
Processing of etoposide metabolite-stabilized topoisomerase II cleavage sites created both breakpoint junctions
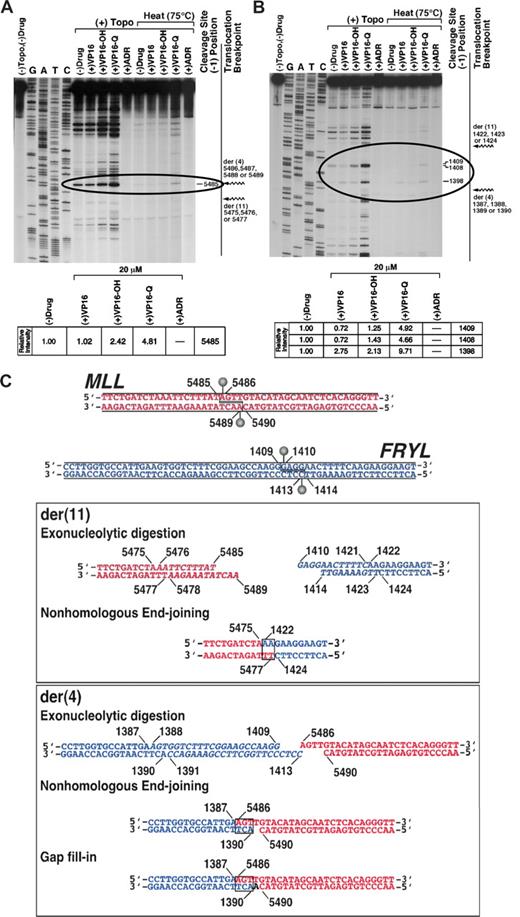
Topoisomerase II in vitro cleavage assays of normal MLL and FRYL sequences focused on cleavage sites within genomic regions deleted by the translocation that could be resolved to form both breakpoint junctions (Figure 4). The assays were performed without drug or with etoposide, etoposide catechol, etoposide quinone, or doxorubicin, which are all of the topoisomerase II poisons29 to which the patient was exposed. The model for topoisomerase II involvement in MLL leukemogenesis entails stabilization of topoisomerase II cleavage complexes by topoisomerase II poisons, which increases cleavage complexes, the direct repair of which forms the translocations.40 The increased cleavage compared with cleavage by topoisomerase II alone indicated that etoposide quinone especially behaved as atopoisomerase II poison at MLL bcr position 5485 and positions 1398, 1408, and 1409 in FRYL intron 50, within the deleted regions (Figure 4A,B, circled). Amounts of cleavage at these sites with 20 μmol/L etoposide, etoposide catechol, etoposide quinone or doxorubicin compared with cleavage without drug are shown (Figure 4A,B). Detection after heating indicated resistance to religation of the cleavage complexes at MLL bcr position 5485 and positions 1398 and 1409 in FRYL intron 50 in the presence of etoposide quinone. In contrast, there was no detectable cleavage at any sites within the deleted region of either gene with doxorubicin at 20 μmol/L. Because doxorubicin has potential mixed effects of topoisomerase II cleavage complex stabilization (ie, activity as a poison) at low concentrations and topoisomerase II catalytic inhibition at high concentrations,31,41 doxorubicin was tested over a range of concentrations from 0.001 to 20 μmol/L. At 0.50 μmol/L, doxorubicin resulted in dose-dependent increased (2.8-fold) cleavage over cleavage without drug at MLL bcr position 5485, consistent with effects of a poison, but doxorubicin at low concentrations did not increase cleavage over cleavage without drug within the deleted region in FRYL (data not shown).
Identification of topoisomerase II cleavage sites in MLL and FRYL that form translocation breakpoint junctions. Substrates contained MLL bcr positions 5382-5570 (A) or positions 1309-1479 in FRYL intron 50 (B). Autoradiographs show cleavage products after 10-minute incubation at 37°C of singly 5′ end-labeled DNA (30 000 cpm) and unlabeled substrate (25 ng, labeled and unlabeled total) with 147 nmol/L human topoisomerase IIα, 1 mmol/L ATP and, where indicated, 20 μmol/L etoposide, etoposide catechol, etoposide quinone, or doxorubicin. Specified reactions were incubated for an additional 10 minutes at 75°C before trapping the covalent complexes (panels A,B). Jagged arrows designating translocation breakpoints (panels A,B far right) bracket the normal homologue of the deleted region in each substrate. Topoisomerase II creates staggered nicks in duplex DNA with 4-base 5′ overhangs40 ; bases at the 5′ side of cleavage (−1 position) on the sense strand were used to designate cleavage site positions (dashes at right of gels). Amounts of cleavage at sites within the deleted regions (circled) were quantified by filmless autoradiographic analysis in (+) Topo reactions with specified compounds without subsequent 75°C incubation relative to cleavage at same sites without any drug (panels A,B bottom). The lanes shown in (A) are from a single large gel that was scanned in multiple sections because of its size. After scanning, the lanes were aligned together in their original positions as they appear in the complete gel. (C) Model for creation of t(4;11) breakpoint junctions by processing of the etoposide metabolite enhanced cleavage sites at MLL bcr position 5485 in intron 8(11) (top, red) and position 1409 in FRYL intron 50 (top, blue). The processing includes exonucleolytic digestion (italic) to form homologous overhangs and create both breakpoint junctions by classic error-prone NHEJ (boxes). In formation of the der(11), positions 5476-5485 on the sense strand and positions 5478-5489 on the antisense strand of MLL bcr and positions 1410-1421 on the sense strand and positions 1414-1423 on the antisense strand of FRYL intron 50 are deleted by exonucleolytic digestion (middle, italic) before NHEJ (middle, box) joins the indicated bases. Exonucleolytic digestion removes positions 1388-1409 on the sense strand and positions 1391-1413 on the antisense strand of FRYL intron 50 (bottom, italic) but all bases in the sense strand of the 4-base 5′ overhang in MLL are retained, and NHEJ (bottom, box) and gap fill-in create the der(4) breakpoint junction.
Identification of topoisomerase II cleavage sites in MLL and FRYL that form translocation breakpoint junctions. Substrates contained MLL bcr positions 5382-5570 (A) or positions 1309-1479 in FRYL intron 50 (B). Autoradiographs show cleavage products after 10-minute incubation at 37°C of singly 5′ end-labeled DNA (30 000 cpm) and unlabeled substrate (25 ng, labeled and unlabeled total) with 147 nmol/L human topoisomerase IIα, 1 mmol/L ATP and, where indicated, 20 μmol/L etoposide, etoposide catechol, etoposide quinone, or doxorubicin. Specified reactions were incubated for an additional 10 minutes at 75°C before trapping the covalent complexes (panels A,B). Jagged arrows designating translocation breakpoints (panels A,B far right) bracket the normal homologue of the deleted region in each substrate. Topoisomerase II creates staggered nicks in duplex DNA with 4-base 5′ overhangs40 ; bases at the 5′ side of cleavage (−1 position) on the sense strand were used to designate cleavage site positions (dashes at right of gels). Amounts of cleavage at sites within the deleted regions (circled) were quantified by filmless autoradiographic analysis in (+) Topo reactions with specified compounds without subsequent 75°C incubation relative to cleavage at same sites without any drug (panels A,B bottom). The lanes shown in (A) are from a single large gel that was scanned in multiple sections because of its size. After scanning, the lanes were aligned together in their original positions as they appear in the complete gel. (C) Model for creation of t(4;11) breakpoint junctions by processing of the etoposide metabolite enhanced cleavage sites at MLL bcr position 5485 in intron 8(11) (top, red) and position 1409 in FRYL intron 50 (top, blue). The processing includes exonucleolytic digestion (italic) to form homologous overhangs and create both breakpoint junctions by classic error-prone NHEJ (boxes). In formation of the der(11), positions 5476-5485 on the sense strand and positions 5478-5489 on the antisense strand of MLL bcr and positions 1410-1421 on the sense strand and positions 1414-1423 on the antisense strand of FRYL intron 50 are deleted by exonucleolytic digestion (middle, italic) before NHEJ (middle, box) joins the indicated bases. Exonucleolytic digestion removes positions 1388-1409 on the sense strand and positions 1391-1413 on the antisense strand of FRYL intron 50 (bottom, italic) but all bases in the sense strand of the 4-base 5′ overhang in MLL are retained, and NHEJ (bottom, box) and gap fill-in create the der(4) breakpoint junction.
Because etoposide catechol and especially etoposide quinone increased topoisomerase II cleavage within the MLL and FRYL deleted regions, and the greatest etoposide quinone stimulated cleavage in these regions was at MLL bcr position 5485 and position 1409 in FRYL intron 50, these cleavage sites (Figure 4C top) were used to develop a model for formation of the breakpoint junctions. Exonucleolytic digestion at both cleavage sites creates 2-base homologies for NHEJ to generate the der(11) breakpoint junction (Figure 4C middle). The 4-base 5′ overhang of the sense strand of the cleavage site in MLL is preserved completely, and exonucleolytic digestion of the FRYL cleavage site creates 3-base homologies for NHEJ and gap fill-in to form the der(4) breakpoint junction (Figure 4C bottom). During the processing of the overhangs from the cleavage, exonucleolytic digestion deletes 10 bases from MLL and 34 bases from FRYL relative to the sense sequences. MLL bcr position 5475 and position 1422 in FRYL intron 50 are joined to form the der(11), and position 1387 in FRYL intron 50 and MLL bcr position 5486 are joined to form the der(4), which is consistent with the breakpoint junction sequences (Figure 2).
Bone marrow cells with MLL-FRYL translocation have reduced expression of genes associated with leukemias with MLL translocations
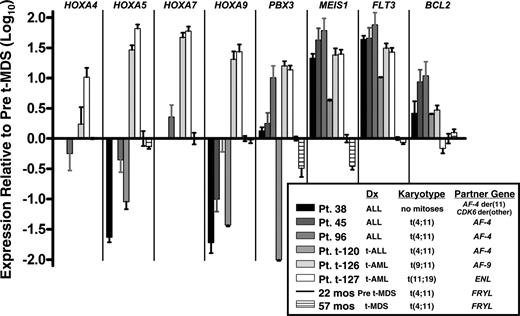
Expression patterns of known MLL fusion oncoprotein target genes and MLL-leukemia associated genes in the patient's marrow from 22 months (before MDS diagnosis) and 57 months after neuroblastoma diagnosis (at MDS diagnosis), which were 100% and 90% replaced by cells with the translocation, respectively, were compared by real-time PCR analysis to de novo and treatment-related acute leukemias with common MLL translocations. There was variability in expression of the HOXA9, HOXA5, and HOXA7 target genes across the sample set. MEIS1 expression was substantially greater in all of the leukemia cases than in the cells with MLL-FRYL. Moreover, MEIS1 expression did not increase at 57 months after neuroblastoma diagnosis, at which time MDS was evident (Figures 1B and 5). The patient's marrow at both time points expressed less FLT3 and generally less BCL2 mRNA compared with cases of de novo and treatment-related acute leukemias with MLL translocations (Figure 5).
Real-time PCR analysis showing different gene expression patterns compared with leukemia cases with MLL translocations. Log10 (Relative Quantity) is 2−ΔΔCT, where the endogenous control is 18S rRNA and the calibration sample (set to 1 for all genes) is bone marrow of patient at 22 months after neuroblastoma diagnosis but before MDS diagnosis, in which 100% of cells contained the translocation by karyotype analysis. No bar appears in graph for any transcript that was undetectable. Error bars indicate standard deviation in triplicate reactions. Treatment-related leukemias are specified as t-ALL or t-AML. Patients 38, 45, and t-120 have been described.35
Real-time PCR analysis showing different gene expression patterns compared with leukemia cases with MLL translocations. Log10 (Relative Quantity) is 2−ΔΔCT, where the endogenous control is 18S rRNA and the calibration sample (set to 1 for all genes) is bone marrow of patient at 22 months after neuroblastoma diagnosis but before MDS diagnosis, in which 100% of cells contained the translocation by karyotype analysis. No bar appears in graph for any transcript that was undetectable. Error bars indicate standard deviation in triplicate reactions. Treatment-related leukemias are specified as t-ALL or t-AML. Patients 38, 45, and t-120 have been described.35
Discussion
These results are of interest for several reasons. The first is that the natural history was followed prospectively from first detection of the translocation to the diagnosis of disease. For 37 months, the patient had no clinical, phenotypic, or morphologic evidence of leukemia, even though the MLL-FRYL clone completely replaced the marrow. Bilineage morphologic abnormalities and increased marrow blasts that surfaced later, together with the clonal MLL translocation eventually met criteria to diagnose secondary MDS. Second is that MLL-FRYL was associated with different gene expression patterns compared with other MLL translocations in leukemia cases. Next is that the translocation became detectable toward the end of an intensive regimen, after a high cumulative dose of oral etoposide and other potentially leukemogenic therapies were used.
FRYL is homologous to D melanogaster furry (Fry), which encodes a protein involved in morphogenesis of cellular extensions.39 Its Mor2 homologue in fission yeast is important for polarity control and cell separation.42 In humans, FRYL encodes a heretofore unknown protein with undetermined function. A case of secondary ALL with the same partner gene occurred 5 years after breast cancer treatment that included mitoxantrone, cyclophosphamide, and radiation and 1 year after radiation for endometrial carcinoma.26 In contrast, the translocation we describe eventually manifested as MDS, not acute leukemia, and conferred a proliferative advantage without any obvious effect on differentiation for greater than 3 years. Thus, not only is MLL-FRYL leukemogenic, but also there is clinical, phenotypic, and morphologic variability in the diseases that it causes.
Although exceptional MLL leukemias occur after longer latency43,44 or present as MDS,45 the latency to MDS was longer than the typical 24- to 30-month latency from epipodophyllotoxin exposure to leukemia with MLL translocations.46,47 The prolonged clonal proliferation without any obvious phenotype may suggest that MLL-FRYL is fundamentally different from typical MLL translocations that affect differentiation.10 This is supported by the low MEIS1 expression before and at secondary MDS diagnosis despite evidence that MLL-FRYL was expressed. HOXA9 and the HOX coregulator gene MEIS1 are usually overexpressed in leukemias with MLL translocations,10 and the HOX coregulator genes MEIS1 and PBX3 are MLL fusion protein targets.48 There was also less expression of other genes frequently altered in leukemias with MLL translocations (eg, FLT310-12,49,50 ), but FLT3 was overexpressed in the secondary ALL with the same translocation.26 Antiapoptotic BCL-2 expression was lower in the secondary MDS relative to most MLL leukemias that were tested. These results suggest that MLL-FRYL conferred a proliferative advantage without a typical MLL leukemia genotype or phenotype, but the differences in gene expression must be interpreted with caution because only a small number of cases were compared.
Recently Meis1 expression levels were shown to inversely correlate with latency in retroviral transplantation models of other MLL leukemias, and Meis1 was shown to mediate effects of MLL fusion proteins on transformation, impaired differentiation, and enhanced self-renewal.51 The demonstration that the leukemogenic effects of other MLL fusion proteins are critically dependent on Meis1 overexpression51 validates the significance of the low MEIS1 expression observed with MLL-FRYL. The protracted latency and unimpaired differentiation are consistent with the dependence of latency and impaired differentiation on Meis1 expression levels in the murine models.51 The reduced PBX3 gene expression observed with MLL-FRYL may be of interest also because PBX proteins function in a complex with MEIS1.51
Latency to leukemia in murine models has suggested that other alterations must be acquired in addition to MLL translocations for leukemia to occur.14 It is also possible that early on there was a more subtle perturbation of hematopoietic differentiation that was inapparent or at an early, reversible stage in a stepwise progression to leukemic transformation,52 because later acquisition of −16, add(18q) in a subpopulation of the cells heralded bilineage dysplasia and increased blasts consistent with secondary MDS.33
Retrospective studies of pediatric cases of treatment-related leukemia showed that the MLL translocation was absent in the bone marrow at primary cancer diagnosis but emerged during treatment,34,53 either early after low cumulative doses of leukemogenic agents34 or later in the treatment (L.J. Raffini, C.P.K., R.J. Whitmarsh, et al. Tracing MLL translocations in secondary leukemias to chemotherapy exposures in multimodality neuroblastoma regimens, manuscript submitted), indicating that treatment causes and does not select for pre-existing translocations. This patient was unlike the prior patients because the translocation was not detected until after a very high cumulative dose of oral etoposide was used. Although genomic breakpoint junction cloning identified a translocation breakpoint hotspot region in treatment-related leukemias in intron 8(11) 3′ in the MLL bcr,30,34,35,54-59 the translocation breakpoints also were not in the hotspot region.
Secondary leukemia cases in which both genomic breakpoint junctions have been characterized indicate precise or near-precise interchromosomal DNA recombinations,30,34,35,54-56 consistent with processing of 4-base staggered double-strand breaks from topoisomerase II cleavage.30,35,55 Likewise, both breakpoint junctions in this translocation could be formed after small deletions and imprecise NHEJ repair at topoisomerase II cleavage sites induced by etoposide quinone, whereas the doxorubicin in the regimen had no activity as a poison within the FRYL deletion.
Why topoisomerase II-mediated DNA double-strand breaks coupled with NHEJ repair remains a lead hypothesis for the MLL translocation mechanism in chemotherapy exposed patients recently was summarized by others and us.40,60 The appearance of secondary leukemias with MLL rearrangements coincident with introducing epipodophyllotoxins into clinical usage, the translocation breakpoint junction sequences, and induction of cleavage sites by drugs at/proximal to translocation breakpoints in experimental systems support this hypothesis. Topoisomerase II is the target of chemotherapeutic topoisomerase II poisons, which stabilize ternary drug/DNA/topoisomerase II cleavage complexes. A review on biochemical mechanisms of translocations concluded that the correlation of translocation breakpoints and sites of topoisomerase II double-strand cleavage in the presence of agents to which patients are exposed confirms that translocations reflect misjoining of exchanged ends of the resultant double-strand breaks60 ; furthermore, general features of the breakpoint junctions including small terminal deletions, splicing of DNA ends at microhomologies, and gap filling are consistent with classic NHEJ repair.60 Evidence for topoisomerase II involvement in MLL translocations in de novo leukemias also recently was summarized.40
Alternative hypotheses on the MLL translocation mechanism have been suggested, one of which involves damage from an apoptotic nuclease,61-63 because exposing hematopoietic cell lines to general cytotoxic drugs, including topoisomerase II poisons, causes apoptotic cleavage in MLL exon 9(12).63,64 This mechanism cannot account for the translocation we describe, translocations in secondary leukemias with both breakpoints in the breakpoint hotspot,30,35,54,56 and other MLL translocations in which both MLL breakpoints are 5′ to the apoptotic cleavage site. Povirk60 also concluded that cleavage associated with abortive apoptosis was unlikely in translocations occurring via highly conservative crossovers within the breakpoint hotspot. When levels of apoptotic cleavage of the bcr were tracked in patients during chemotherapy, the rearrangement and panhandle PCR product in the one observed case of secondary acute myelogenous leukemia (AML) suggested that the apoptotic cleavage site was not involved in the translocation.65 Homologous recombination between Alu sequences may occur in MLL tandem duplications, a small subset of MLL rearrangements.66 Heptamer- and nonamer-like se-quences suggested a potential role of the VDJ recombinase in some de novo cases.67
The timing of emergence of the translocation after the oral etoposide in the patient we report and repair of etoposide metabolite-induced topoisomerase II cleavage sites to form the breakpoint junctions may be consistent with the recently observed increased risk of treatment-related leukemia when semicontinuous or continuous oral etoposide schedules resulting in cumulative etoposide doses of more than 6 g/m2 were used.68 However, because etoposide was also used early in the treatment, it is possible that the translocation occurred earlier but remained below the sensitivity of the PCR until later on.
In addition, other elements in the regimen may have contributed to the ultimate evolution of the clone with the translocation into MDS. The regimen included alkylating agent treatment, the other major class of leukemogenic drugs.47 If the MLL translocation was an earlier event than indicated by the PCR, it is possible that the alkylators played a role in the subclone evolution. Autologous stem cell transplantation itself carries risk of secondary AML, which is higher after transplantation of peripheral blood stem cells harvested after chemotherapy and cytokines than after bone marrow transplantation without priming.69
The hematopoietic growth factor GM-CSF was used later in the treatment. The function of cytokine receptors in Janus tyrosine kinase/signal transducer and activator of transcription (JAK/STAT) signaling70 raises questions about GM-CSF as a potential contributor to the eventual MDS. Several studies have evaluated hematopoietic growth factors, especially granulocyte cell-stimulating factor (GCSF), as risk factors for secondary leukemia/MDS. GCSF was implicated as a possible risk factor for the form of secondary AML resulting from topoisomerase II poisons after intensive pediatric ALL treatment regimens.71 Analyses in breast cancer72,73 and severe congenital neutropenia populations74 concluded that GCSF contributes to an increased risk. However, no increased risk was found among healthy donors administered GCSF for peripheral blood stem cell mobilization and the Research on Adverse Drug Events and Reports project concluded that evidence supporting a causal relationship is unclear.75
The 13-cis-retinoic acid in the neuroblastoma regimen seems an unlikely contributor because previous cytogenetic studies did not detect secondary chromosomal abnormalities in marrows of patients treated for acute promyelocytic leukemia (APL) with all-trans retinoic acid76 and leukemia is not among the life-threatening complications of 13-cis-retinoic acid in dermatologic treatments.77
This patient kindles new questions about the positive predictive value of MLL-FRYL and MLL translocations in general as biomarkers of disease. Depending on the partner gene, epipodophyllotoxin-related leukemias with MLL translocations usually are monocytic or myelomonocytic AML,78,79 but they can present as other AML subtypes, ALL, or MDS.46,80,81 The recent developments in this patient indicate that the protracted bone marrow proliferation by the MLL-FRYL clone was a harbinger of secondary MDS in a heterogeneous spectrum. The lack of clinical evidence of leukemia in the patient for so long despite replacement of the marrow by morphologically and phenotypically normal cells with the t(4;11)(p12;q23) suggests that MLL-FRYL increases cell proliferation without an obvious effect on differentiation. The low MEIS1 expression suggests that, unlike other MLL fusion oncoproteins, MEIS1 is not a downstream target of MLL-FRYL and that there is additional heterogeneity in the pathways to MLL leukemogenesis.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Jo Ann Byl for preparing topoisomerase IIα, Ian Blair for providing etoposide metabolites, Andrew Lee for analyzing sequence data, Junhyong Kim for assisting with data interpretation and Michael Cleary and Piu Wong for helpful discussion. The opinions in this article are ours and do not necessarily represent those of the Pennsylvania Department of Health.
This work was supported by National Institutes of Health grants R01-CA85469, R01-CA77683, and in part by a grant from the Pennsylvania Department of Health, Joshua Kahan Fund, Friends of the Joseph Claffey Fund.
National Institutes of Health
Authorship
Contribution: B.W.R. designed and performed research, analyzed data, and wrote the manuscript. N.-K.V.C. analyzed data and wrote the manuscript. C.P.K. performed research. S.C.J. and J.K.C. analyzed data. N.O. contributed vital new reagents. C.A.F. designed research, analyzed data, and wrote the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Carolyn A. Felix, Division of Oncology, Leonard and Madlyn Abramson Pediatric Research Center, Room 902B, The Children's Hospital of Philadelphia, 3615 Civic Center Blvd, Philadelphia, PA 19104-4318; e-mail: felix@email.chop.edu.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal