Abstract
Microchimerism (MC), defined as the persistence of allogeneic cells at low concentrations, is well documented in transfused trauma patients. We hypothesized that genetic polymorphisms linked to cytokine production could contribute to trauma-induced immune modulation and development of microchimerism after transfusion of trauma patients. We used high-throughput SYBR-green-based genotyping of single nucleotide polymorphisms (SNPs) to characterize 59 transfused trauma patients, with MC (n = 30) and without MC (n = 29), for 4 functionally significant SNPs: TNF (−308), IL 10 (−1082), IFNG (+874), and TGFB1 (+915). We then compared likelihood for development of MC and the magnitude of immune suppression among subjects with and without these selected immune response SNPs. We identified a significant association between TNF (−308A) SNP and both development of MC and diminished immune responsiveness. Hence predisposing genetic factors may explain, in part, why only a subset of trauma patients develops transfusion-associated microchimerism.
Introduction
Microchimerism (MC) is the long-term persistence of a population of allogeneic cells within an individual and is associated with pregnancy, transplantation, and blood transfusion.1-7 We have reported that up to 10% of transfused trauma patients develop long-term (≥ 2 years) MC at levels as high as 3% to 5% of all circulating white blood cells (WBCs),6 whereas MC has not been observed after transfusion in other clinical settings where it might be expected, such as HIV8 and pediatric cardiac surgery (M.P.B., T.-H.L., W.F.R., unpublished data, October 1999). In addition to transfusion-associated MC, fetal-maternal MC occurs naturally but has also been implicated in the pathogenesis of several autoimmune diseases in adult women.9
Biologic conditions and mechanisms underlying the development of transfusion-associated MC (TA-MC) remain poorly understood. Curiously, in limited studies to date, HLA similarity does not appear to be necessary.4 We have presented evidence that the magnitude of short-term immune suppression associated with severe trauma and the degree of donor-recipient immunologic reactivity correlate with development of TA-MC.10 Because only a subset of massively transfused trauma patients develop long-term TA-MC, heterogeneity in the development of tolerance presumably also exists. Cytokine single nucleotide polymorphisms (SNPs) have been investigated in relation to solid organ allotransplant rejection with associations reported for tumor necrosis factor α (TNF) (−308A), interleukin-10 (IL 10) (−1082G), interferon γ (IFNG) (+874T), and transforming growth factor β1 (TGFB1) (+915C).11,12 These cytokines act as critical mediators of inflammation, are important in both adaptive and innate immune responses, and have been found in increased quantity in individuals with the above SNPs [except for TGFB1 (+915C), which is associated with decreased levels of TGFB1].13-15 Because these cytokines are involved in complex ways with dysregulated immune responses, we hypothesized that polymorphisms in these genes may also influence the likelihood for development of TA-MC. We investigated the association between the development of TA-MC and specific SNPs in cytokine genes by comparing the frequency of common SNPs in individuals who either developed or failed to develop TA-MC after transfusion for severe traumatic injury.
Methods
Patients and samples
Between September 18, 2000, and February 7, 2002, we enrolled a cohort of 63 transfused trauma patients at the University of California, Davis, Medical Center (UCDMC). TA-MC results6,16 and immunologic results10 from this cohort have been reported elsewhere. For the current study, we analyzed samples from all 59 of the 63 transfused trauma patients for whom there was an available cryopreserved sample.16 The study design was reviewed and approved by the Institutional Review Board at the University of California, Davis after subjects provided written informed consent in accordance with the Declaration of Helsinki.
SNP polymerase chain reaction genotyping
DNA extraction was performed as previously reported.16 Following our previously described protocol,17 SNP genotyping assays were optimized using DNA from random blood donor samples of known cytokine genotype. The primers were designed with the allele-differentiating base at the 3′ end of one primer. The primer pairs we used are as follows: ATAGGTTTTGAGGGGCATGG (TNF (−308) lower producing SNP forward primer), ATAGGTTTTGAGGGGCATGA (TNF (−308) higher producing SNP forward primer), GCCTTAGCCTCGTCCCTC (TNF (−308) reverse primer), CTACTAAGGGCTTCTTTGGGAA (IL 10(−1082) lower producing SNP forward primer), CTACTAAGGGCTTCTTTGGGAG (IL 10(−1082) higher producing SNP forward primer), GTTGAAGAAGGTGGGGTAGA (IL 10(−1082) reverse primer), GGTGCTGACGCCTGGCCC (TGFB1(+915) lower producing SNP forward primer), GGTGCTGACGCCTGGCCG (TGFB1(+915) higher producing SNP forward primer), CTCCGGTAGGCGCCGGTC (TGFB1(+915) reverse primer), TATTCTTACAACACAAAATCAAATCA (IFNG(+874) lower producing SNP forward primer), TATTCTTACAACACAAAATCAAATCT (IFNG(+874) higher producing SNP forward primer), and CGAGCATATTATTATGGGATG (IFNG(+874) reverse primer).
No mismatches were introduced in the primers. Annealing temperatures were 60°C, 60°C, 70°C, and 52°C, respectively. The results were assessed based on cycle threshold (Ct) differences between parallel amplifications with polymorphism-specific primers. Samples were tested in duplicate, and the average Ct was used for further analysis. The difference in Ct between sample DNA amplified with polymorphism-specific primers yields the delta Ct (high producing polymorphism Ct − low producing polymorphism Ct). A delta Ct greater than + 5 was assigned as the cutoff for designation of those patients homozygous for the low producing polymorphism. Heterozygous patients were identified when the delta Ct ranged from −2 to +2 and results indicating that the patient was homozygous for the high producing polymorphism were designated by a delta Ct less than −5. No samples had an ambiguous delta Ct between −5 and −2 or +2 and +5. Genotyped control samples representing the various combinations of polymorphisms were evaluated by delta Ct analysis, which identified the correct genotype with perfect accuracy (data not shown). Our experience with this amplification strategy in the study of extremely low-level MC allowed us to be confident that assay results represent the major (recipient) leukocyte population and not the genotype of any minor (donor) cell population that would be present in these patient samples at less than 0.1%.17,18 Thus, this technique is well suited to any targeted investigation of SNP status in patients who have recently undergone transfusion or transplantation or who have been pregnant.
Validation of polymerase chain reaction SNP status by direct sequencing
For assay validation, the regions flanking each cytokine SNP were amplified and sequenced directly. For TNF, we used forward primer TNF G4(F)GCCCCTCCCAGTTCTAGTTC and reverse primer TNF G6(R)GGATACCCCTCACACTCCC to amplify a product of 160 base pairs. For IL 10, we used forward primer IL 10G2(F)AATCCAAGACAACACTAC and the reverse primer IL 10G4(R)TCTGTGGCTGGAGTCTAAAG to amplify a 100 base pair product. For IFNG, we used forward primer IFNGG3(F)TCTATTACATCTACTGTGCC and a reverse primer IFNGG5(R)GCTGTTATAATTATAGCTGTC to amplify a 153 base pair product. For TGFB1, we used forward primer TGFB1G6(F)CGCGCTCTCGGCAGTGCC and a reverse primer TGFB1G3 (R)GGCGAGCCGCAGCTTGGAC to amplify a 180 base pair product. Sequencing was carried out as previously described.17
Statistical analysis
Each subject was classified as positive (n = 30) or negative (n = 29) for MC based on prior published results.16 Because it is associated with detection of less than or equal to 2 HLA class II alleles, TA-MC appears to involve only a single donor in cases we have encountered to date and correlates with neither the amount of blood transfused nor the injury severity. Each subject was also classified as positive or negative for each of the 4 cytokine polymorphisms. Data were evaluated using Fisher exact test, and a 2-sided P value was calculated using SAS Software (Cary, NC). For immunologic comparisons, we reanalyzed previously published data10 from these trauma patients characterizing the phytohemaglutinin (PHA) responsiveness of their lymphocytes and the intensity of recipient-to-donor mixed lymphocyte culture (MLC) with respect to the new SNP genotypic data. A stimulation index (SI) was calculated for both PHA and MLC. The SI is the ratio of scintillation counts per minute of experimental cultures relative to the mean counts per minute for control cultures. The SI difference associated with a particular cytokine SNP was determined using linear regression with clustering on recipients. Although injury severity has not been associated with probability for development of TA-MC in our limited data, it is associated with the degree of consequent change in subject immunologic responsiveness as measured by donor-to-recipient MLC. Therefore, we adjusted the data for Injury Severity Score, an indicator of the severity of anatomic injury, ranging from 0 (no injury) to 75 (uniformly fatal injury), and the SI difference we report is the change in stimulation index attributable to SNP status, after adjusting for Injury Severity Score.
Results and discussion
TNFα is a pleiotropic cytokine able to produce multiple phenotypic and functional effects. Importantly, TNFα has a marked capacity to induce apoptosis, so it is biologically plausible that overproduction of TNFα could lead to decreased immune responsiveness after trauma, a condition we think may be necessary but not sufficient for TA-MC to become established.
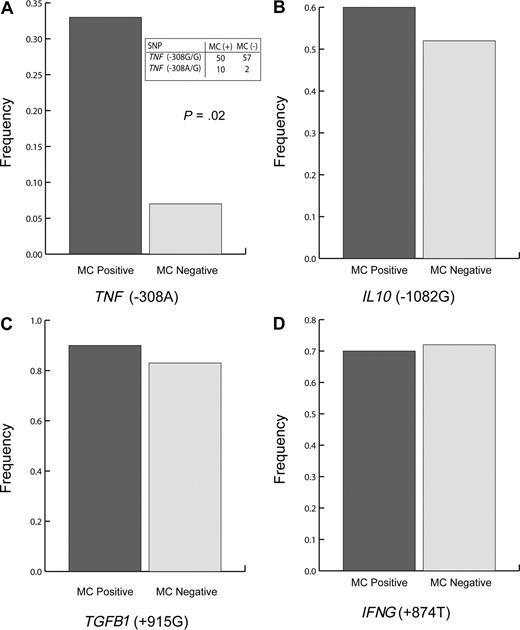
Frequencies of the higher producing polymorphisms [TNF (−308A), IL 10 (−1082G), TGFB1 (+915G), and IFNG (+874T)] in our test population were 10%, 37%, 93%, and 46%, respectively; no subject was homozygous for the overproducing TNF (−308A) SNP (Figure 1A). Presence of the TNF (−308A) SNP was associated with trends toward lower MLC responsiveness and reduced lymphocyte responsiveness to PHA (Table 1). The TNF (−308A) polymorphism was also significantly associated with the development of MC (Figure 1A), suggesting that individuals with the TNF (−308A) SNP experience immune suppression sufficient to allow for at least short-term tolerance to transfused allogeneic cells.
TNF (308A) polymorphism is associated with microchimerism in transfused trauma patients (P = .02). Polymorphism frequency of (A) TNF (−308A), (B) IL 10 (−1082G), (C) TGFB1 (+915G), and (D) IFNG (+874T) SNPs. Dark bars represent transfused trauma patients who were classified as MC positive (n = 30), and light bars represent transfused trauma patients who were classified as MC negative (n = 29). Two-sided Fisher exact test was used. Panel A includes an inset table depicting the relevant raw data for TNF (308) polymorphism.
TNF (308A) polymorphism is associated with microchimerism in transfused trauma patients (P = .02). Polymorphism frequency of (A) TNF (−308A), (B) IL 10 (−1082G), (C) TGFB1 (+915G), and (D) IFNG (+874T) SNPs. Dark bars represent transfused trauma patients who were classified as MC positive (n = 30), and light bars represent transfused trauma patients who were classified as MC negative (n = 29). Two-sided Fisher exact test was used. Panel A includes an inset table depicting the relevant raw data for TNF (308) polymorphism.
TNF (−308A) polymorphism is associated with reduced immune responsiveness, as measured by recipient to donor direction mixed lymphocyte culture (MLC), and is possibly also associated with reduced lymphocyte mitogen responsiveness (PHA)
| SNP . | MLC . | PHA . | ||
|---|---|---|---|---|
| SI difference, 95% CI* . | P† . | SI difference, 95% CI* . | P‡ . | |
| TNF (−308A) | −4.2 (−8.2 to −0.1) | 0.04 | −8.1 (−17.9 to 1.6) | .10 |
| IL 10 (−1082G) | 1.2 (−3.5 to 6.0) | 0.60 | −3.5 (−11.5 to 4.5) | .38 |
| TGFB1 (+915C) | −1.1 (−6.3 to 4.1) | 0.53 | −3.1 (−11.9 to 5.6) | .47 |
| IFNG (+874T) | 1.4 (−3.2 to 6.1) | 0.68 | −0.1 (−11.6 to 11.8) | .98 |
| SNP . | MLC . | PHA . | ||
|---|---|---|---|---|
| SI difference, 95% CI* . | P† . | SI difference, 95% CI* . | P‡ . | |
| TNF (−308A) | −4.2 (−8.2 to −0.1) | 0.04 | −8.1 (−17.9 to 1.6) | .10 |
| IL 10 (−1082G) | 1.2 (−3.5 to 6.0) | 0.60 | −3.5 (−11.5 to 4.5) | .38 |
| TGFB1 (+915C) | −1.1 (−6.3 to 4.1) | 0.53 | −3.1 (−11.9 to 5.6) | .47 |
| IFNG (+874T) | 1.4 (−3.2 to 6.1) | 0.68 | −0.1 (−11.6 to 11.8) | .98 |
Stimulation index of SNP, adjusted for Injury Severity Score.
P value calculated using linear regression, with clustering on recipients.
P value calculated using linear regression.
Numerous cytokine SNPs have been studied in relation to immune function, and strong associations have been established with the outcome of solid organ and hematopoietic stem cell transplantation,12,19,20 as well as with infection (viral, bacterial, and parasitic), shock, and autoimmune diseases.21 Mira et al reported an association of the higher producing TNF (−308A) SNP with septic shock susceptibility and mortality,22 and Imahara et al linked this SNP to increased risk of severe sepsis in trauma patients.23 Because our studies associate this SNP with development of MC and its antecedent reduced immune responsiveness after injury, there is need for further investigation into the relationship between specific cytokine responses and patient outcomes after transfusion for traumatic injury. In addition, given the proximity of the TNF (−308) polymorphism to HLA-B and HLA-DR loci, the role of HLA similarity in predicting the development of TA-MC (we have not observed this correlation to date) and the potential for linkage disequilibrium of TNF and HLA must also be explored further.24 Finally, although this polymorphism is now linked to magnitude of transfusion-induced immune suppression and propensity to develop TA-MC, its low sensitivity (17%) and negative predictive value (53%) for predicting which trauma patients will develop TA-MC suggest minimal diagnostic utility as a single test. A moderately high specificity (97%) and a modest positive predictive value (83%), on the other hand, support the relevance of the association described in this report, but, because it is likely that this immune pathway only explains a proportion of the overall determinants of TA-MC, there is need for more research to define other contributing host genetic, epigenetic, blood component, and donor-recipient compatibility factors.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by grants from the National Heart Lung and Blood Institute, Special Center of Research in Transfusion Medicine (P50-HL-54 476, M.P.B.), and the National Institutes of Health (R01-HL-083388, M.P.B.).
National Institutes of Health
Authorship
Contribution: R.M.G. designed and performed research, analyzed data, and wrote and revised the paper; T-H.L. designed research; G.H.U. analyzed data and helped revise the paper; W.F.R. analyzed data and helped revise the paper; L.W. performed research; D.C. performed research; and M.P.B. designed research, analyzed data, and helped revise the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Michael P. Busch, Blood Systems Research Institute, 270 Masonic Avenue, San Francisco, CA 94118; e-mail: mbusch@bloodsystems.org.