Abstract
Vascular endothelial and myeloid cells have been proposed to originate from a common precursor cell, the hemangioblast. The mechanism of endothelial and myeloid cell specification and differentiation is poorly understood. We have previously described the endothelial-specific zebrafish Ets1-related protein (Etsrp), which was both necessary and sufficient to initiate vasculogenesis in the zebrafish embryos. Here we identify human Etv2/ER71 and mouse ER71 proteins as functional orthologs of Etsrp. Overexpression of mouse ER71 and Etsrp caused strong expansion of hemangioblast and vascular endothelial lineages in a zebrafish embryo. In addition, we show that etsrp is also required for the formation of myeloid but not erythroid cells. In the absence of etsrp function, the number of granulocytes and macrophages is greatly reduced. Etsrp overexpression causes expansion of both myeloid and vascular endothelial lineages. Analysis of mosaic embryos indicates that etsrp functions cell autonomously in inducing myeloid lineage. We further demonstrate that the choice of endothelial versus myeloid fate depends on a combinatorial effect of etsrp, scl, and alk8 genes.
Introduction
Hematopoiesis, the differentiation of hematopoietic stem cells into various blood cell lineages, and vasculogenesis, the differentiation of endothelial cell progenitors into vascular endothelial cells, are closely related in many different organisms. Endothelial and hematopoietic cells emerge in the embryo in close proximity and time, suggesting the possibility of a common progenitor, the hemangioblast.1 In the extraembryonic visceral yolk sac of a mouse embryo, vascular plexus and blood islands are closely associated with each other and share expression of multiple genes such as flk1, flt1, tie1, tie2, and others.2 Differentiating embryonic stem cells contain blast colony-forming cells which can differentiate into both hematopoietic and endothelial cells in vitro.3 In the zebrafish, which recently emerged as an excellent model system for the studies of vertebrate hematopoiesis and vasculogenesis/angiogenesis, both primitive hematopoietic and endothelial progenitors originate within lateral plate mesoderm, later forming an intermediate cell mass region, equivalent to the mouse blood islands.4,5 The zebrafish cloche mutants are deficient in both vascular endothelial and hematopoietic lineages.6 Zebrafish vascular endothelial and hematopoietic cell progenitors also share expression of common markers such as a basic helix-loop-helix transcription factor scl/tal17,8 and an ETS domain transcription factor fli1.9 Overexpression of scl can induce both hematopoietic and endothelial markers7,8,10 whereas knockdown of scl results in defective hematopoietic and endothelial development.11,12 Single-cell labeling in a zebrafish gastrula embryo demonstrated that individual cells can give rise to both erythroid and endothelial cells,13 providing a definitive evidence for the existence of hemangioblasts in vivo.
In the zebrafish, the anterior lateral plate mesoderm gives rise to the progenitors of myeloid cells and head vessels while the posterior lateral plate mesoderm gives rise to the primitive erythroid cells, axial, and intersegmental blood vessels.14 Two recent studies have demonstrated that the interplay between an early myeloid-specific transcription factor pu.1/spi1 and an erythroid-specific gata1 determines the choice between myeloid and erythroid fates in the progenitor cells.15,16 Pu.1 is first expressed in both myeloid and erythroid cell progenitors, and later its erythroid-specific expression is repressed by gata1, ensuring that pu.1 expression remains restricted to the myeloid cells within the anterior lateral mesoderm. Scl signaling is critical for myeloid development, as scl-deficient embryos lack pu.1-expressing cells.12 BMP receptor Alk8 signaling functions in parallel to scl, also providing an instructive input for myelopoiesis.17 However, the mechanism of transition from the putative hemangioblast to the myeloid and endothelial lineages is still unknown. Although a recent study demonstrated the existence of the common erythroid-endothelial progenitor cells,13 myeloid-endothelial progenitor cells have not been analyzed in this study. Therefore, the definitive evidence for the existence of a common myeloid-endothelial hemangioblast progenitor cell is still missing.
We have recently described a novel ets1-related protein, etsrp.18 Etsrp expression is localized to the presumptive vascular progenitor cells from the early somitogenesis stages. Knockdown of etsrp results in the complete lack of circulation. Angioblasts in etsrp-deficient embryos do not differentiate, fail to migrate toward midline, and do not express other vascular markers. Scl expression is strongly down-regulated in the anterior lateral mesoderm in etsrp-morpholino (MO)–injected embryos (morphants) while its posterior erythroid-specific expression is not affected. Consistent with this, erythroid lineage is not affected in etsrp morphants. Overexpression of etsrp RNA is sufficient to induce expression of endothelial and hemangioblast markers, including scl, in many different regions of an embryo. These data show that etsrp is both necessary and sufficient for the initiation of vasculogenesis in a zebrafish embryo. Although this study clearly established etsrp as a critical regulator of zebrafish vasculogenesis, it was not clear whether etsrp function is conserved in other vertebrates, including mammals.
In the current study, we demonstrate by the phylogeny analysis that zebrafish etsrp is related to the mouse ER71 and human etv2/ER71 genes. We show that mouse ER71 is functionally related to Etsrp by performing overexpression of both proteins in zebrafish embryos. We also further investigate etsrp function within the putative hemangioblast cells. We show that etsrp is both necessary and sufficient to initiate the myeloid cell formation, in addition to the endothelial lineage. We demonstrate that etsrp function is specific to the anterior but not the posterior hemangioblasts, thus providing the first distinction between the 2 pools. We also show that as the endothelial and myeloid lineages separate, etsrp is excluded from the myeloid lineage, and remains restricted to the endothelial cell precursors. Finally, we show that etsrp-expressing cells can give rise to both myeloid and endothelial lineages, supporting the existence of a common myeloid-endothelial cell progenitor, the hemangioblast.
Methods
Microinjection of MOs
In all experiments, unless otherwise noted, 8 ng to 10 ng etsrp-specific MO1 and MO2 mixture (4-5 ng each) was injected at the 1- to 2-cell stage.18 Typically, 2 independent experiments were performed, with at least 15 to 20 embryos per experiment analyzed. Penetrance of observed phenotypes was greater than 90%, unless otherwise noted. Other morpholinos used: 1.2 ng alk8 MO: ACAACTCCTCAAGTGACTCTCAGCG (Open Biosystems, Huntsville, AL)19 ; scl MO: GCTCGGATTTCAGTTTTTCCATCAT (Open Biosystems)18 ; 8 ng pu.1 MO: GATATACTGATACTCCATTGGTGGT15 (kind gift of J.P. Kanki, Dana-Farber Cancer Institute, Boston, MA); 7 ng gata1-ATG MO: CTGCAAGTGTAGTATTGAAGATGTC16 (kindly donated by L. Zon, Harvard Medical School, Boston, MA).
Overexpression and epistasis experiments
etsrp mRNA (75-150 pg) was injected into the zebrafish embryos at the 1- to 8-cell stages.18 Approximately 100 pg scl mRNA8 and 10 pg CA-alk8 mRNA19 (kindly donated by M. Hammerschmidt, Max Planck Institute for Immunobiology, Freiburg, Germany) was used in the overexpression and epistasis experiments. etsrp DNA microinjection construct was made by subcloning full-length etsrp cDNA into the HindIII site of pXeX.20 Circular etsrp-XeX plasmid (25-75 pg) was injected into the blastomere at the 1-cell stage. hER71 coding sequence was cloned into pCMV-SPORT6 vector (Open Biosystems). hER71 DNA (75-100 pg) was injected into the flk1-GFP zebrafish embryos at the 1-cell stage. mER71 coding sequence has been subcloned into pcDNA3.1/V5-His vector (Invitrogen, Carlsbad, CA). mER71 DNA (75 pg) was injected in overexpression experiments. For hEts1 RNA injections, hEts1 ORF (kindly provided by P. Oettgen, Beth Israel Deaconess Medical Center, Boston, MA) has been subcloned into the SpeI site of pT3TS,21 subsequently linearized with BamHI and transcribed with T3 mMessage Machine kit (Ambion, Austin, TX). For hEts1 DNA injections, 200 pg hEts1 cDNA in pSG5neo vector (Stratagene, La Jolla, CA; kindly provided by R. Forough, Texas A&M University, College Station, TX) was injected.
In situ hybridization
In situ hybridization was performed as described.22 For the 2-color conventional in situ hybridization, the same protocol was followed except that embryos were incubated with 2 probes, etsrp-labeled with fluorescein-UTP (Roche, Indianapolis, IN), and the other cDNA labeled with digoxygenin-UTP (Roche). Following the first color (blue) development, embryos were fixed in the 4% paraformaldehyde/phosphate-buffered saline (PBS) solution for 1 hour, washed in PBS with 2% bovine serum albumin and 2% Tween 20 (PBT),22 incubated with anti–fluorescein antibody conjugated with alkaline phosphatase (Roche) overnight at 4°C, washed in PBT, and developed using 5-Bromo-4-chloro-3-indolyl β-D-galactopyranoside (X-gal; Sigma-Aldrich, St Louis, MO) and iodonitrotetrazolium chloride (INT; Sigma-Aldrich) as the substrate for red color. To detect fluorescein-dextran and pu.1 localization, a fluorescent 2-color in situ protocol was followed (J. Schoeneback, B. Keegan, and D. Yelon, unpublished). Briefly, fixed embryos were hybridized with DIG-labeled pu.1 probe at 65°C, washed in 0.2× saline-sodium citrate (SSC), blocked in 1× blocking reagent (Roche), incubated with 1:500 anti-DIG POD (Roche), washed in PBT, incubated with DNP-tyramide at 1:50 (Perkin Elmer, Waltham, MA), washed in PBT, blocked in 1× blocking reagent, incubated with anti-DNP POD at 1:500 (Perkin Elmer), washed in PBT, incubated with Cy3-tyramide at 1:25 (Perkin Elmer), sequentially washed in PBT, 1% H2O2, PBT, 0.1 M pH 2.2 glycine, PBT, blocked in 1× blocking reagent, incubated with anti-FITC POD at 1:1000 (Roche), washed in PBT, incubated with FITC-tyramide (Perkin Elmer), washed in PBT, and imaged as described below in “Image processing and analysis.” The following probes were used: etsrp23 ; pu.124 ; gata125 ; flk126 ; scl8 ; mpx27 (made by subcloning mpx cDNA into the pCR4 vector; Invitrogen; kindly provided by J. Larson, University of Minnesota, Minneapolis/St Paul); lcp128 (made by subcloning lcp1 cDNA into the pCR4 vector; kindly provided by J. Larson).
Transplantation
Donor embryos (wt or flk1-GFP homozygous) were injected with a mixture of etsrp RNA (100 pg) and fluorescein isothiocyanate-dextran (2 ng; molecular weight [Mw] 2 MDa; Sigma-Aldrich) or tetramethyl rhodamine isothiocyanate-dextran (2 ng; Mw 2 MDa; Sigma-Aldrich) into the blastomere at the 1-cell stage. Recipient embryos (wt or flk1-GFP) were injected with 7.5 ng to 8 ng etsrp MO1 and MO2 1:1 mixture at the 1- to 2-cell stage. Embryos were transferred to Danieau solution and manually dechorionated prior to transplantation. Ten to 50 cells were transplanted at the sphere to 30% epiboly stages by using capillary needles and adjusting balance pressure of PLI-100 microinjector (Harvard Apparatus, Holliston, MA) to move cells up and down the needle. Embryos were fixed at the 8- to 10-somite stages and analyzed for pu.1 expression and the presence for fluorescein. Two-color conventional or fluorescent in situ hybridization protocol was followed to detect DIG-labeled pu.1 and fluorescein (only pu.1 probe was used).
Image processing and analysis
Tetramethylrhodamine isothiocyanate (TRITC)–labeled live flk1-GFP embryos or FITC-labeled pu.1-Cy3 hybridized embryos were imaged using a monochrome CCD camera (Hamamatsu C4742-95) on a compound microscope (Axioplan; Zeiss, Thornwood, NY) equipped with rhodamine and green fluorescent protein (GFP) filter sets. To image stained embryos after in situ, they were either positioned in 1× PBS on the agarose-coated dishes, or they were manually deyolked and mounted in 50% glycerol (2-color in situ) or araldite (single-color). Images were taken using a color digital CCD camera (Axiocam; Zeiss) and a dissecting (Stemi SV11; Zeiss) or a compound microscope (Axioskop 2; Zeiss) with ×5, ×10, or ×20 objective. Typically, frames in different focus planes were manually combined using Adobe Photoshop to yield the maximum clarity image. During image analysis of transplanted embryos, color levels, hue, and saturation were adjusted for the whole image. Color of the transplanted cells was analyzed using the eyedropper tool in Photoshop, averaged from 7 to 10 samplings, and normalized for the intensity in red channel. The following color ratios were calculated for the cells 1 to 5 (Figure 6H) channels red/green/blue: (cell 1) 1 : 1.02 ± 0.03 : 1.28 ± 0.12; (cell 2) 1 : 0.74 ± 0.05 : 0.56 ± .05; (cell 3) 1 : 0.54 ± 0.10 : 0.59 ± 0.11; (cell 4) 1 : 0.77 ± 0.05 : 0.93 ± 0.09; (cell 5) 1 : 0.89 ± 0.04 : 1.03 ± 0.05. The ratios were used to select representative colors shown in the rectangles in Figure 6H.
Results
mER71 and hEtv2/ER71 proteins are functional orthologs of zebrafish Etsrp
The previous study suggested that zebrafish Etsrp may be evolutionary related to mammalian Ets1 proteins.18 However, the homology between Etsrp and Ets1 proteins is limited to the ETS DNA binding domain. Other domains such as Transactivation and Pointed domains, present in other Ets1 family members, are not recognizable within the Etsrp sequence. To identify a potential Etsrp ortholog in other vertebrates, we performed phylogenetic analysis of the sequences and protein structure of different Ets family members. ER71 proteins from different vertebrates, including medaka (Oryzias latipes), stickleback (Gasterosteus aculeatus), frog (Xenopus laevis), mouse, and humans appeared as potential orthologs of Etsrp (Figure 1). Mouse ER71 was originally isolated as a testis-specific factor 1,29,30 but its embryonic expression has not been previously investigated. Etsrp sequence is 57% and 61%, respectively, identical to its medaka and stickleback homologs, while it displays a lower identity of 26% (37% similarity) to the human ER71 protein and 26% identity (36% similarity) to the mouse ER71 (Figure 1A). Etsrp and hER71 proteins are 71% similar within the ETS domain region, whereas homology is much lower within the rest of the sequence. Interestingly, similar to Etsrp, other vertebrate ER71 proteins have no recognizable structural domains outside the ETS domain. Recent functional studies determined a potent transactivation domain present within the mouse ER71 N-terminal region.30 Syntenic analysis shows that vertebrate etsrp/ER71 chromosomal regions are highly conserved (Figure 1C).
Etsrp is a functional ortholog of the mammalian ER71 subfamily. (A) Alignment of zebrafish Etsrp, medaka, stickleback, mouse, and human ER71 amino acid sequences. Identical and similar amino acids are labeled in red and blue, respectively. Etsrp and hER71 share 71% homology within the ETS DNA-binding domain (underlined in gray). GenBank accession numbers used for the analysis are as follows: human ER71 (O00321), mouse ER71 (NP_031985), and zebrafish ETSRP (AAY89037). Medaka ER71 (ENSORLP00000019929) and stickleback ER71 (ENSGACP00000016315) are Ensembl predictions. (B) Phylogenetic analysis of zebrafish Etsrp and its closest human, mouse, frog, and fish homologs. The phylogenetic tree is built using the Neighbor Joining method. Length of horizontal branches is proportional to the evolutionary distance between the protein molecules. GenBank accession numbers used for the analysis are as follows: human ER71 (O00321), mouse ER71 (NP_031985), zebrafish ETSRP (AAY89037), human ETS1 (NP_005229), mouse ETS1 (NP_035938), frog ETS1 (NP_001081621), zebrafish ETS1a (NP_001017558), human ETS2 (NP_005230), mouse ETS2 (NP_035939), frog ETS2 (NP_001081007), zebrafish ETS2 (NP_001018874), human FLI1 (NP_002008), mouse FLI1 (NP_032052), zebrafish FLI1a (NP_571423), and zebrafish FLI1b (NP_001008780). Medaka ER71 (ENSORLP00000019929), stickleback ER71 (ENSGACP00000016315), fugu ETS1 (SINFRUP00000163510), and medaka ETS1 (ENSORLP00000016939) are Ensembl predictions. NTI Vector (Invitrogen) has been used to build the alignment and the phylogenetic tree. (C) Chromosomal location of the zebrafish etsrp, medaka, stickleback, mouse, and human ER71 genes. Numbers alongside the chromosomal regions of interest correspond to the actual physical distances (Mb). Etsrp/ER71 genes are highlighted in red. (D-J) Mouse ER71 and zebrafish Etsrp overexpression causes ectopic expression of hemangioblast marker scl and vascular endothelial marker flk1. flk1-GFP transgenic embryos were injected with 75 pg of either Etsrp or mER71 DNA at the 1-cell stage and analyzed at 8- to 10-somite stages. Relative to uninjected embryos (D,G), both Etsrp (E) and mER71 (F,H) result in the ectopic induction of scl when examined by in situ hybridization. Flk1 expression was also induced by ER71 injections as revealed by ectopic GFP expression (J) relative to uninjected controls (I). Panels G to J are lateral views with anterior to the left. Images were taken using Zeiss CV11 stereomicroscope, Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision, Waltham, MA). Magnification: 12× (D-F); 60× (G-J).
Etsrp is a functional ortholog of the mammalian ER71 subfamily. (A) Alignment of zebrafish Etsrp, medaka, stickleback, mouse, and human ER71 amino acid sequences. Identical and similar amino acids are labeled in red and blue, respectively. Etsrp and hER71 share 71% homology within the ETS DNA-binding domain (underlined in gray). GenBank accession numbers used for the analysis are as follows: human ER71 (O00321), mouse ER71 (NP_031985), and zebrafish ETSRP (AAY89037). Medaka ER71 (ENSORLP00000019929) and stickleback ER71 (ENSGACP00000016315) are Ensembl predictions. (B) Phylogenetic analysis of zebrafish Etsrp and its closest human, mouse, frog, and fish homologs. The phylogenetic tree is built using the Neighbor Joining method. Length of horizontal branches is proportional to the evolutionary distance between the protein molecules. GenBank accession numbers used for the analysis are as follows: human ER71 (O00321), mouse ER71 (NP_031985), zebrafish ETSRP (AAY89037), human ETS1 (NP_005229), mouse ETS1 (NP_035938), frog ETS1 (NP_001081621), zebrafish ETS1a (NP_001017558), human ETS2 (NP_005230), mouse ETS2 (NP_035939), frog ETS2 (NP_001081007), zebrafish ETS2 (NP_001018874), human FLI1 (NP_002008), mouse FLI1 (NP_032052), zebrafish FLI1a (NP_571423), and zebrafish FLI1b (NP_001008780). Medaka ER71 (ENSORLP00000019929), stickleback ER71 (ENSGACP00000016315), fugu ETS1 (SINFRUP00000163510), and medaka ETS1 (ENSORLP00000016939) are Ensembl predictions. NTI Vector (Invitrogen) has been used to build the alignment and the phylogenetic tree. (C) Chromosomal location of the zebrafish etsrp, medaka, stickleback, mouse, and human ER71 genes. Numbers alongside the chromosomal regions of interest correspond to the actual physical distances (Mb). Etsrp/ER71 genes are highlighted in red. (D-J) Mouse ER71 and zebrafish Etsrp overexpression causes ectopic expression of hemangioblast marker scl and vascular endothelial marker flk1. flk1-GFP transgenic embryos were injected with 75 pg of either Etsrp or mER71 DNA at the 1-cell stage and analyzed at 8- to 10-somite stages. Relative to uninjected embryos (D,G), both Etsrp (E) and mER71 (F,H) result in the ectopic induction of scl when examined by in situ hybridization. Flk1 expression was also induced by ER71 injections as revealed by ectopic GFP expression (J) relative to uninjected controls (I). Panels G to J are lateral views with anterior to the left. Images were taken using Zeiss CV11 stereomicroscope, Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision, Waltham, MA). Magnification: 12× (D-F); 60× (G-J).
To test if mammalian ER71 and zebrafish Etsrp proteins are functionally conserved, we injected DNA encoding mouse and human ER71 into early zebrafish embryos. Overexpression of both mER71 and hER71 resulted in the strong expansion of hemangioblast marker scl and vascular endothelial flk1-GFP expression, similar to the effect of etsrp expression (Figure 1D-J; data not shown). On the other hand, overexpression of related human Ets1 DNA and RNA caused no apparent effect on early development and no increase in flk1-GFP expression (data not shown). These results argue that ER71 proteins, similar to Etsrp, can initiate vasculogenesis in the zebrafish embryos, therefore Etsrp and ER71 but not Ets1 proteins are functional orthologs.
Etsrp is necessary and sufficient for the formation of myeloid cells
In the previous study, we showed that scl expression in the anterior lateral mesoderm is nearly completely missing in etsrp morphants.18 Scl knockdown has been previously shown to affect the myeloid lineage.12 Therefore, we investigated if myeloid cells were affected in etsrp morphants. L-plastin (lcp1) expressing macrophages28 and neutrophils31 and myeloid peroxidase (mpx) expressing heterophilic granulocytes27,32 were nearly completely absent in etsrp morphants (Figure 2A-D). Expression of an early myeloid marker pu.124 was strongly down-regulated in the anterior lateral plate mesoderm, whereas the posterior erythroid-specific pu.1 expression was not significantly affected (Figure 2E,F). These results show that etsrp function is required for the formation of myeloid lineage, in addition to its previously demonstrated requirement for endothelial lineage.18 Control 5-base mismatch morpholino caused no noticeable defects in embryonic development, including myeloid, hemangioblast, and endothelial marker expression,18 confirming the specificity of the observed phenotypes (Figure S1, available on the Blood website; see the Supplemental Materials link at the top of the online article; data not shown).
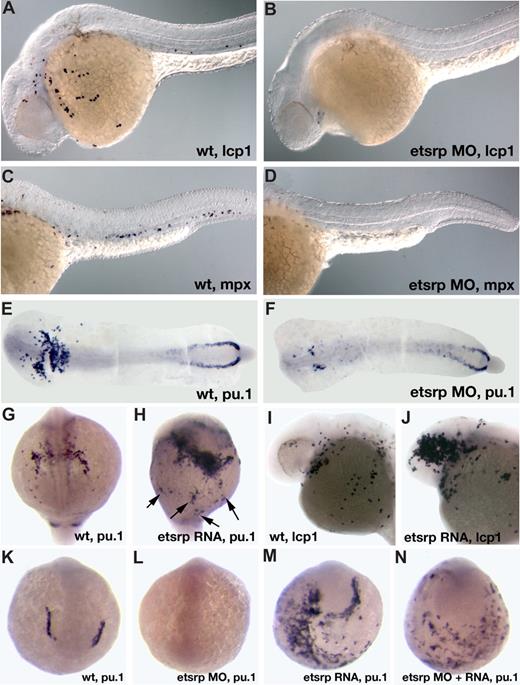
Etsrp is necessary and sufficient for the formation of myeloid cells. (A-F) Knockdown of Etsrp results in the nearly complete absence of myeloid cells as analyzed by in situ hybridization. Anterior is to the left. (A,C,E) Control uninjected embryos. (B,D,F) Etsrp morphants, injected with 12 ng to 15 ng of etsrp MO1 and etsrp MO2 in a 1:1 mixture. (A,B) L-plastin (lcp1) expression at 24 hours postfertilization (hpf). Note that lcp1-expressing macrophages are nearly completely absent in etsrp morphants (B). (C,D) mpx expression at 24 hpf. Note that mpx-expressing neutrophils are nearly completely absent in etsrp morphants (D). (E,F) pu.1 expression at the 16-somite stage. Embryos have been flat-mounted with the yolk removed. Note that the anterior myeloid-specific pu.1 expression is severely reduced in panel F, whereas posterior erythroid-specific expression is not significantly affected. (G-J) Etsrp RNA overexpression induces ectopic myeloid cell formation. (G,I) Control uninjected embryos. (H,J) etsrp RNA-overexpressing embryos. (G,H) pu.1 expression at the 16-somite stage, anterior view (G), ventro-lateral view (H). Note the strong expansion of pu.1-expressing cells, some of which are located ectopically (H,  ). (I,J) lcp1 expression at 24 hpf. Note the increase in the number of lcp1-expressing macrophages in panel J. (K-N) Etsrp RNA with missing MO-binding sites can restore pu.1 expression in etsrp morphants. Embryos are at the 8-somite stage; (K,L) anterior view; (M,N) anterior-ventral view. (K) Control uninjected embryo; (L) 10 ng etsrp MO2-injected embryo; (M) 100 pg etsrp RNA-injected embryo; (N) embryo coinjected with 10 ng etsrp MO2 and 100 pg etsrp RNA. Images were taken using Axioskop2 and 10×/0.30 NA dry objective (Zeiss) (A-F; I,J) or CV11 stereomicroscope (Zeiss) (G,H,K-N), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A-D); 60× (E-N).
). (I,J) lcp1 expression at 24 hpf. Note the increase in the number of lcp1-expressing macrophages in panel J. (K-N) Etsrp RNA with missing MO-binding sites can restore pu.1 expression in etsrp morphants. Embryos are at the 8-somite stage; (K,L) anterior view; (M,N) anterior-ventral view. (K) Control uninjected embryo; (L) 10 ng etsrp MO2-injected embryo; (M) 100 pg etsrp RNA-injected embryo; (N) embryo coinjected with 10 ng etsrp MO2 and 100 pg etsrp RNA. Images were taken using Axioskop2 and 10×/0.30 NA dry objective (Zeiss) (A-F; I,J) or CV11 stereomicroscope (Zeiss) (G,H,K-N), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A-D); 60× (E-N).
Etsrp is necessary and sufficient for the formation of myeloid cells. (A-F) Knockdown of Etsrp results in the nearly complete absence of myeloid cells as analyzed by in situ hybridization. Anterior is to the left. (A,C,E) Control uninjected embryos. (B,D,F) Etsrp morphants, injected with 12 ng to 15 ng of etsrp MO1 and etsrp MO2 in a 1:1 mixture. (A,B) L-plastin (lcp1) expression at 24 hours postfertilization (hpf). Note that lcp1-expressing macrophages are nearly completely absent in etsrp morphants (B). (C,D) mpx expression at 24 hpf. Note that mpx-expressing neutrophils are nearly completely absent in etsrp morphants (D). (E,F) pu.1 expression at the 16-somite stage. Embryos have been flat-mounted with the yolk removed. Note that the anterior myeloid-specific pu.1 expression is severely reduced in panel F, whereas posterior erythroid-specific expression is not significantly affected. (G-J) Etsrp RNA overexpression induces ectopic myeloid cell formation. (G,I) Control uninjected embryos. (H,J) etsrp RNA-overexpressing embryos. (G,H) pu.1 expression at the 16-somite stage, anterior view (G), ventro-lateral view (H). Note the strong expansion of pu.1-expressing cells, some of which are located ectopically (H,  ). (I,J) lcp1 expression at 24 hpf. Note the increase in the number of lcp1-expressing macrophages in panel J. (K-N) Etsrp RNA with missing MO-binding sites can restore pu.1 expression in etsrp morphants. Embryos are at the 8-somite stage; (K,L) anterior view; (M,N) anterior-ventral view. (K) Control uninjected embryo; (L) 10 ng etsrp MO2-injected embryo; (M) 100 pg etsrp RNA-injected embryo; (N) embryo coinjected with 10 ng etsrp MO2 and 100 pg etsrp RNA. Images were taken using Axioskop2 and 10×/0.30 NA dry objective (Zeiss) (A-F; I,J) or CV11 stereomicroscope (Zeiss) (G,H,K-N), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A-D); 60× (E-N).
). (I,J) lcp1 expression at 24 hpf. Note the increase in the number of lcp1-expressing macrophages in panel J. (K-N) Etsrp RNA with missing MO-binding sites can restore pu.1 expression in etsrp morphants. Embryos are at the 8-somite stage; (K,L) anterior view; (M,N) anterior-ventral view. (K) Control uninjected embryo; (L) 10 ng etsrp MO2-injected embryo; (M) 100 pg etsrp RNA-injected embryo; (N) embryo coinjected with 10 ng etsrp MO2 and 100 pg etsrp RNA. Images were taken using Axioskop2 and 10×/0.30 NA dry objective (Zeiss) (A-F; I,J) or CV11 stereomicroscope (Zeiss) (G,H,K-N), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A-D); 60× (E-N).
We have previously shown that overexpression of etsrp RNA was sufficient to induce strong ectopic scl expression.18 Etsrp RNA also induced ectopic pu.1 expression (Figure 2G,H) and caused an increase in the number of lcp1-expressing macrophages and neutrophils (Figure 2I,J). Interestingly, while the total of 67% of etsrp-overexpressing embryos displayed ectopic pu.1, 60% of the injected embryos also had reduced or completely absent endogenous pu.1 expression; 51% displayed both effects at the same time (Table 1). Etsrp RNA that lacked etsrp-MO binding sites induced ectopic pu.1 expression in etsrp morphants, demonstrating that ectopic pu.1 cells are not merely mislocalized endogenous pu.1 cells (Figure 2K-N, Table 1). In the same experiment, etsrp RNA also caused ectopic expression of an endothelial marker flk1 (vegfr2)26 in etsrp morphants, consistent with our previous results (data not shown; Sumanas and Lin18 ). Etsrp overexpression can therefore induce cells to initiate myeloid or endothelial development. Endogenous pu.1 may become down-regulated in etsrp-overexpressing embryos, as those cells undertake endothelial fate.
Effect of etsrp overexpression and knockdown on pu.1 expression at the 8- to 10-somite stages
| . | Normal or slightly distorted pu.1 pattern . | Normal endogenous and ectopic pu.1 expression . | Reduced or absent pu.1 expression . | Reduced or absent endogenous and ectopic pu.1 expression . |
|---|---|---|---|---|
| Control (n > 50) | 100 | 0 | 0 | 0 |
| etsrp MO (n = 40) | 5 ± 5 | 0 | 95 ± 5 | 0 |
| etsrp RNA (n = 57) | 24 ± 11 | 16 ± 1 | 9 ± 9 | 51 ± 19 |
| etsrpMO + RNA (n = 56) | 1 ± 1 | 0 | 27 ± 1 | 72 ± 2 |
| . | Normal or slightly distorted pu.1 pattern . | Normal endogenous and ectopic pu.1 expression . | Reduced or absent pu.1 expression . | Reduced or absent endogenous and ectopic pu.1 expression . |
|---|---|---|---|---|
| Control (n > 50) | 100 | 0 | 0 | 0 |
| etsrp MO (n = 40) | 5 ± 5 | 0 | 95 ± 5 | 0 |
| etsrp RNA (n = 57) | 24 ± 11 | 16 ± 1 | 9 ± 9 | 51 ± 19 |
| etsrpMO + RNA (n = 56) | 1 ± 1 | 0 | 27 ± 1 | 72 ± 2 |
Etsrp RNA causes ectopic pu.1 expression in control embryos and etsrp morphants and also results in the reduction of endogenous etsrp expression. Results are percentage averages from 2 independent experiments.
Etsrp affects both hematopoiesis and vasculogenesis in the anterior but not the posterior region
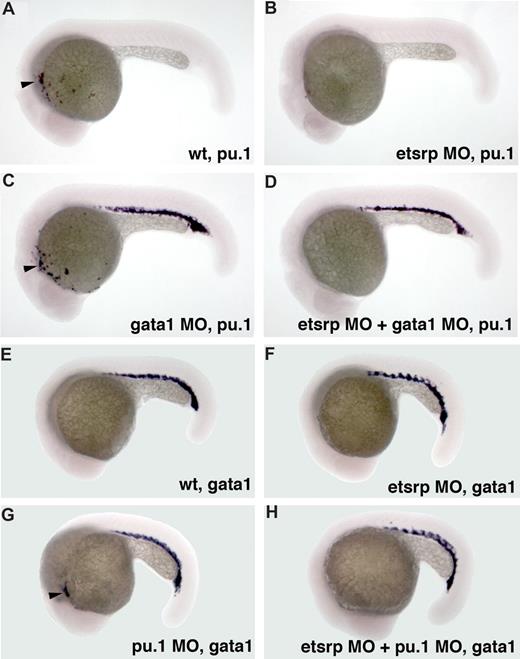
We have previously shown that etsrp knockdown and overexpression did not significantly affect formation of erythroid cells, including erythroid-specific expression of gata118 (Figure S2). We investigated whether etsrp function is limited to the myeloid cells within anterior lateral plate mesoderm. In wild-type embryos, pu.1 is expressed in both erythroid and myeloid progenitors by the 10-somite stage, and its erythroid expression disappears during later somitogenesis stages.24 Knockdown of gata1 has been reported to cause erythroid cells to undertake myeloid fate resulting in the persistence of posterior pu.1 expression15,16 (Figure 3C). Coinjection of etsrp and gata1 MOs resulted in the loss of pu.1 expression in the anterior but not the posterior region (Figure 3D), supporting etsrp requirement for the formation of anterior but not posterior myelo-erythroid progenitors. Knockdown of pu.1 has been reported to result in the myeloid cells undertaking erythroid fate and expressing gata1 in the anterior region15 (Figure 3G). These anterior gata1-expressing cells were absent in the etsrp and pu.1 double morphants (Figure 3H), consistent with etsrp affecting myelo-erythroid progenitors in the anterior region.
Etsrp affects both hematopoiesis and vasculogenesis in the anterior but not the posterior region. (A-D) Posterior ectopic pu.1 expression is independent of etsrp function as analyzed by in situ hybridization at 22 hpf. (A) Control uninjected embryo; (B) embryo injected with etsrp MOs; (C) gata1 MO-injected embryo; (D) gata1 MO and etsrp MO coinjected embryo. Note that ectopic pu.1 expressed in the erythroid cells in gata1 morphants (C) is unaffected in the double gata1/etsrp morphants (D). Anterior myeloid-specific pu.1 expression ( , A,C) is missing in etsrp morphants (B,D). (E-H) Anterior ectopic gata1 expression is dependent on etsrp function as analyzed by the in situ hybridization at 20 hpf to 21 hpf. (E) Control uninjected embryo; (F) embryo injected with etsrp MOs; (G) pu.1 MO-injected embryo; (H) etsrp and pu.1 MOs coinjected embryo. Note that the ectopic myeloid-specific anterior gata1 expression in pu.1 morphants (
, A,C) is missing in etsrp morphants (B,D). (E-H) Anterior ectopic gata1 expression is dependent on etsrp function as analyzed by the in situ hybridization at 20 hpf to 21 hpf. (E) Control uninjected embryo; (F) embryo injected with etsrp MOs; (G) pu.1 MO-injected embryo; (H) etsrp and pu.1 MOs coinjected embryo. Note that the ectopic myeloid-specific anterior gata1 expression in pu.1 morphants ( , G) is absent in double etsrp/pu.1 morphants (H). Posterior erythroid gata1 expression is not affected in etsrp morphants. Images were taken using Axioskop2 and 5×/0.15 NA dry objective, Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 75×.
, G) is absent in double etsrp/pu.1 morphants (H). Posterior erythroid gata1 expression is not affected in etsrp morphants. Images were taken using Axioskop2 and 5×/0.15 NA dry objective, Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 75×.
Etsrp affects both hematopoiesis and vasculogenesis in the anterior but not the posterior region. (A-D) Posterior ectopic pu.1 expression is independent of etsrp function as analyzed by in situ hybridization at 22 hpf. (A) Control uninjected embryo; (B) embryo injected with etsrp MOs; (C) gata1 MO-injected embryo; (D) gata1 MO and etsrp MO coinjected embryo. Note that ectopic pu.1 expressed in the erythroid cells in gata1 morphants (C) is unaffected in the double gata1/etsrp morphants (D). Anterior myeloid-specific pu.1 expression ( , A,C) is missing in etsrp morphants (B,D). (E-H) Anterior ectopic gata1 expression is dependent on etsrp function as analyzed by the in situ hybridization at 20 hpf to 21 hpf. (E) Control uninjected embryo; (F) embryo injected with etsrp MOs; (G) pu.1 MO-injected embryo; (H) etsrp and pu.1 MOs coinjected embryo. Note that the ectopic myeloid-specific anterior gata1 expression in pu.1 morphants (
, A,C) is missing in etsrp morphants (B,D). (E-H) Anterior ectopic gata1 expression is dependent on etsrp function as analyzed by the in situ hybridization at 20 hpf to 21 hpf. (E) Control uninjected embryo; (F) embryo injected with etsrp MOs; (G) pu.1 MO-injected embryo; (H) etsrp and pu.1 MOs coinjected embryo. Note that the ectopic myeloid-specific anterior gata1 expression in pu.1 morphants ( , G) is absent in double etsrp/pu.1 morphants (H). Posterior erythroid gata1 expression is not affected in etsrp morphants. Images were taken using Axioskop2 and 5×/0.15 NA dry objective, Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 75×.
, G) is absent in double etsrp/pu.1 morphants (H). Posterior erythroid gata1 expression is not affected in etsrp morphants. Images were taken using Axioskop2 and 5×/0.15 NA dry objective, Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 75×.
Scl functions downstream of etsrp in the myeloid but not endothelial lineage
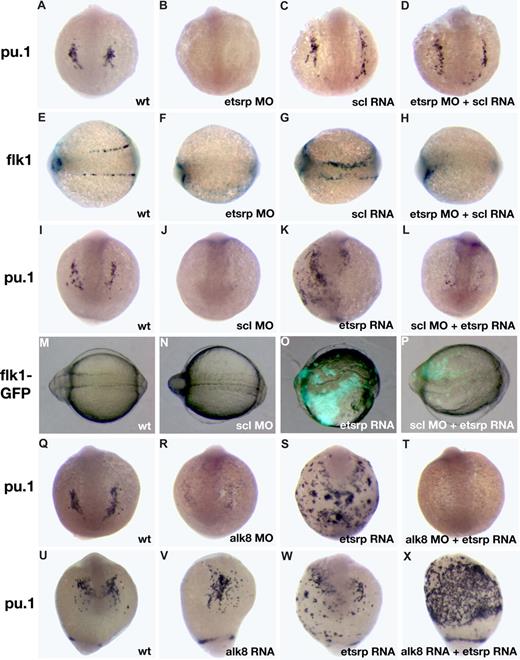
Scl has been previously implicated in myelopoiesis as well as vasculogenesis.11,12 We studied the interaction of etsrp and scl in myeloid and vascular development. We have previously shown that etsrp overexpression induces strong ectopic scl expression.18 Scl overexpression also enhanced etsrp expression, which was limited mostly to the somitic mesoderm (Figure S3; Figure 5G,I). Scl knockdown had no apparent effect on etsrp expression whereas etsrp morphants had strongly reduced scl expression in the anterior and trunk domains.18 Addition of scl RNA restored the endogenous pattern of pu.1 expression in etsrp morphants (Figure 4A-D; Table S1). In the sibling embryos from the same experiment, scl RNA failed to rescue endothelial flk1 expression in etsrp morphants, although it was sufficient to expand flk1 expression in control embryos (Figure 4E-H), consistent with our previously published data.18 As reported previously, pu.1 expression was severely reduced or absent in scl morphants (Figure 4I-J).11,12 Etsrp RNA failed to rescue pu.1 expression in scl morphants (Figure 4K,L). In the sibling embryos from the same experiment, etsrp induced ectopic flk1-GFP expression in scl morphants, although to a smaller extent than etsrp overexpression alone (Figure 4M-P). These experiments argue that scl functions downstream of etsrp in the myeloid induction. However, in the vascular induction etsrp is the critical factor, and scl may only modify etsrp expression or function but is not absolutely required. Overexpression of pu.1 mRNA was also sufficient to rescue lcp1 expression in etsrp morphants (data not shown).
Analysis of interaction between etsrp and scl and alk8 signaling pathways. (A-H) scl RNA can rescue myeloid but not vascular cell formation in etsrp morphants as evident from pu.1 and flk1 expression analysis at the 10-somite stage. (A-D) pu.1 expression, anterior view; (E-H) flk1 expression, dorsal view, anterior to the left. (A,E) Control uninjected embryos; (B,F) embryos injected with etsrp MOs; (C,G) scl RNA-injected embryos; (D,H) embryos coinjected with etsrp MOs and scl RNA. Note that the myeloid-specific pu.1 expression is restored in etsrp MO and scl RNA coinjected embryos (D). Also note that the vascular-specific flk1 expression is absent in etsrp MO and scl RNA coinjected embryos in the same experiment (H). (I-P) etsrp RNA can rescue vascular but not myeloid cell formation in scl morphants as evident from pu.1 and flk1 expression analysis at the 8-somite stage. (I-L) pu.1 expression as analyzed by in situ hybridization, anterior view; (M-P) GFP fluorescence in flk1-GFP transgenic embryos, dorsal view, anterior is to the left. (I,M) Control uninjected embryo; (J,N) scl MO-injected embryo; (K,O) etsrp RNA-injected embryo; (L,P) scl MO and etsrp RNA coinjected embryo. Note that etsrp RNA fails to rescue myeloid-specific pu.1 expression in scl morphants (L). Etsrp RNA can restore vascular flk1-GFP expression in the same experiment (P). Flk1-GFP fluorescence in wild-type embryos and scl morphants (M,N) is much weaker and not apparent under the same exposure. (Q-T) etsrp RNA fails to rescue pu.1 expression in alk8 morphants. pu.1 expression analyzed at the 10-somite stage, anterior view, except for panel S, which is anterior-ventral. (Q) Control uninjected embryo; (R) alk8 MO-injected embryo; (S) etsrp RNA-injected embryo; (T) alk8 MO and etsrp RNA coinjected embryo. (U-X) etsrp and constitutively active CA-alk8 RNA synergize in inducing pu.1 expression at the 14-somite stage. Anterior-ventral views except for panel X, which is the ventrolateral view, anterior is to the top. (U) Control uninjected embryo; (V) CA-alk8 RNA-injected embryo; (W) etsrp RNA-injected embryo; (X) CA-alk8 RNA and etsrp RNA coinjected embryo. Images were taken using CV11 stereomicroscope (Zeiss), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 60×.
Analysis of interaction between etsrp and scl and alk8 signaling pathways. (A-H) scl RNA can rescue myeloid but not vascular cell formation in etsrp morphants as evident from pu.1 and flk1 expression analysis at the 10-somite stage. (A-D) pu.1 expression, anterior view; (E-H) flk1 expression, dorsal view, anterior to the left. (A,E) Control uninjected embryos; (B,F) embryos injected with etsrp MOs; (C,G) scl RNA-injected embryos; (D,H) embryos coinjected with etsrp MOs and scl RNA. Note that the myeloid-specific pu.1 expression is restored in etsrp MO and scl RNA coinjected embryos (D). Also note that the vascular-specific flk1 expression is absent in etsrp MO and scl RNA coinjected embryos in the same experiment (H). (I-P) etsrp RNA can rescue vascular but not myeloid cell formation in scl morphants as evident from pu.1 and flk1 expression analysis at the 8-somite stage. (I-L) pu.1 expression as analyzed by in situ hybridization, anterior view; (M-P) GFP fluorescence in flk1-GFP transgenic embryos, dorsal view, anterior is to the left. (I,M) Control uninjected embryo; (J,N) scl MO-injected embryo; (K,O) etsrp RNA-injected embryo; (L,P) scl MO and etsrp RNA coinjected embryo. Note that etsrp RNA fails to rescue myeloid-specific pu.1 expression in scl morphants (L). Etsrp RNA can restore vascular flk1-GFP expression in the same experiment (P). Flk1-GFP fluorescence in wild-type embryos and scl morphants (M,N) is much weaker and not apparent under the same exposure. (Q-T) etsrp RNA fails to rescue pu.1 expression in alk8 morphants. pu.1 expression analyzed at the 10-somite stage, anterior view, except for panel S, which is anterior-ventral. (Q) Control uninjected embryo; (R) alk8 MO-injected embryo; (S) etsrp RNA-injected embryo; (T) alk8 MO and etsrp RNA coinjected embryo. (U-X) etsrp and constitutively active CA-alk8 RNA synergize in inducing pu.1 expression at the 14-somite stage. Anterior-ventral views except for panel X, which is the ventrolateral view, anterior is to the top. (U) Control uninjected embryo; (V) CA-alk8 RNA-injected embryo; (W) etsrp RNA-injected embryo; (X) CA-alk8 RNA and etsrp RNA coinjected embryo. Images were taken using CV11 stereomicroscope (Zeiss), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 60×.
A recent study demonstrated that the alk8 signaling provides a necessary input for myelopoiesis, apparently functioning in parallel to the scl pathway.17 We tested if etsrp RNA could induce ectopic pu.1 in alk8 morphants. As shown in Figure 4Q-T, alk8 is required for etsrp RNA to induce ectopic pu.1 expression. Overexpression of constitutively active alk8 (CA-alk8) RNA caused mild ventralization of injected embryos, and the number of myeloid cells was not changed significantly. Coinjection of CA-alk8 and etsrp RNAs resulted in the dramatic expansion of myeloid lineage (Figure 4U-X). These results support the idea that etsrp and alk8 signaling are both necessary parallel inputs during myeloid formation.
Etsrp and pu.1 expression are mutually exclusive
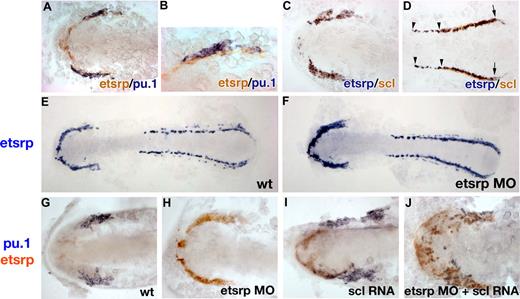
To understand better how overexpression of endothelial-specific etsrp induces myeloid pu.1 expression, we investigated expression of etsrp, pu.1, and scl in greater detail by the 2-color in situ hybridization. Etsrp expression first appears at the 1- to 2-somite stage18 and precedes pu.1 expression, which is first apparent at the 6-somite stage.24 Etsrp- and pu.1-expressing cells are positioned immediately adjacent to each other, but their expression does not overlap at the 6-somite stage, soon after pu.1 expression first appears (Figure 5A,B). In contrast, etsrp and scl expression overlap nearly perfectly at the 6-somite stage in the anterior region (Figure 5C). In the posterior region, etsrp expression extends further into the trunk; scl and etsrp expression start to separate as scl expression becomes restricted to the erythroid cells (Figure 5D).
Analysis of etsrp, pu.1, and scl expression by the 2-color in situ hybridization. Flat-mounted embryos, anterior is to the left. (A, B) Etsrp (red) and pu.1 (blue) expression in the anterior region of a flat-mounted embryo at the 6-somite stage. Panel B is a higher magnification of panel A. Note that pu.1-expressing cells lie immediately adjacent to etsrp-expressing cells but expression of the 2 markers does not overlap. (C,D) Etsrp (blue) and scl (red) expression in the anterior (C) and posterior (D) regions of a flat-mounted embryo at the 6-somite stage. Note that the 2 markers completely overlap in panel C while in panel D the trunk region contains only etsrp-expressing cells ( ); scl expression partially overlaps with etsrp in the tail region where scl is restricted to erythroid cells during later stages (
); scl expression partially overlaps with etsrp in the tail region where scl is restricted to erythroid cells during later stages ( and the right
and the right  ). (E,F) Etsrp RNA expression expands into the myeloid region in etsrp morphants injected with Etsrp translation-blocking MOs. Etsrp expression in control uninjected embryos (E) and etsrp morphants (F) at the 9-somite stage. Note the more intense and expanded etsrp expression in panel F. (G-J) scl RNA restores pu.1 expression in etsrp morphants with pu.1 and etsrp-expressing cells intermingled. Two-color in situ hybridization analysis for pu.1 (blue) and etsrp (red) expression at the 9- to 10-somite stage. Only the anterior part of an embryo is shown. (G) Control uninjected embryo; (H) etsrp MO-injected embryo; (I) scl RNA-injected embryo; (J) etsrp MOs and scl RNA coinjected embryo. Etsrp staining is very weak in the control embryos because of the short staining time, which was the same for all experimental batches. Note that etsrp morphants in panel H have absent pu.1 expression and strongly up-regulated and expanded etsrp expression. scl RNA-injected embryos (I) display up-regulated etsrp expression. pu.1-expressing cells are intermingled with etsrp-expressing cells in panel J but they do not overlap. Images were taken using Axioskop2 and 10×/0.30 NA (A; C-J) or 20×/0.50 NA (B) dry objectives (Zeiss), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A,C,D,G-J); 200× (B); 75× (E,F).
). (E,F) Etsrp RNA expression expands into the myeloid region in etsrp morphants injected with Etsrp translation-blocking MOs. Etsrp expression in control uninjected embryos (E) and etsrp morphants (F) at the 9-somite stage. Note the more intense and expanded etsrp expression in panel F. (G-J) scl RNA restores pu.1 expression in etsrp morphants with pu.1 and etsrp-expressing cells intermingled. Two-color in situ hybridization analysis for pu.1 (blue) and etsrp (red) expression at the 9- to 10-somite stage. Only the anterior part of an embryo is shown. (G) Control uninjected embryo; (H) etsrp MO-injected embryo; (I) scl RNA-injected embryo; (J) etsrp MOs and scl RNA coinjected embryo. Etsrp staining is very weak in the control embryos because of the short staining time, which was the same for all experimental batches. Note that etsrp morphants in panel H have absent pu.1 expression and strongly up-regulated and expanded etsrp expression. scl RNA-injected embryos (I) display up-regulated etsrp expression. pu.1-expressing cells are intermingled with etsrp-expressing cells in panel J but they do not overlap. Images were taken using Axioskop2 and 10×/0.30 NA (A; C-J) or 20×/0.50 NA (B) dry objectives (Zeiss), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A,C,D,G-J); 200× (B); 75× (E,F).
Analysis of etsrp, pu.1, and scl expression by the 2-color in situ hybridization. Flat-mounted embryos, anterior is to the left. (A, B) Etsrp (red) and pu.1 (blue) expression in the anterior region of a flat-mounted embryo at the 6-somite stage. Panel B is a higher magnification of panel A. Note that pu.1-expressing cells lie immediately adjacent to etsrp-expressing cells but expression of the 2 markers does not overlap. (C,D) Etsrp (blue) and scl (red) expression in the anterior (C) and posterior (D) regions of a flat-mounted embryo at the 6-somite stage. Note that the 2 markers completely overlap in panel C while in panel D the trunk region contains only etsrp-expressing cells ( ); scl expression partially overlaps with etsrp in the tail region where scl is restricted to erythroid cells during later stages (
); scl expression partially overlaps with etsrp in the tail region where scl is restricted to erythroid cells during later stages ( and the right
and the right  ). (E,F) Etsrp RNA expression expands into the myeloid region in etsrp morphants injected with Etsrp translation-blocking MOs. Etsrp expression in control uninjected embryos (E) and etsrp morphants (F) at the 9-somite stage. Note the more intense and expanded etsrp expression in panel F. (G-J) scl RNA restores pu.1 expression in etsrp morphants with pu.1 and etsrp-expressing cells intermingled. Two-color in situ hybridization analysis for pu.1 (blue) and etsrp (red) expression at the 9- to 10-somite stage. Only the anterior part of an embryo is shown. (G) Control uninjected embryo; (H) etsrp MO-injected embryo; (I) scl RNA-injected embryo; (J) etsrp MOs and scl RNA coinjected embryo. Etsrp staining is very weak in the control embryos because of the short staining time, which was the same for all experimental batches. Note that etsrp morphants in panel H have absent pu.1 expression and strongly up-regulated and expanded etsrp expression. scl RNA-injected embryos (I) display up-regulated etsrp expression. pu.1-expressing cells are intermingled with etsrp-expressing cells in panel J but they do not overlap. Images were taken using Axioskop2 and 10×/0.30 NA (A; C-J) or 20×/0.50 NA (B) dry objectives (Zeiss), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A,C,D,G-J); 200× (B); 75× (E,F).
). (E,F) Etsrp RNA expression expands into the myeloid region in etsrp morphants injected with Etsrp translation-blocking MOs. Etsrp expression in control uninjected embryos (E) and etsrp morphants (F) at the 9-somite stage. Note the more intense and expanded etsrp expression in panel F. (G-J) scl RNA restores pu.1 expression in etsrp morphants with pu.1 and etsrp-expressing cells intermingled. Two-color in situ hybridization analysis for pu.1 (blue) and etsrp (red) expression at the 9- to 10-somite stage. Only the anterior part of an embryo is shown. (G) Control uninjected embryo; (H) etsrp MO-injected embryo; (I) scl RNA-injected embryo; (J) etsrp MOs and scl RNA coinjected embryo. Etsrp staining is very weak in the control embryos because of the short staining time, which was the same for all experimental batches. Note that etsrp morphants in panel H have absent pu.1 expression and strongly up-regulated and expanded etsrp expression. scl RNA-injected embryos (I) display up-regulated etsrp expression. pu.1-expressing cells are intermingled with etsrp-expressing cells in panel J but they do not overlap. Images were taken using Axioskop2 and 10×/0.30 NA (A; C-J) or 20×/0.50 NA (B) dry objectives (Zeiss), Axiocam color camera (Zeiss, model 412-312) and Openlab 4.0 software (Improvision). Magnification: 100× (A,C,D,G-J); 200× (B); 75× (E,F).
Because etsrp and pu.1 are expressed in neighboring but not the same cells, we tested the hypothesis that etsrp and pu.1 may negatively regulate each other's expression. As we described earlier, knockdown of Etsrp translation resulted in the enhanced etsrp RNA expression, suggesting the presence of a negative autoregulatory loop.18 Expansion of etsrp expression in etsrp morphants appeared to extend into the region where the myeloid cells would have normally formed, which no longer expressed pu.1 (Figure 5E-H). As shown in Figure 4A-D, scl RNA could restore pu.1 expression in etsrp morphants. We analyzed pu.1 and etsrp expression in etsrp MO and scl RNA-injected embryos. Scl RNA and etsrp MO coinjected embryos displayed scattered pu.1 cells, which were embedded within the expanded domain of etsrp-expressing cells (Figure 5J). Noticeably, expression of etsrp and pu.1 still did not overlap in these embryos, resulting in the salt-and-pepper pattern of intermingled pu.1- and etsrp-expressing cells. This experiment shows that etsrp expression expands into the myeloid domain in etsrp morphants. It also shows that etsrp and pu.1 expression is mutually exclusive even in the absence of Etsrp protein function.
We tested if pu.1 itself could possibly down-regulate etsrp expression in the myeloid precursors. However, pu.1 RNA overexpression and MO knockdown did not significantly affect etsrp expression (data not shown).
Etsrp functions cell autonomously in myeloid and endothelial lineages
As we have shown so far, etsrp overexpression results in the induction of both vascular and myeloid markers. We hypothesized that etsrp induces common myeloid-endothelial precursor cells, hemangioblasts, which then differentiate into myeloid and/or endothelial cells. To test whether etsrp functions cell autonomously in myeloid and endothelial induction, we traced the lineage of etsrp-expressing cells by performing cell transplantation experiments. To test etsrp function in endothelial induction, flk1-GFP donor embryos were injected with the mixture of tetramethylrhodamine isothiocyanate (TRITC)–conjugated dextran and etsrp RNA. Cells from donor embryos were transplanted into etsrp morphants at the beginning of epiboly, which were imaged for the presence of TRITC and flk1-GFP expression at the 8- to 10-somite stages (Figure 6A). In 2 independent experiments, 24 of 61 successfully transplanted embryos displayed flk1-GFP–expressing cells. In every single case, all flk1-GFP–expressing cells also contained TRITC label, arguing that they originated from etsrp-expressing cells (Figure 6B-D).
Etsrp-expressing precursor cells give rise to both vascular endothelial and myeloid lineages. (A) Diagram of transplantation experiment. Donor embryos were injected at the 1-cell stage with etsrp RNA and TRITC-dextran (B-D) or FITC-dextran (E-H), while recipient embryos were injected with 7.5 ng etsrp MO1/MO2 mixture. Cells were transplanted at the beginning of epiboly. (B-D) flk1-GFP–expressing cells are a subset of etsrp RNA/TRITC-labeled cells. The embryo is at the 8-somite stage, lateral view, anterior to the left. (B) TRITC-filter image. Only transplanted cells are visible. (C) GFP-filter image. Only flk1-GFP–expressing cells are visible. (D) Overlay of the TRITC, GFP, and transmitted light DIC images. Cells where GFP and TRITC fluorescence overlaps are in yellow ( point to some of these cells). Note that every GFP-expressing cell has also TRITC fluorescence. (E-H) pu.1-expressing cells originate from etsrp-expressing cells, transplanted from etsrp RNA-overexpressing embryos into etsrp morphants. Etsrp RNA was coinjected with fluorescein-labeled dextran; pu.1 expression and fluorescein presence was analyzed by 2-color fluorescent (E-G) or conventional (H) in situ hybridization at the 8- to 10-somite stages. (E-G) Anterior-lateral view of the same embryo, dorsal is up. (E) FITC-filter image. Only transplanted cells are visible. (F) pu.1 expression as detected by tyramide-Cy3 amplification, visualized through the rhodamine channel filter. Note the ectopically located pu.1-expressing cells (
point to some of these cells). Note that every GFP-expressing cell has also TRITC fluorescence. (E-H) pu.1-expressing cells originate from etsrp-expressing cells, transplanted from etsrp RNA-overexpressing embryos into etsrp morphants. Etsrp RNA was coinjected with fluorescein-labeled dextran; pu.1 expression and fluorescein presence was analyzed by 2-color fluorescent (E-G) or conventional (H) in situ hybridization at the 8- to 10-somite stages. (E-G) Anterior-lateral view of the same embryo, dorsal is up. (E) FITC-filter image. Only transplanted cells are visible. (F) pu.1 expression as detected by tyramide-Cy3 amplification, visualized through the rhodamine channel filter. Note the ectopically located pu.1-expressing cells ( ) and 3 remaining endogenous pu.1-expressing cells that are located bilaterally within the anterior lateral mesoderm (
) and 3 remaining endogenous pu.1-expressing cells that are located bilaterally within the anterior lateral mesoderm ( ). (G) Overlay of FITC, Cy3, and transmitted light images. Note that all 3 ectopic pu.1-expressing cells contain FITC label (
). (G) Overlay of FITC, Cy3, and transmitted light images. Note that all 3 ectopic pu.1-expressing cells contain FITC label ( ) while the endogenous pu.1 cells do not (
) while the endogenous pu.1 cells do not ( ). (H) A posterior region from an embryo containing multiple pu.1 and fluorescein-positive cells. Embryo has been flat-mounted to show dorsal, lateral, and ventral tissues. (1) Endogenous pu.1-expressing cells in the posterior lateral mesoderm. (2) Fluorescein-labeled transplanted cells. (3-5) Double pu.1 and fluorescein-positive cells. Average color for each cell group is shown in the boxes below the figure (“Methods”). Images were taken using Axioplan2 and 10×/0.30 NA (A;C-G) (Zeiss), Axiocam color camera (Zeiss, model 412-312) (H) or monochrome C4742-95 camera (B-G) (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab 4.0 software (Improvision). Magnification: 100× (B-G); 300× (H).
). (H) A posterior region from an embryo containing multiple pu.1 and fluorescein-positive cells. Embryo has been flat-mounted to show dorsal, lateral, and ventral tissues. (1) Endogenous pu.1-expressing cells in the posterior lateral mesoderm. (2) Fluorescein-labeled transplanted cells. (3-5) Double pu.1 and fluorescein-positive cells. Average color for each cell group is shown in the boxes below the figure (“Methods”). Images were taken using Axioplan2 and 10×/0.30 NA (A;C-G) (Zeiss), Axiocam color camera (Zeiss, model 412-312) (H) or monochrome C4742-95 camera (B-G) (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab 4.0 software (Improvision). Magnification: 100× (B-G); 300× (H).
Etsrp-expressing precursor cells give rise to both vascular endothelial and myeloid lineages. (A) Diagram of transplantation experiment. Donor embryos were injected at the 1-cell stage with etsrp RNA and TRITC-dextran (B-D) or FITC-dextran (E-H), while recipient embryos were injected with 7.5 ng etsrp MO1/MO2 mixture. Cells were transplanted at the beginning of epiboly. (B-D) flk1-GFP–expressing cells are a subset of etsrp RNA/TRITC-labeled cells. The embryo is at the 8-somite stage, lateral view, anterior to the left. (B) TRITC-filter image. Only transplanted cells are visible. (C) GFP-filter image. Only flk1-GFP–expressing cells are visible. (D) Overlay of the TRITC, GFP, and transmitted light DIC images. Cells where GFP and TRITC fluorescence overlaps are in yellow ( point to some of these cells). Note that every GFP-expressing cell has also TRITC fluorescence. (E-H) pu.1-expressing cells originate from etsrp-expressing cells, transplanted from etsrp RNA-overexpressing embryos into etsrp morphants. Etsrp RNA was coinjected with fluorescein-labeled dextran; pu.1 expression and fluorescein presence was analyzed by 2-color fluorescent (E-G) or conventional (H) in situ hybridization at the 8- to 10-somite stages. (E-G) Anterior-lateral view of the same embryo, dorsal is up. (E) FITC-filter image. Only transplanted cells are visible. (F) pu.1 expression as detected by tyramide-Cy3 amplification, visualized through the rhodamine channel filter. Note the ectopically located pu.1-expressing cells (
point to some of these cells). Note that every GFP-expressing cell has also TRITC fluorescence. (E-H) pu.1-expressing cells originate from etsrp-expressing cells, transplanted from etsrp RNA-overexpressing embryos into etsrp morphants. Etsrp RNA was coinjected with fluorescein-labeled dextran; pu.1 expression and fluorescein presence was analyzed by 2-color fluorescent (E-G) or conventional (H) in situ hybridization at the 8- to 10-somite stages. (E-G) Anterior-lateral view of the same embryo, dorsal is up. (E) FITC-filter image. Only transplanted cells are visible. (F) pu.1 expression as detected by tyramide-Cy3 amplification, visualized through the rhodamine channel filter. Note the ectopically located pu.1-expressing cells ( ) and 3 remaining endogenous pu.1-expressing cells that are located bilaterally within the anterior lateral mesoderm (
) and 3 remaining endogenous pu.1-expressing cells that are located bilaterally within the anterior lateral mesoderm ( ). (G) Overlay of FITC, Cy3, and transmitted light images. Note that all 3 ectopic pu.1-expressing cells contain FITC label (
). (G) Overlay of FITC, Cy3, and transmitted light images. Note that all 3 ectopic pu.1-expressing cells contain FITC label ( ) while the endogenous pu.1 cells do not (
) while the endogenous pu.1 cells do not ( ). (H) A posterior region from an embryo containing multiple pu.1 and fluorescein-positive cells. Embryo has been flat-mounted to show dorsal, lateral, and ventral tissues. (1) Endogenous pu.1-expressing cells in the posterior lateral mesoderm. (2) Fluorescein-labeled transplanted cells. (3-5) Double pu.1 and fluorescein-positive cells. Average color for each cell group is shown in the boxes below the figure (“Methods”). Images were taken using Axioplan2 and 10×/0.30 NA (A;C-G) (Zeiss), Axiocam color camera (Zeiss, model 412-312) (H) or monochrome C4742-95 camera (B-G) (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab 4.0 software (Improvision). Magnification: 100× (B-G); 300× (H).
). (H) A posterior region from an embryo containing multiple pu.1 and fluorescein-positive cells. Embryo has been flat-mounted to show dorsal, lateral, and ventral tissues. (1) Endogenous pu.1-expressing cells in the posterior lateral mesoderm. (2) Fluorescein-labeled transplanted cells. (3-5) Double pu.1 and fluorescein-positive cells. Average color for each cell group is shown in the boxes below the figure (“Methods”). Images were taken using Axioplan2 and 10×/0.30 NA (A;C-G) (Zeiss), Axiocam color camera (Zeiss, model 412-312) (H) or monochrome C4742-95 camera (B-G) (Hamamatsu Photonics, Hamamatsu City, Japan) and Openlab 4.0 software (Improvision). Magnification: 100× (B-G); 300× (H).
To test whether etsrp functions cell autonomously in inducing myeloid lineage, donor embryos were injected with a mixture of fluorescein-labeled dextran and etsrp RNA; cells from the donor embryos were transplanted into etsrp morphants at the beginning of epiboly, which were assayed for pu.1 expression and fluorescein presence at the 8- to 10-somite stages. Overall, among 127 successfully transplanted embryos, 40 ectopically located pu.1-expressing cells were detected. Although most etsrp morphant recipient embryos had no endogenous pu.1 expression in the anterior region and only posterior erythroid-specific pu.1 stripes were present, occasional embryos displayed a few pu.1-expressing cells in the anterior lateral mesoderm. These cells, positioned in the location of endogenous pu.1 expression, were not counted in this experiment. Among the 40 ectopic pu.1 cells in the transplanted embryos, 39 also displayed the presence of fluorescein and one could not be reliably determined, as assayed by fluorescent or conventional 2-color in situ hybridization (Figure 6E-H). These data argue that the pu.1-expressing myeloid progenitor cells originate from etsrp-expressing cells, which down-regulate etsrp expression as they undertake the myeloid fate.
Discussion
In this study, we show that Etsrp function is evolutionary conserved and identify mammalian ER71 proteins as functional orthologs of Etsrp. Both phylogeny and chromosomal synteny analysis argues that Etsrp and ER71 proteins are homologous to each other. In addition, mammalian ER71 and zebrafish Etsrp but not mammalian Ets1 proteins result in a similar phenotype when overexpressed in zebrafish embryos. Similarly, overexpression of mouse ER71 in embryonic stem cells causes induction of multiple hemangioblast and vascular endothelial markers.33 Mouse ER71 expression domains include yolk blood islands and major vessels,33 supporting the idea that Etsrp is related to ER71 protein family. Thus it appears that ER71/Etsrp proteins are critical to induce vascular endothelial cell fate in undifferentiated progenitor cells in different vertebrates.
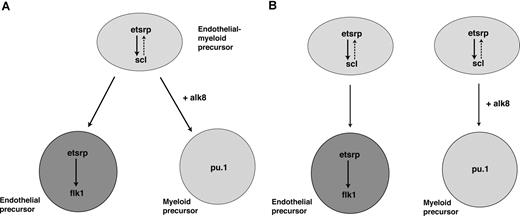
In addition, we demonstrate that etsrp is not only a critical factor that initiates vasculogenesis, but it also participates in the induction of myelopoiesis. In the absence of etsrp, myeloid cells were nearly completely absent, whereas overexpression of etsrp resulted in the formation of ectopic myeloid cells. We show that etsrp regulates myeloid development by functioning through transcription factor scl, a known regulator of myeloid as well as erythroid and vascular development. We also show that etsrp-expressing cells give rise to both vascular endothelial and myeloid cells, with etsrp becoming localized to the endothelial cells and excluded from myeloid progenitors. These data suggest the following 2 possibilities. First, Etsrp functions within the common myeloid-endothelial progenitor, hemangioblast cells in the anterior part of an embryo (Figure 7A). Alternatively, there may be 2 separate pools of Etsrp-expressing cells giving rise to endothelial and myeloid lineages (Figure 7B). Although at present we cannot distinguish between the 2 models, we favor the first model, based on the recent study demonstrating the existence of posterior hemangioblast.13 Thus we propose a model where etsrp is necessary and sufficient for the up-regulation of scl in hemangioblasts, positioned within the anterior lateral plate mesoderm (Figure 7). Etsrp function within hemangioblasts is necessary for their further differentiation into both endothelial and myeloid lineages. As hemangioblasts divide, giving rise to angioblasts and myeloid precursors, etsrp becomes localized to the endothelial progenitor cells and excluded from the myeloid precursors. Scl appears to support etsrp function within angioblasts, either by up-regulating etsrp transcription and/or functioning together with etsrp to up-regulate expression of its target genes such as flk1.
Proposed models for etsrp function within the anterior lateral mesoderm. (A) Etsrp induces scl in the hemangioblast cells, which give rise to both vascular endothelial and myeloid precursors. As hemangioblasts divide, etsrp expression becomes restricted to endothelial cells where it is both necessary and sufficient for vasculogenesis. (B) Alternatively, there are 2 separate pools of etsrp-expressing endothelial and myeloid precursors. As the cells initiate myeloid marker expression, they down-regulate etsrp expression.
Proposed models for etsrp function within the anterior lateral mesoderm. (A) Etsrp induces scl in the hemangioblast cells, which give rise to both vascular endothelial and myeloid precursors. As hemangioblasts divide, etsrp expression becomes restricted to endothelial cells where it is both necessary and sufficient for vasculogenesis. (B) Alternatively, there are 2 separate pools of etsrp-expressing endothelial and myeloid precursors. As the cells initiate myeloid marker expression, they down-regulate etsrp expression.
It is currently not clear how etsrp expression is excluded from the myeloid cells. All the experimental data support the idea that etsrp and pu.1 expression are mutually exclusive. This mutually exclusive expression pattern is maintained even in the absence of Etsrp protein, as seen in the etsrp morphants injected with scl RNA. It is possible that pu.1 directly or indirectly represses etsrp transcription in the myeloid cells, although our data do not support this model as etsrp expression does not change significantly upon pu.1 down-regulation or overexpression. Alternatively, a different signaling pathway may be involved in maintaining this relationship. Interestingly, etsrp itself appears to repress pu.1 expression as etsrp RNA-overexpressing embryos often have the endogenous pu.1 down-regulated or even totally absent. This may help to ensure that pu.1 is never expressed in the endothelial cells.
It is also not known how cells make a choice between the endothelial and myeloid fates. Our results support the idea that continued etsrp expression drives cells toward endothelial fate. In contrast, the combination of etsrp and alk8 signaling17 combined with the subsequent etsrp down-regulation is sufficient to direct multiple cells toward myeloid fate. It is possible that additional as-yet-undiscovered pathways participate in the cross-talk between endothelial and myeloid cells and influence this fate decision.
Until now, it has been generally accepted that the anterior and posterior hemangioblasts are equivalent in their potential. For example, the myeloid-forming anterior hemangioblasts can give rise to erythroid cells in the absence of pu.1, and the erythroid-forming posterior hemangioblasts can give rise to myeloid cells in the absence of gata1.15,16 Etsrp is the first known gene to specifically affect anterior hemangioblasts, while in the posterior region it functions in the angioblasts but does not affect hematopoietic cells. The most likely explanation is that in the anterior region etsrp is expressed within the hemangioblast cells, whereas the posterior etsrp expression is limited to the angioblasts. It is possible that related Ets-domain proteins such as Ets134 may play a similar role in the posterior hemangioblasts; it has been demonstrated that etsrp, ets1, fli1, and fli1b genes can function somewhat redundantly.35 Interestingly, etsrp overexpression resulted in the expansion of hemangioblasts, which differentiate into endothelial and myeloid, but not erythroid cells. Possible explanations are that etsrp induces hemangioblasts in both regions but they behave differently in the posterior region, or etsrp induces hemangioblasts in the anterior but not the posterior region. Scl has been previously shown to be necessary for erythroid, myeloid, and to certain extent, endothelial development.11,12 Scl overexpression also induces erythroid, myeloid, and endothelial lineages.7,8 Etsrp induces strong ectopic scl expression while scl can feedback and induce etsrp expression as well, although this induction is limited to the portions of lateral, somitic and intermediate mesoderm. Apparently, etsrp-induced scl-expressing cells cannot differentiate into erythroid lineage, either because etsrp presence directs them toward endothelial fate or additional genes up-regulated by etsrp ensure endothelial fate of these cells, even after etsrp expression is lost or down-regulated (which happens in a subset of etsrp-expressing cells).
Transplantation experiments show that etsrp-expressing cells can give rise to both endothelial and myeloid lineages. A relatively low number of transplanted cells (less than 1%) displayed pu.1 expression. It is possible that not all etsrp-expressing cells have the ability to differentiate into both lineages. There may be a certain bias in the transplantation experiments, (eg, the transplanted cells do not end up in the myeloid-competent region often enough). We noticed that the ectopic pu.1-expressing cells in the overexpression and the transplantation experiments were often located in the ventral and almost never in the dorsal side of an embryo, whereas flk1-expressing cells were located mostly at the dorsal side, possibly reflecting different myeloid and endothelial competences within distinct regions of an embryo. Not all etsrp-expressing cells differentiated into endothelial cells at the time of analysis, which may also reflect different competence to undertake endothelial fate, or simply a delay between the etsrp expression and flk1 induction. Although our experiments show that etsrp-expressing cells can give rise to both lineages, thus supporting the hemangioblast idea, additional experiments are needed to convincingly demonstrate the existence of a common endothelial-myeloid progenitor.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This research was supported by grants from the National Institutes of Health (R01 DK54508, T32 HL069766, and R25 CA098010).
National Institutes of Health
Authorship
Contribution: S.S. designed and performed experiments, analyzed data, and wrote the paper; G.G. and Y.Z. performed experiments and analyzed data; C.P. and K.C. contributed reagents; S.L. designed experiments and analyzed data.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Saulius Sumanas, Cincinnati Children's Hospital Medical Center, Division of Developmental Biology, ML7007, 3333 Burnet Ave, Cincinnati, OH 45229; e-mail: saulius.sumanas@cchmc.org; or Shuo Lin, University of California, Los Angeles, Department of Molecular, Cell and Developmental Biology, 621 C Young Dr South, Los Angeles, CA 90095; e-mail: shuolin@ucla.edu.