Abstract
The restricted immunoglobulin (Ig) repertoire found in B-cell chronic lymphocytic leukemia (CLL) implies a role for antigen(s) in the leukemogenesis. The nature of the antigens has, however, not been characterized, although examples of autoantigens have been demonstrated. We have analyzed a panel of 28 CLL cell lines and primary cultures, producing monoclonal Ig with different Ig heavy-chain variable region gene usage and mutational status, including several complementarity determining region 3 homology subset members. Using mass-spectrometry, immunoassays, or protein macroarrays, we have discovered novel antigens binding to CLL Igs. These antigens included cytoskeletal proteins vimentin, filamin B, and cofilin-1, but also phosphorylcholine-containing antigens (eg, Streptococcus pneumoniae polysaccharides and oxidized low-density lipoprotein [oxLDL]). Additional new antigens identified were cardiolipin and proline-rich acidic protein-1. Remarkably, these antigens represent molecular motifs exposed on apoptotic cells/blebs and bacteria, and several CLL Igs bound to apoptotic Jurkat cells. In conclusion, these intriguing data, showing a limited target structure recognition, indicate that CD5+ CLL B cells are derived from a cell compartment that produces “natural antibodies,” which may be instrumental in elimination and scavenging of apoptotic cells and pathogenic bacteria.
Introduction
B-cell chronic lymphocytic leukemia (CLL) cells show gene expression profiles similar to memory B cells1 as well as a surface membrane phenotype reminiscent of activated and antigen-experienced B cells.2 CLL can be divided into subgroups with either unmutated or mutated Ig heavy-chain variable (IGHV) genes, where unmutated IGHV (IGHVUM) genes are associated with a more aggressive clinical course than mutated IGHV (IGHVM) genes.3,4 Biased use of certain IGHV genes, foremost IGHV1-69, IGHV3-07, IGHV3-21, and IGHV4-34, has been reported.4-6 Recently, more than 110 subgroups of CLL have been identified that showed remarkably similar IGHV and Ig light-chain variable (IGLV) gene rearrangements with homologous complementarity determining region 3 (CDR3) amino acid sequences among members of the respective groups.7,8 These findings of “stereotyped” B-cell antigen receptors (BCRs) in CLL subsets imply a role for specific antigens in leukemogenesis.9,10
Which antigens do the CLL-derived Abs then bind to? The fact that CLL patients often present with autoimmune phenomena including autoimmune hemolytic anemia and autoimmune thrombocytopenia indicates an immune dysregulation associated with the malignancy. However, antierythrocyte, antiplatelet, anti–double-stranded DNA, and anti-ia Abs found in these patients are polyclonal and, by definition, not produced by the malignant clone. Interestingly, recent studies have shown that several monoclonal recombinant CLL-derived Abs, each representing the malignant clone in subsets with stereotyped BCRs, bind to cytoplasmic and nuclear autoantigens.11
Previous attempts to isolate Ig from malignant CLL clones have been hampered by minute amounts of Ig secreted in vitro, contaminations of a small number of normal nonmalignant B cells, and transient short survival times in vitro, impeding a correct verification of clonal origin. Recombinant Abs (often monomeric) from CLL clones have been successfully used,11 but the difference between recombinant Ab and native pentameric IgM may present drawbacks regarding conclusion based on avidity/affinity of the mAb.
The aim of the present study was to characterize in detail the antigens in CLL, first using cell lines12 derived from the neoplastic CLL clone by Epstein-Barr virus (EBV) transformation,13 and second using primary ex vivo CLL cultures. Unexpectedly, the specificities of this panel of 28 CLL Abs showed limited target structure recognition on apoptotic cells and bacteria, and we identify here several previously unpublished (auto)antigens. Our findings may give new insight into the origin and clonal expansion of these leukemic B cells.
Methods
Cell lines
EBV-transformed CLL cell lines representing the malignant clone were previously established in our laboratory and by colleagues (Table 1). Their CLL origin was reconfirmed by one or several of the following methods: karyotyping, fluorescent in situ hybridization (FISH) analysis, fluorescence-activated cell sorting (FACS) phenotype, Ig gene sequencing, and microsatellite analysis, verifying that the cells were neoplastic clones (A.L.M, E.H., A.R. et al, unpublished data, November 2007). A recombinant monovalent IGHV3-21M mAb (rIGHV3-21M/subset-2) expressing IGH/IGL sequences14 of the stereotyped IGHV3-21 subset 2 was also included in this study (for details, see Document S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Primary CLL cultures were established from peripheral blood mononuclear cells (PBMCs) of CLL patients consecutively attending the Hematology Clinic, Linköping University Hospital, after informed consent, under the guidelines from the Linköping University Hospital ethics committee approval D.no.02-459, in compliance with the Declaration of Helsinki. The CLL cell lines and cultures, FM55M2 human melanoma, HepG2 hepatoma (no. 85011430; ECACC, Salisbury, United Kingdom) and Jurkat T cell line (no. 88042803; ATCC, Manassas, VA) were cultivated in RPMI 1640 medium (Invitrogen, Paisley, United Kingdom) with 10% FCS (InVitrogen) with additives. Rat smooth muscle cells (SMCs) were kindly provided by Dr Hans Arnqvist (Linköping, Sweden). Human fibroblasts, Ag1523, were cultivated in MEM (Invitrogen) obtained from Coriell Institute for Medical Research (Camden, NJ). Monocyte derived macrophages were prepared from PBMCs from healthy volunteers by Ficoll-Hypaque (Pharmacia Biotech AB, Uppsala, Sweden). Plastic adherence-purified monocytes were cultured in DMEM (Invitrogen) with 20% human AB+ serum. The rIGHV3-21M Ab was produced in CHO cells grown in 20% RPMI 1640 + 80% CHO-S-SFMII with 2% FBS (Invitrogen).
CLL cell lines used in this study
| Name . | Ig produced . | IGHV (CDR3 subset) . |
|---|---|---|
| I83 | IgMλ | IGHV3-30M |
| HG3 | IgMλ | IGHV1-2UM |
| Wa | IgMκ | IGHV3-30.3UM (subset 32) |
| CII | IgMλ | IGHV1-69UM (subset 5) |
| 232B4 | IgGκ | IGHV3-48M |
| PGA1 | IgGκ | IGHV4-39M |
| EHEB | IgG | IGHV1-18M |
| AIII | IgMκ | IGHV4-59 |
| rIGHV3–21* | IgMλ | IGHV3-21M (subset 2) |
| Name . | Ig produced . | IGHV (CDR3 subset) . |
|---|---|---|
| I83 | IgMλ | IGHV3-30M |
| HG3 | IgMλ | IGHV1-2UM |
| Wa | IgMκ | IGHV3-30.3UM (subset 32) |
| CII | IgMλ | IGHV1-69UM (subset 5) |
| 232B4 | IgGκ | IGHV3-48M |
| PGA1 | IgGκ | IGHV4-39M |
| EHEB | IgG | IGHV1-18M |
| AIII | IgMκ | IGHV4-59 |
| rIGHV3–21* | IgMλ | IGHV3-21M (subset 2) |
Additional details and references to cell lines are given in Table S1.
M indicates mutated; UM, unmutated.
Recombinant CLL Ab produced in CHO cells (for details, see Document S1).
Immunohistochemistry and tissue microarrays
Tissue microarray slides (SuperBioChips Laboratories, Seoul, South Korea) containing paraffin-embedded human tissue samples (n = 59) were used for antigen-specificity screening according to the manufacturer's protocol. IgM from CLL cell lines was used with biotinylated goat anti–human IgM (Sigma-Aldrich, St Louis, MO) and streptavidin-HRP conjugate (Zymed Labs, San Francisco, CA). The slides were developed with DAB substrate and counterstained in Mayer hematoxylin solution (Histolab, Göteborg, Sweden). Formalin-fixed, paraffin-embedded sections from human tonsils were used for confirmatory immunohistochemical analysis.
Immunofluorescence
CLL IgMs were used for detection of autoAb reactivity in 5-μm unfixed cryostat sections of rat liver, stomach, and kidney tissues. The binding was visualized by FITC–rabbit antihuman μ-chain Ab (Dako, Glostrup, Denmark). Extended screening was performed on FM55M2, Jurkat, HepG2 (Immunoconcepts, Sacramento, CA), Ag1523, and rat SMCs. The cells were fixed in 4% paraformaldehyde and permeabilized in 0.1% saponin. CLL IgM binding was visualized using FITC–rabbit antihuman μ-chain Ab (Dako) or goat anti–human IgM Ab-Alexa594 (Molecular Probes, Eugene, OR). Anti-TNFα mAb (Innogenetics, Gent, Belgium), anti-CD3 mAb (Dako), and anti-SMA mAb (Sigma-Aldrich) were used as positive controls for FM55M2, Jurkat, and rat SMCs, respectively. Human IgM antimyelin P0 mAb15 was used as a negative control for all cell types. The cells were mounted in Gel/Mount medium with antifading agents (BioMeda, Foster City, CA) and examined in a Zeiss Axiovert 200M inverted fluorescence microscope (Carl Zeiss, Heidelberg, Germany), equipped with a 63×/1.2 NA Plan Apochromat water objective. Images were collected with an AxioCam MRm CCD camera (Carl Zeiss) using Zeiss AxioVision software (Carl Zeiss). Final presentation and mounting of the images were performed using Adobe Photoshop version 7.0 (Adobe Systems, San Jose, CA).
Western blots and protein macroarrays
IgM/IgG mAbs from the CLL cell lines were used for further autoAb specificity screening. Total protein (30 μg) from FM55M2 or HepG2 cell lysates was separated by sodium dodecyl sulfate–polyacrylamide gel electrophoresis (SDS-PAGE) and analyzed in Western blots. AutoAb screening of CLL IgM mAbs was performed on RZPD GmbH (German Resource Center for Genome Research, Berlin, Germany) and produced high-density protein macroarray membranes representing E coli–expressed proteins (n = 38 016) from a human fetal brain cDNA library. Colonies were grown on the membranes and IPTG stimulated. Cells were lysed to release the recombinant protein, which was immobilized on the membrane as spots. Primary Abs were incubated overnight at + 4°C, and secondary Ab rabbit anti–human IgM/G/A-HRP (Dianova, Hamburg, Germany) was incubated for 2 hour at room temperature and developed by chemiluminescence (Attophos; Roche Diagnostics, Mannheim, Germany). Secondary Ab–alone controls were included in each assay and low-intensity binding patterns were regarded as negative. Binding quality/strength was graded ++, +, or +/−.
Affinity purification of I83 and HG3 mAb-specific antigens
The I83 IgM-specific antigen(s) were isolated by Sieze X protein G immunoprecipitation (IP) kit (Pierce Biotechnology, Rockford, IL) according to the manufacturer's protocol, including anti-IgM/IgM cross-linking to protein G by disuccinimidyl suberate. Purified proteins were separated by SDS-PAGE. IP was not required in the case of HG3, since the HG3-specific antigen was adequately separated on the gel. For in-gel digestion, the Coomassie-stained proteins were excised from the gel, destained, and trypsin digested (Sequencing Grade Modified Trypsin, 2.5 μg/mL; Promega, Madison, WI) as previously described.16
MALDI-TOF-MS
Equal volumes of sample and saturated α-cyano-4-hydroxycinnamic acid in 70% (vol/vol) acetonitrile with 0.3% (vol/vol) trifluoroacetic acid were mixed and applied on the target plate. Matrix-assisted laser desorption/ionization time-of-flight mass spectometry (MALDI-TOF-MS) spectra were acquired on a Voyager-DE Pro (Applied Biosystems, Foster City, CA) and recorded using the instrument settings recommended by Applied Biosystems. The obtained monoisotopic peptide masses were used to search databases (ProteinProspector),17 allowing a peptide mass accuracy of 100 ppm and one partial cleavage. Carbamido-methylation modification of cysteine was considered.
Peptide sequencing
Peptide sequencing by electrospray ionization tandem MS (ESI-MS/MS) was performed on a hybrid mass spectrometer API Q-STAR Pulsar i (Applied Biosystems) equipped with a nanoelectrospray ion source (MDS Protana, Odense, Denmark). Collision-induced decomposition of selected peptides was performed using the instrument settings recommended by Applied Biosystems with manual operation of collision energy during the spectra acquisition.
Antigen specificity reconfirmed in ELISA and Western blots
I83 vimentin specificity was reconfirmed by Western blot analysis using purified human recombinant vimentin (no. 62115; Progen Biotechnik, Heidelberg, Germany) and by vimentin–enzyme-linked immunosorbent assay (ELISA). IgM in cell culture medium from several CLL patients was tested along with the I83 IgM in the ELISA assay. The plates were coated with 0.5 μg/mL recombinant vimentin in PBS overnight at 4°C and then blocked with 3% BSA in PBS for 4 hours at room temperature (RT). Primary Abs were incubated overnight at 4°C and secondary polyclonal rabbit anti–human IgM HRP-conjugated Ab (Dako) for 1 hour at room temperature (RT). The plate was washed 3 times with 0.9% NaCl with 0.05% Tween-20 between each step. OPD-substrate tablets (Dako) and H2O2 were used for development and detection of vimentin-binding IgM. The antivimentin clone V9 mAb (Dako) was used as positive control and the human IgM antimyelin P0 mAb as negative control. The primary Abs were tested at 7.8 nM to 500 nM. We also used an anti-CCP ELISA (CCP2 RA scan; EuroDiagnostica, Arnhem, The Netherlands) for detection of I83 IgM reactivity with citrullinated (vimentin) epitopes. Samples were tested in triplicates in 3 separate experiments.
Binding of CLL IgMs to Streptococcus pneumoniae polysaccharides
CLL mAbs were screened for S pneumoniae capsular polysaccharide binding using a multiplex bead assay including serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F. Three separate experiments were performed. For detailed description, see Document S1.
Flow cytometric analysis
Macrophages were cultured for 6 days in 20% human serum, followed by 2 days of serum deprivation. The cells were stained for vimentin, using the I83 mAb and V9 mAb. Human IgM or mouse IgG1 (Dako) was used as negative control. Rabbit anti–human IgM–FITC or goat anti–mouse IgG1–FITC (Dako) was used as secondary reagent. Analysis was performed in a FACS Calibur flow cytometer (BD Biosciences, Mountain View, CA).
Vimentin expression on CLL surface membrane
For membrane vimentin analysis, CLL cells were incubated with V9 mAb, developed with anti–mouse Ig-Alexa594 conjugate (Molecular Probes). Differential interference contrast illumination was used for overlay immunofluorescence images.
LDL isolation and modifications
Low-density lipoprotein (LDL) was isolated by sequential ultracentrifugation from pooled healthy donors and oxidized by exposure to copper ions (10 μM) at 37°C for 24 hours. MDA modification was performed as previously described (Document S1).
Chemiluminescent ELISA for mAb binding to oxLDL
Ab binding to various antigens was performed as described in detail in Document S1 using antigen-coated MicroFluor plates (Dynatech Laboratories, Chantilly, VA), primary Ab (< 3 μg/mL), and alkaline phosphatase–labeled goat anti-IgM or anti-IgG (Sigma-Aldrich) and LumiPhos530 (Lumigen, Southfield, MI) as luminescent substrate. Five to 11 separate experiments were performed.
CLL mAb binding to apoptotic Jurkat cells
Apoptotic human Jurkat cells were prepared by exposure to UV light (51 mJ/cm2) after which they were further cultured 18 hours before use. The apoptotic cells were incubated with the primary human mAbs (0.1 to 1.0 μg/mL) for 45 minutes at 4°C in the presence or absence of 100 μg/mL MDA-LDL as specific competitor, followed by incubation with FITC-labeled anti–human-IgM (or IgG) secondary Ab for 30 minutes at 4°C. After staining, cells were double-stained with 7-AAD and analyzed using a FACSCalibur (BD Biosciences). Results were validated using camptothecin-exposed Jurkat cells.
Results
CLL cell line–derived mAbs bind autoantigens
The CLL cell lines used in this study represent EBV-transformed neoplastic clones as evidenced by IGH rearrangements, IGHV sequences, or phenotypic markers (Table 1; Table S1). The cell lines showed different IGHV gene use and mutational status; and some of them belonged to recently described subsets with stereotyped CDR3s. The subset numbering is according to Stamatopoulos et al7 and Murray et al.8 The CLL cells secreted mAbs in the concentration range of 0.1 to 3 μg/mL per 3 d/107 cells.
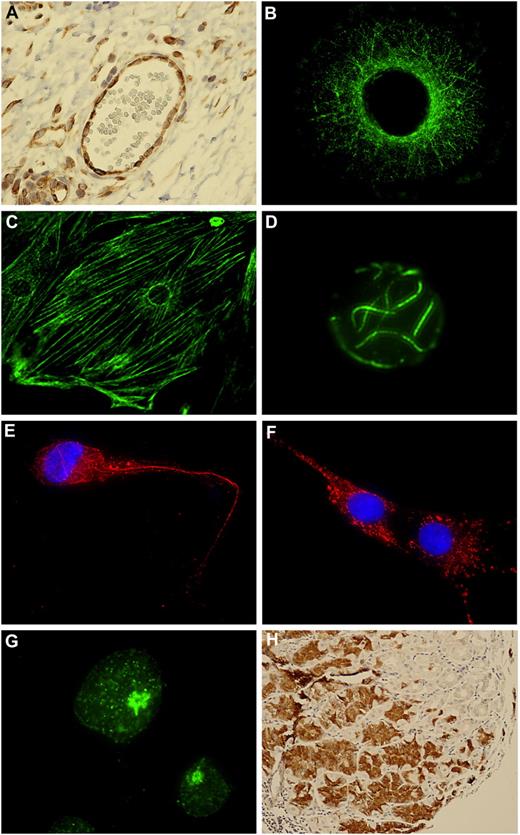
Prior to proteomic analysis, we analyzed the mAbs for possible autoantigen binding in a cell panel including vascular smooth muscle cells, FM55M2 melanoma cells, HepG2 hepatocellular carcinoma, Ag1523 fibroblasts, and Jurkat T-cell leukemia. Autoreactivity with various binding profiles was observed for the CLL mAbs. The binding pattern of the I83/IGHV3-30M mAb is illustrated in the color micrograph images: human lymph node blood vessel lining endothelial cells (Figure 1A), human malignant melanoma FM55M2 cells (Figure 1B), rat smooth muscle cells (Figure 1C), and Jurkat human T-cell leukemia cells (Figure 1D). The intracellular cytoskeletal pattern (Figure 1B-D) resembles intermediate filaments. The 2 HG3/IGHV1-2UM and Wa/IGHV3-30.3UM/subset-32 mAbs revealed cytoskeletal and cytoplasmic patterns, respectively, in HepG2 cells (Figure 1E,F), whereas AIII/IGHV4-59M showed a nuclear dotted pattern in Jurkat cells (Figure 1G).
CLL mAbs bind to autoantigens. (A) I83/IGHV3-30UM IgM binding to lymph node (tonsil) thin paraffin section visualized by immunohistochemical staining. (B) I83 mAb binding to FM55M2 melanoma cells. (C) I83 mAb binding to rat aortic smooth muscle cells. (D) I83 mAb binding to Jurkat T cells. (E) HG3/IGHV1-2UM IgM binding to HepG2 cells. (F) Wa/IGHV3-30.3UM/subset-32 IgM binding to HepG2 cells. (G) AIII/IGHV4-59M IgM binding to Jurkat T cells. (H) rIGHV3-21M/subset-2 IgM binding to stomach chief cells. Secondary Ab for immunohistochemical staining was anti–IgM-biotin followed by streptavidin-HRP. Secondary Ab for immunofluorescence was anti–IgM-FITC or anti–IgM-Alexa594.
CLL mAbs bind to autoantigens. (A) I83/IGHV3-30UM IgM binding to lymph node (tonsil) thin paraffin section visualized by immunohistochemical staining. (B) I83 mAb binding to FM55M2 melanoma cells. (C) I83 mAb binding to rat aortic smooth muscle cells. (D) I83 mAb binding to Jurkat T cells. (E) HG3/IGHV1-2UM IgM binding to HepG2 cells. (F) Wa/IGHV3-30.3UM/subset-32 IgM binding to HepG2 cells. (G) AIII/IGHV4-59M IgM binding to Jurkat T cells. (H) rIGHV3-21M/subset-2 IgM binding to stomach chief cells. Secondary Ab for immunohistochemical staining was anti–IgM-biotin followed by streptavidin-HRP. Secondary Ab for immunofluorescence was anti–IgM-FITC or anti–IgM-Alexa594.
In tissue microarrays based on thin sections of paraffin-embedded human tissues (n = 59), Wa/IGHV3-30.3UM/subset-32 IgM revealed multiple binding patterns (not shown), thus classified as polyreactive. The rIGHV3-21M/subset-2 mAb reacted with stomach chief cells (Figure 1H) and pancreatic exocrine glands, but showed no reactivity with the cell lines represented in Figure 1A-G. The CII/IGHV1-69UM/subset-5 mAb also bound stomach chief cells, whereas no cell line reactivity was found.
Vimentin and filamin B autoantigens identified in mass spectrometry
The mAbs I83/IGHV3-30M and HG3/IGHV1-2UM that showed distinct cytoskeletal patterns (Figure 1B-E) were analyzed in detail by mass spectrometry (MS). These 2 IgM clones showed single bands of 54 kDa and 277 kDa MW in Western blots (Figure 2A,B). Affinity-chromatography isolation of I83/IGHV3-30M–reacting proteins from melanoma cell extracts on protein G anti-IgM/I83 IgM matrix revealed the 54-kDa immunoreactive moiety, and additional 39-kDa and 17-kDa moieties (Figure 2A left panel). Trypsin digest peptides from gel-excised bands were analyzed by matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS and electrospray ionization tandem MS (ESI-MS/MS). The MS peptide profile of the immunoreactive 54-kDa protein is shown in Figure 2A right panel. The HG3/IGHV1-2UM IgM immunoreactive protein of 277 kDa was excised from SDS-PAGE–separated total cell extract of HepG2 cells, and analyzed in a similar fashion (Figure 2B).
Mass spectrometry (MS) and immunoassay identification of CLL mAb-binding antigens. (A) Purification and identification of I83/IGHV3-30M IgM-specific proteins by MS. FM55M2 melanoma cell extract was affinity purified by I83 IgM–protein G beads. The photographs show parallel lanes from SDS-PAGE (left panel), one transferred for Western blot and one lane was Coomassie stained. A single 54-kDa protein was found in Western blot. The Coomassie-stained gel show 3 major proteins, 54 kDa, 39 kDa, and 17 kDa. These were excised and digested with trypsin and analyzed by ESI-MS/MS. The doubly charged molecular ion with m/z = 786.4, obtained from the 54-kDa antigen, was selected and subjected to ESI-MS/MS. Peptide sequencing by ESI-MS/MS identified the I83 IgM-specific antigen (54 kDa) as vimentin and the protein of 39 kDa as aldolase A. The detected b (N-terminal) and y (C-terminal) fragment ions are shown in the spectrum, and the complete peptide sequences are shown above each spectrum with the numbers corresponding to the b and y ions. (B) Purification and identification HG3/IGHV1-2UM IgM specific antigen. (C) Vimentin-ELISA performed on I83/IGHV3-30M IgM and Abs from 48-hour ex vivo CLL cultures. (D) Binding of Wa/IGHV3-30.3UM/subset-32 CLL IgM to S pneumoniae polysaccharides in luminex assay. Multiplex beads were coated with 1 of 7 pneumococcal capsular polysaccharides. The median fluorescent intensity (MFI) of IgM binding to each of 7 serotype-coated multiplex beads is shown against 4 doubling dilutions.
Mass spectrometry (MS) and immunoassay identification of CLL mAb-binding antigens. (A) Purification and identification of I83/IGHV3-30M IgM-specific proteins by MS. FM55M2 melanoma cell extract was affinity purified by I83 IgM–protein G beads. The photographs show parallel lanes from SDS-PAGE (left panel), one transferred for Western blot and one lane was Coomassie stained. A single 54-kDa protein was found in Western blot. The Coomassie-stained gel show 3 major proteins, 54 kDa, 39 kDa, and 17 kDa. These were excised and digested with trypsin and analyzed by ESI-MS/MS. The doubly charged molecular ion with m/z = 786.4, obtained from the 54-kDa antigen, was selected and subjected to ESI-MS/MS. Peptide sequencing by ESI-MS/MS identified the I83 IgM-specific antigen (54 kDa) as vimentin and the protein of 39 kDa as aldolase A. The detected b (N-terminal) and y (C-terminal) fragment ions are shown in the spectrum, and the complete peptide sequences are shown above each spectrum with the numbers corresponding to the b and y ions. (B) Purification and identification HG3/IGHV1-2UM IgM specific antigen. (C) Vimentin-ELISA performed on I83/IGHV3-30M IgM and Abs from 48-hour ex vivo CLL cultures. (D) Binding of Wa/IGHV3-30.3UM/subset-32 CLL IgM to S pneumoniae polysaccharides in luminex assay. Multiplex beads were coated with 1 of 7 pneumococcal capsular polysaccharides. The median fluorescent intensity (MFI) of IgM binding to each of 7 serotype-coated multiplex beads is shown against 4 doubling dilutions.
Sixteen of 17 peptides from the MALDI-TOF-MS analysis of the I83/IGHV3-30M–reactive 54-kDa protein band matched vimentin. In the case of the 39-kDa protein (Figure 2A), 8 of 15 peptides matched aldolase A (Table S3). The 17-kDa protein (Figure 2A) did not generate any peptide signals. The HG3/IGHV1-2UM–specific protein of 277 kDa generated 20 major peptides, of which 15 matched filamin B (Table S2). The matched peptides covered 47% and 10% of vimentin and filamin B, respectively. The proteins identified by MALDI-TOF-MS were confirmed by sequencing the aa 411-424 peptide of the 54 kDa protein, and the aa 537-551, 658-673, 823-836, 932-946, 2401-2415, and 2465-2484 peptides of the 277-kDa protein (Table S2). Hence, the I83/IGHV3-30M–specific antigen of 54 kDa was identified as vimentin, and the HG3/IGHV1-2UM–specific protein of 277 kDa as filamin B.
The specificities of I83/IGHV3-30M and HG3/IGHV1-2UM were verified with recombinant proteins or specific mouse mAbs. Furthermore, mAbs from 19 primary ex vivo CLL cultures (Table 2) were obtained by PMA/ionophore stimulation for 48 hours (a mean value of 0.2 μg/mL Ig was released) and analyzed by vimentin-ELISA. I83/IGHV3-30M showed a robust dose-response binding to vimentin, whereas none of the ex vivo CLL Abs were positive (Figure 2C), which was expected since none of the primary CLL cultures expressed the same IGHV gene as the I83/IGHV3-30M clone did. The V9 mouse antivimentin mAb served as an external assay control. The antivimentin mAb I83/IGHV3-30M did not react with citrullinated vimentin epitope as analyzed in a specific immunoassay (data not shown).
Primary ex vivo CLL cultures used in this study
| Patient ID . | Sex/age, y . | Stage, Binet . | Ig produced . | IGHV (CDR3 subset)* . |
|---|---|---|---|---|
| p239 | M/74 | A | IgM | IGHV1-69UM (subset 6) |
| p241 | M/63 | C | IgM | IGHV3-21UM |
| p457 | M/57 | B | IgM | IGHV1-18UM (subset 1) |
| p467 | F/61 | A | IgG | IGHV4-34M |
| p497 | M/73 | C | IgM | IGHV1-2UM (subset 1) |
| p498 | M/62 | B | IgM | IGHV3-49UM |
| p524 | M/68 | C | IgM | IGHV3-11UM (subset 61) |
| p571 | M/58 | A | IgG | IGHV4-4M |
| p781 | F/58 | B | IgM | IGHV3-11UM (subset 61) |
| p814 | M/68 | C | IgM | IGHV3-66UM |
| p825 | F/71 | B | IgM | IGHV3-73UM |
| ID-5 | F/66 | A | IgG | IGHV3-33M |
| ID-6 | M/65 | C | IgM | IGHV1-69UM |
| ID-7 | F/54 | B | IgM | IGHV1-69UM (subset 6) |
| ID-11 | M/56 | C | IgM | IGHV3-21UM (subset 2) |
| ID-13 | M/70 | A | IgM | ND |
| ID-15 | M/50 | C | IgM | IGHV3-20UM |
| ID-20 | M/78 | B | IgM | IGHV4-34M |
| ID-23 | M/75 | C | IgM | IGHV3-21M (subset 2)‡ |
| p417† | M/69 | A | — | IGHV4-30UM |
| p432† | M/58 | A | — | IGHV1-69UM |
| p494† | M/71 | C | — | IGHV3-48UM (subset 23) |
| p562† | F/61 | A | — | IGHV4-59M |
| Patient ID . | Sex/age, y . | Stage, Binet . | Ig produced . | IGHV (CDR3 subset)* . |
|---|---|---|---|---|
| p239 | M/74 | A | IgM | IGHV1-69UM (subset 6) |
| p241 | M/63 | C | IgM | IGHV3-21UM |
| p457 | M/57 | B | IgM | IGHV1-18UM (subset 1) |
| p467 | F/61 | A | IgG | IGHV4-34M |
| p497 | M/73 | C | IgM | IGHV1-2UM (subset 1) |
| p498 | M/62 | B | IgM | IGHV3-49UM |
| p524 | M/68 | C | IgM | IGHV3-11UM (subset 61) |
| p571 | M/58 | A | IgG | IGHV4-4M |
| p781 | F/58 | B | IgM | IGHV3-11UM (subset 61) |
| p814 | M/68 | C | IgM | IGHV3-66UM |
| p825 | F/71 | B | IgM | IGHV3-73UM |
| ID-5 | F/66 | A | IgG | IGHV3-33M |
| ID-6 | M/65 | C | IgM | IGHV1-69UM |
| ID-7 | F/54 | B | IgM | IGHV1-69UM (subset 6) |
| ID-11 | M/56 | C | IgM | IGHV3-21UM (subset 2) |
| ID-13 | M/70 | A | IgM | ND |
| ID-15 | M/50 | C | IgM | IGHV3-20UM |
| ID-20 | M/78 | B | IgM | IGHV4-34M |
| ID-23 | M/75 | C | IgM | IGHV3-21M (subset 2)‡ |
| p417† | M/69 | A | — | IGHV4-30UM |
| p432† | M/58 | A | — | IGHV1-69UM |
| p494† | M/71 | C | — | IGHV3-48UM (subset 23) |
| p562† | F/61 | A | — | IGHV4-59M |
All patients were untreated at time of blood sampling.
M indicates mutated CLL; UM, unmutated CLL; ND, not determined; and — not applicable.
Cells from these patients were included only in the vimentin analysis (Figure 4C).
ID-23 has a 9-codon–long CDR3, but with IGHJ3 usage.
S pneumoniae polysaccharide-reacting CLL Abs
Reports on vimentin cross-reactivity of Ab to streptococcal antigens18 prompted us to test our CLL Abs for binding to S pneumoniae capsular polysaccharides (serotypes 4, 6B, 9V, 14, 18C, 19F, and 23F) by both solid- and fluid-based assays (ELISA and luminex assay). Non–serotype-restricted binding was demonstrated by Wa/IGHV3-30.3UM/subset-32 and ID-6/IGHV1-69UM. ID-20/IGHV4-34M showed a type 14–restricted reactivity. The Wa/IGHV3-30.3UM/subset-32 mAb dose-response binding to multiple polysaccharides is shown in Figure 2D. The remaining CLL mAbs included in Table 1 were negative (not shown).
Oxidized LDL and phosphorylcholine molecular motifs on apoptotic cells recognized by CLL mAbs
CLL Abs have previously been described as “natural Abs.”19-21 Interestingly, natural Abs in mice that bind S pneumoniae capsular polysaccharides were found to cross-react with moieties on oxidized LDL (oxLDL).22 Therefore, we wanted to further characterize the binding specificities of our CLL mAbs to the 2 most commonly used models of oxLDL (LDL oxidized by copper ions: CuOx-LDL; and LDL modified by malondialdhyde: MDA-LDL). Figure 3 shows results from a direct Ab-binding assay yielding both positive and negative CLL mAbs binding to MDA-LDL, CuOx-LDL, native-LDL, pneumococcal cell wall polysaccharides (CPSs), and phosphorylcholine-modified BSA (PC-BSA). Each of these 6 CLL IgM clones positive for binding to oxLDL had their own unique binding pattern: Wa/IGHV3-30.3UM/subset-32 and p241/IGHV3-21UM showed broad reactivity including both CuOx-LDL and MDA-LDL, and also native LDL; I83/IGHV3-30M reacted only with CuOxLDL and MDA-LDL, but not with native LDL. Interestingly, 2 of the patients, p457/IGHV1-18UM/subset-1 and p497/IGHV1-2UM/subset-1, belonging to the recently described HCDR3 homology subset 1 including IGHV genes of the same clan (IGHV1-2/IGHV1-3/IGHV1-18, IGHV5-a, IGHV7-4-1),8,14 reacted with MDA-LDL only. The specificity of the binding was verified in competition assays (Figure 3), which demonstrate that only MDA-LDL was able to specifically compete the binding of the clones. Recombinant vimentin was also included in the assays as a positive control and it demonstrated high specificity in competing for the I83/IGHV3-30M and Wa/IGHV3-30.3UM/subset-32 IgM clones (Figure 3). The oxLDL experiments were confirmed in 5 to 11 separate experiments.
CLL mAb binding to oxLDL determined in chemiluminescent ELISA. The 2 most frequently used models of oxLDL—CuOxLDL and MDA-LDL—were used. Abs against phosphorylcholine-BSA (PC-BSA), cell wall pneumococcal polysaccharides (CPSs), native LDL (nLDL), and BSA were also tested. Competition ELISA was performed for the oxLDL-reactive CLL mAb clones. B/B0 indicates the ratio between bound (cpm) and bound (cpm) without competitor.
CLL mAb binding to oxLDL determined in chemiluminescent ELISA. The 2 most frequently used models of oxLDL—CuOxLDL and MDA-LDL—were used. Abs against phosphorylcholine-BSA (PC-BSA), cell wall pneumococcal polysaccharides (CPSs), native LDL (nLDL), and BSA were also tested. Competition ELISA was performed for the oxLDL-reactive CLL mAb clones. B/B0 indicates the ratio between bound (cpm) and bound (cpm) without competitor.
Natural Abs to oxidized cardiolipin binds to oxLDL, apoptotic cells, and atherosclerotic lesions.23 Cardiolipin is found in membranes of bacteria, in the inner membrane of mitochondria and in plasma LDL. It was of interest, therefore, to analyze the CLL mAbs for cardiolipin. The results revealed that one of the clones (I83/IGHV3-30M) had a dose-response binding, whereas the others were negative (data not shown).
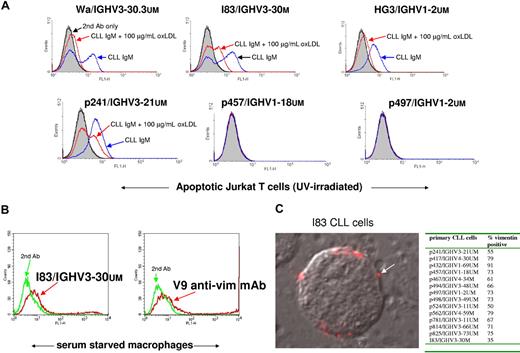
Oxidation moieties for natural Abs are not only found in oxLDL, but also in apoptotic cells.24 Therefore, the binding of CLL IgM to apoptotic cells was investigated. Figure 4A shows that the 4 mAbs binding to oxLDL (Wa/IGHV3-30.3UM/subset-32, I83/IGHV3-30M, HG3/IGHV1-2UM, p241/IGHV3-21UM) also bound to apoptotic Jurkat cells (apoptosis induced by UV light exposure). The specificity was verified by addition of 100 μg/mL oxLDL (MDA-LDL) into the incubation medium, which almost totally abolished the mAb binding to the apoptotic cells (Figure 4A). In addition to UV light, camptothecin, a topoisomerase I inhibitor, was also used for apoptosis induction in experiments confirming UV-induced apoptosis results (data not shown). In these experiments, annexin V–phycoerythrin (PE) or 7-amino-actinomycin D (7-ADD) was used to separate apoptotic from nonapoptotic cells during flow cytometry.
Binding of CLL Abs to apoptotic cells, macrophages, and membrane vimentin expression on live CLL cells. (A) CLL mAbs bind apoptotic Jurkat T cells. Jurkat T cells were UV-light exposed (51 mJ/cm2) for apoptosis induction. After 18 hours, CLL mAbs were incubated for 45 minutes followed by FITC–anti-IgM or FITC–anti-IgG. Blue line indicates the CLL mAb binding to apoptotic cells in the absence of the competitor, and the red line is in the presence of the competitor, 100 μg/mL MDA-LDL. Black line indicates secondary Ab only, and filled gray curve indicates cells only. (B) I83 IgM reacts with vimentin expressed on surface membrane of human macrophages. Purified macrophages cultured in 20% human serum for 6 days followed by 2 days of serum deprivation and analysis in FACS. Red line indicates cells binding the mAb, and green line indicates cells with secondary Ab only. (C) I83 CLL cells show distinct surface membrane staining (arrow) with antivimentin V9 mouse mAb. Secondary Ab was anti–mouse IgG–Alexa594. Overlay image with differential interference contrast. The insert table shows data from immunofluorescence analysis of membrane vimentin on 14 different primary CLL cells.
Binding of CLL Abs to apoptotic cells, macrophages, and membrane vimentin expression on live CLL cells. (A) CLL mAbs bind apoptotic Jurkat T cells. Jurkat T cells were UV-light exposed (51 mJ/cm2) for apoptosis induction. After 18 hours, CLL mAbs were incubated for 45 minutes followed by FITC–anti-IgM or FITC–anti-IgG. Blue line indicates the CLL mAb binding to apoptotic cells in the absence of the competitor, and the red line is in the presence of the competitor, 100 μg/mL MDA-LDL. Black line indicates secondary Ab only, and filled gray curve indicates cells only. (B) I83 IgM reacts with vimentin expressed on surface membrane of human macrophages. Purified macrophages cultured in 20% human serum for 6 days followed by 2 days of serum deprivation and analysis in FACS. Red line indicates cells binding the mAb, and green line indicates cells with secondary Ab only. (C) I83 CLL cells show distinct surface membrane staining (arrow) with antivimentin V9 mouse mAb. Secondary Ab was anti–mouse IgG–Alexa594. Overlay image with differential interference contrast. The insert table shows data from immunofluorescence analysis of membrane vimentin on 14 different primary CLL cells.
Human red blood cells (rbcs) were investigated for CLL mAb binding before and after UV light–induced apoptosis. None of the CLL mAbs reacted with viable or apoptotic rbcs (data not shown).
Based on previous reports that activated macrophages secrete vimentin, and expose it on the extracellular side of the plasma membrane,25 we further analyzed I83/IGHV3-30M IgM binding to serum-starved macrophages. Flow cytometry analysis of I83/IGHV3-30M IgM revealed binding to macrophage-plasma membrane (Figure 4B) similar in strength to the V9 antivimentin mouse mAb that was used as a positive control. The surface membrane vimentin expression was also investigated on 14 freshly thawed primary CLL cells. All CLL cells revealed surface membrane vimentin, exemplified with CLL I83 cells stained with antivimentin V9 mAb (Figure 4C).
Proline-rich acidic protein-1 (PRAP-1) and cofilin-1 are novel autoantigens identified in protein macroarrays
Protein macroarrays including 38 016 E coli–expressed proteins (in duplicates) derived from a human fetal brain cDNA library (complete or partial ORF clones) were developed with CII/IGHV1-69UM/subset-5, 232B4/IGHV3-48M, and rIGHV3–21M/subset-2 CLL mAbs (Figure 5). Secondary Ab–alone controls were included in each assay and regarded as negative. CII/IGHV1-69UM/subset-5 bound to 5/38 016 proteins; 232B4/IGHV3-48M bound to 4/38 016 proteins; and rIGHV3-21M/subset-2 bound to 4/38 016 proteins. Table 3 shows the detailed macroarray data including low-intensity binding reactions. The CII/IGHV1-69UM/subset-5 mAb reacted with 6 different clones, of which 2 reacted with PRAP-1, and one with Ig lambda–like polypeptide 1 isoform b precursor at medium intensity. The mAbs 232B4/IGHV3-48M and rIGHV3-21M/subset-2 bound to cofilin-1 with strong intensity, and medium intensity binding was observed for cleavage and polyadenylation specific factor 5. Figure 5 shows the digital image of a macroarray filter with the CII/IGHV1-69UM/subset-5, 232B4/IGHV3-48M, and rIGHV3-21M/subset-2 mAb patterns. Macroarray data were validated with purified His-tagged E coli–produced PRAP-1 or cofilin-1. CII/IGHV1-69UM/subset-5 showed specific binding to PRAP-1 in ELISA. The binding was dose-response inhibited by preincubation with recombinant PRAP-1. 232B4/IGHV3-48M and rIGHV3-21M/subset-2 mAbs showed binding to recombinant cofilin-1 only in complex with E coli extracts (IPTG-induced for cofilin-1 expression); there was no binding to E coli extracts alone or other E coli–expressing proteins (data not shown).
Three CLL mAb-binding patterns in protein macroarray of human fetal brain recombinant proteins (n = 38 016). Digital images from protein macroarray performed on CII/IGHV1-69UM/subset-5 mAb, 232B4/IGHV3-48M mAb, and rIGHV3-21M/subset-2 mAb. The membranes were incubated with CLL mAbs overnight at + 4°C followed by anti–human IgM/A/G-HRP secondary Ab using Lumiphos substrate for development. The E coli–expressed human fetal brain proteins are positioned in duplicates in specific patterns. Arrows show examples of duplicate positive reactions. Secondary Ab reactivities, nonduplicate, and low-avidity bindings were excluded.
Three CLL mAb-binding patterns in protein macroarray of human fetal brain recombinant proteins (n = 38 016). Digital images from protein macroarray performed on CII/IGHV1-69UM/subset-5 mAb, 232B4/IGHV3-48M mAb, and rIGHV3-21M/subset-2 mAb. The membranes were incubated with CLL mAbs overnight at + 4°C followed by anti–human IgM/A/G-HRP secondary Ab using Lumiphos substrate for development. The E coli–expressed human fetal brain proteins are positioned in duplicates in specific patterns. Arrows show examples of duplicate positive reactions. Secondary Ab reactivities, nonduplicate, and low-avidity bindings were excluded.
Summary of protein macroarray data (human fetal brain cDNA library) from the CLL mAb screening
| CLL antibody/protein . | Quality . | Clone ID . |
|---|---|---|
| CII/IGHV1-69UM subset-5 | ||
| Proline-rich acidic protein 1 | + | MPMGp800H11522 |
| Proline-rich acidic protein 1 | + | MPMGp800L07522 |
| Igλ-like polypeptide 1 isoform b precursor | + | MPMGp800L22599 |
| Cofilin 1 (nonmuscle) | ± | MPMGp800F21524 |
| Mannosidase, alpha, class 2C, member 1 | ± | MPMGp800E09593 |
| Predicted: similar to tripartite motif protein 11 | ± | MPMGp800F21600 |
| 232B4/IGHV3-48M | ||
| Cofilin 1 (nonmuscle) | ++ | MPMGp800F21524 |
| Cleavage and polyadenylation specific factor 5 | ± | MPMGp800N01597 |
| Predicted: similar to tripartite motif protein 11 | ± | MPMGp800F21600 |
| Igλ-like polypeptide 1 isoform b precursor | ± | MPMGp800L22599 |
| rIGHV3-21M subset-2 | ||
| Cofilin 1 (nonmuscle) | ++ | MPMGp800F21524 |
| Cleavage and polyadenylation specific factor 5 | + | MPMGp800N01597 |
| Signal peptide peptidase-like 2B isoform 2 | ± | MPMGp800M20598 |
| Igλ-like polypeptide 1 isoform b precursor | ± | MPMGp800L22599 |
| CLL antibody/protein . | Quality . | Clone ID . |
|---|---|---|
| CII/IGHV1-69UM subset-5 | ||
| Proline-rich acidic protein 1 | + | MPMGp800H11522 |
| Proline-rich acidic protein 1 | + | MPMGp800L07522 |
| Igλ-like polypeptide 1 isoform b precursor | + | MPMGp800L22599 |
| Cofilin 1 (nonmuscle) | ± | MPMGp800F21524 |
| Mannosidase, alpha, class 2C, member 1 | ± | MPMGp800E09593 |
| Predicted: similar to tripartite motif protein 11 | ± | MPMGp800F21600 |
| 232B4/IGHV3-48M | ||
| Cofilin 1 (nonmuscle) | ++ | MPMGp800F21524 |
| Cleavage and polyadenylation specific factor 5 | ± | MPMGp800N01597 |
| Predicted: similar to tripartite motif protein 11 | ± | MPMGp800F21600 |
| Igλ-like polypeptide 1 isoform b precursor | ± | MPMGp800L22599 |
| rIGHV3-21M subset-2 | ||
| Cofilin 1 (nonmuscle) | ++ | MPMGp800F21524 |
| Cleavage and polyadenylation specific factor 5 | + | MPMGp800N01597 |
| Signal peptide peptidase-like 2B isoform 2 | ± | MPMGp800M20598 |
| Igλ-like polypeptide 1 isoform b precursor | ± | MPMGp800L22599 |
Ab-binding quality/strengths are ++, +, and ±.
Figure 6 summarizes the CLL mAb specificities found in this study. Each CLL mAb-binding specificity is allocated to autoantigen/molecular motifs found on apoptotic cells or allocated to molecular motifs present on bacteria.
Summary of CLL mAb-binding specificities found in this study. Each CLL mAb-binding specificity is allocated to autoantigen/molecular motifs found on apoptotic cells or allocated to molecular motifs present on bacteria.
Summary of CLL mAb-binding specificities found in this study. Each CLL mAb-binding specificity is allocated to autoantigen/molecular motifs found on apoptotic cells or allocated to molecular motifs present on bacteria.
Specific antigen exposure increases CLL viability
We assessed whether specific antigen exposure had any consequences for CLL viability in growth factor withdrawal–induced apoptosis. I83 cells, with an IGHV3-30M antivimentin BCR, were exposed to solid-phase protein (0.5 μg vimentin/100 μL adsorbed to Nunc Maxisorb microplates [Rochester, NY]) for 48 hours in RPMI 1640 with 0.5% FCS. Flow analysis for annexin V–FITC and PI showed that control unstimulated cells incubated in medium alone had 59.0% viability after 48 hours, whereas in vimentin-exposed cultures 66.3% were viable. Anti-IgM–exposed cells (1.0 μg/100 μL) showed 66.9% viability. The experiment was repeated twice, also showing increased viability in vimentin-stimulated cultures. The effect was not seen in cultures of p498/IGHV3-49UM cells.
Discussion
The novel and previously unpublished finding of this study is that several CLL Ab clones react with molecular motifs present on apoptotic cells and bacteria. The molecular structures identified were vimentin, filamin B, cofilin-1, PRAP-1, phosphorylcholine, cardiolipin, oxLDL, and S pneumoniae polysaccharides. The CLL mAb-specificity patterns, in particular the observed antibacterial and anti-oxLDL antigens, are reminiscent of mouse natural Ab reactivities described in autoimmune diseases,26 and in clearance of senescent cells and pathogenic bacteria.22,23,27 Notably, microbial associations have been reported in several B-cell lymphomas,28-30 and recently it was suggested that respiratory tract infections increase the risk of CLL.31
CD5+ CLL B lymphocytes have previously been reported to produce natural autoAbs.11,19,32,33 Although less well defined in humans compared with mice, CD5+ B cells are considered to constitute a self-replenishing B-cell subset producing “natural IgM Abs” characterized by lack of somatic hypermutation, low affinity for microbes (eg, S pneumoniae), and autoreactivity. The unmutated CLL mAbs found in this study are polyreactive, thus resembling natural Abs. The mutated CLL clones tended to have a more restricted binding profile (eg, type 14 S pneumoniae polysaccharides and cofilin-1; Figure 6), confirming previous findings by Martin et al20,21 and recent studies by Herve et al11 showing that when in vitro–produced recombinant mutated nonautoreactive CLL mAbs were reverted to their original unmutated counterpart, they encoded polyreactive and autoreactive Abs.
Oxidation of LDL generates a wide variety of neoself determinants that have been shown to lead to a variety of cellular and humoral immune responses, which are important steps in early events of atherosclerosis.34 Although these neoself structures occur in oxLDL particles, such oxidation-specific epitopes can also be found in other proteins and phospholipids. Furthermore, oxLDL can induce humoral immune responses in vivo, and both IgG and IgM autoantibodies specifically binding to these oxidation-specific epitopes are found in plasma of animals and humans.34 Of particular relevance is the recent discovery that mice have “natural” germline Abs binding to oxidation-specific epitopes present on oxLDL, on apoptotic cells, as well as on certain microbial antigens.24,35 These earlier observations on mice together with the new findings of the present paper on human CLL clones further validate the concept that natural Abs, in both mice and humans, exhibit a remarkably conserved repertoire, and that they are typically regarded as polyreactive so that they bind to a number of self- or foreign antigens. This was evident especially among those CLL clones demonstrating binding to oxLDL. This broad reactivity pattern of innate Abs can be considered a requirement for the rapid and immediate recognition and protection against a variety different types of invading pathogens.22 On the other hand, the binding property to apoptotic cells may also represent physiological “housekeeping” roles in the recognition and removal of senescent cells, cell debris, and other self-antigens of the natural Abs.
It is intriguing that several of the autoantigens found in this study have functional association with microbial infections and/or apoptotic cell removal. Vimentin is a type III cytoskeletal protein and intermediate filament that recently was found to have functions on the extrafacial side of the plasma membrane,25 and was secreted as a stress response after bacterial insult.25,36 Vimentin is exposed on apoptotic primary T cells, oxLDL-binding macrophages, neutrophils, and platelets.37,38 Cell membrane–exposed vimentin was recently shown to be a port of entry for group A streptococci.39 Modified citrullinated vimentin (the “Sa” antigen), is an “eat me” signal for phagocytes, but can also generate autoAbs as demonstrated in patients with rheumatoid arthritis (RA).40 In this study, we found that the I83/IGHV3-30M and Wa/IGHV3-30.3UM/subset-32 mAbs reacted with vimentin (Figure 3) and showed binding to apoptotic Jurkat cells (Figure 4A). The I83/IGHV3-30M mAb was also found to bind to vimentin epitopes on serum-starved macrophages (Figure 4B), but lacked binding to the citrullinated vimentin epitope (data not shown). Antiphosphorylcholine Abs cross-react with vimentin,41 indicating that natural Abs may have a dual recognition for effective opsonization and removal of vimentin-coated microbes.
Filamin B was detected in Western blots, isolated, and identified in MS after SDS-PAGE separation (Figure 2B). Filamin A and filamin B are actin cross-linking filamentous proteins that are engaged in more than 40 molecular interactions such as with TRAF2, androgen receptors, dopamine receptors, and Smad.42 Filamin B is also exposed on the outer surface membrane and bind human natural IgM Abs that induce apoptosis in human neuroblastoma cells.43 Salmonella type III secretion effector proteins target filamin for successful invasion of epithelial cells.44 In this study, the HG3/IGHV1-2UM anti–filamin B mAb reacted with oxLDL epitopes and apoptotic Jurkat cells. The apoptotic cell binding was competed by oxLDL (MDA-LDL) in a dose-dependent fashion (Figure 4A).
Proline-rich acidic protein-1 (PRAP-1) is an epigenetically regulated protein preferentially found in epithelial cells of the gastrointestinal tract and uterus.45 It is up-regulated 30-fold and secreted during induction of terminal differentiation/apoptosis as a stress response protein.45 The biologic function is not known, but parallel studies reveal binding to bacterial surfaces of PRAP-1 (A.R. et al, unpublished data, November 2007), analogous to proline-rich antibacterial peptides (binding Gram-negative bacteria).46
Cofilin-1 is an actin-binding and depolarizing protein that colocalizes with Arp2/3 and the Wiskott-Aldrich syndrome protein (WASp) complex in apoptotic blebs during programmed cell death.47 WASp is required for efficient phagocytosis of apoptotic cells. During infection of epithelial cells by Gram-negative bacteria (eg, Salmonella, Chlamydia, Listeria), cofilin is being “hijacked” for a successful takeover of the cytoskeletal rearrangements.48 CLL clones 232B4/IGHV3-48M and rVH3-21M/subset-2 showed binding to cofilin-1 (Figure 5), but revealed no binding to oxLDL epitopes or apoptotic Jurkat cells (Figures 3,4). This observed lack of binding may have several explanations, including posttranslational modifications such as phosphorylation/dephosphorylation of cofilin-1 at Ser3 (epitope conformational changes), restricted cofilin-1 expression to specific time intervals, cell type–dependent expression, cryptic epitopes, low avidity, and others. The CLL anti–cofilin-1–binding pattern renders further studies on the epitope structure, expression in multiple cell types, and time-kinetic studies necessary.
One of the low-affinity reactions found in the protein macroarray (Table 3) is worth mentioning: cleavage and polyadenylation specific factor 5 (CPSF5) reacted with CLL mAb 232B4/IGHV3-48M. CPSF5 interacts directly with U2 snRNP,49 a nuclear complex structure, which is known to generate autoAbs in systemic lupus erythematosus (SLE) patients (anti-Sm, etc).50 Clearance of apoptotic cells is of outstanding importance for a balanced cellular homeostasis and involves a complex network of scavenger, recognition, decorating, and bridging molecules.51,52 In CLL, the CD5+ CLL cells are considered to be self-renewing, and our data revealing BCR–to–self-antigen recognition (on apoptotic cells) may have serious consequences if dysregulated. For example, overexpressed apoptotic molecular motifs (ie, membrane-exposed vimentin, as shown in Figure 4C and reported by others53,54 ) may generate uncontrolled (BCR)–proliferative signaling. This hypothesis, however, is not proven by our data and requires further studies. We suggest that these studies should include experiments that consider activation and proliferation signals generated by specific antigens alone or antigens exposed on apoptotic cells and bacteria, as well as signaling via CD3855 and its interaction with CD31, involved in “tethering” apoptotic cells.56 In addition, antiapoptotic signals in the proliferative environment such as stromal cell–derived thioredoxin,57 Bcl-2 overexpression,58 or DAPK1 down-regulation59 may contribute to a vicious circle.
In conclusion, the most important finding of this study is that molecular patterns on apoptotic cells were identified as neoautoantigen, specifically binding to CLL Igs encoded by the various IGHV genes. These results constitute a proof-of-principle that natural Abs previously described only in mice are also present in humans. The dual-recognition of the CLL mAbs is of interest in that they react with both bacterial components (S pneumoniae polysaccharides and phosphorylcholine) or self-antigens that have been reported to interact with microbes (vimentin, filamin B, cofilin-1) on one hand and motifs exposed on apoptotic blebs on the other hand (vimentin, PRAP-1, oxLDL, cardiolipin). Together with recent findings that pneumonial infections increase the risk of CLL,31 and preliminary observations on antibacterial CLL Abs,60 our data point at the possibility that bacterial infections in synergy with neo–self-antigen/apoptotic cells may drive promotion of CLL by continuously triggering the BCR, thus making the B-cell population vulnerable to tumor-transforming events.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors express their sincere thanks to Drs Kenneth Nilsson and Eva Klein for valuable discussion on the results of this study. Many thanks to Dr Shing Chuan Hooi, Singapore, for the generous gift of anti–PRAP-1 and recombinant PRAP-1, and also to Dr Johan Akter Hossain for cell culture work, Ulrik Lindquist for ELISA determination, Drs Alexander Vener and Maria Hansson for advice with MS, Dr Per Hultman for advice on immunohistologic evaluations, Drs Gunnar Juliusson and Karin Karlsson for help with recruiting patients, and to Marina Johnson for pneumococcal capsular polysaccharide–binding studies.
This work was supported by the Academy of Finland, Finnish Foundation for Cardiovascular Research and Sigrid Juselius foundation (S.H.); Special Trustees of Great Ormond St National Health Service Trust (H.B.); the Swedish Research Council, Swedish East Gothia Cancer Foundation, Linköping University Hospital Funds, and Swedish Cancer Association no. 3171-B04-16XBB.GSD.
Authorship
Contribution: A.L.M. and E.H. planned and performed research, analyzed data, and wrote the paper; E.S., A.S., H.B., C.D., G.T., E.B., and O.S. performed research and analyzed data; K.W. performed research; R.R. analyzed data and wrote the paper; S.H. performed research, analyzed data, and wrote the paper; A.R. designed and supervised the research, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anders Rosén, Department of Clinical and Experimental Medicine, Division of Cell Biology, Linköping University, SE-58185 Linköping, Sweden; e-mail: anders.rosen@ibk.liu.se.
References
Author notes
A.L.M. and E.H. contributed equally to this study.