Abstract
Chromosomal aberrations (CAs) have emerged as important pathogenetic and prognostic factors in plasma cell disorders. Using interphase fluorescence in situ hybridization (FISH) analysis, we evaluated CAs in a series of 75 patients with amyloid light chain amyloidosis (AL) as compared with 127 patients with monoclonal gammopathy of unknown significance (MGUS). We investigated IgH translocations t(11;14), t(4;14), and t(14;16) as well as gains of 1q21, 11q23, and 19q13 and deletions of 8p21, 13q14, and 17p13, detecting at least one CA in 89% of the patients. Translocation t(11;14) was the most frequent aberration in AL, with 47% versus 26% in MGUS (P = .03), and was strongly associated with the lack of an intact immunoglobulin (P < .001), thus contributing to the frequent light chain subtype in AL. Other frequent aberrations in AL included deletion of 13q14 and gain of 1q21, which were shared by MGUS at comparable frequencies. The progression to multiple myeloma (MM) stage I was paralleled by an increased frequency of gain of 1q21 (P = .001) in both groups. Similar branching patterns were observed in an oncogenetic tree model, indicating a common mechanism of underlying karyotypic instability in these plasma cell disorders.
Introduction
Systemic amyloid light chain amyloidosis (AL) is characterized by the deposition of immunoglobulin light chains as amyloid fibrils in different organs, where they form toxic protein aggregates. The underlying disease is a plasma cell disorder, overwhelmingly a monoclonal gammopathy.1 The predilection for certain tissues is attributed to some extent to the respective clonality isotype of the light chain.2-5 Whereas the clonal light chain repertoire has been intensively studied in AL, the pathogenetic role of chromosomal aberrations (CAs) has only been addressed in a few reports based on small patient cohorts so far.6-9
This is different in multiple myeloma (MM), a malignant plasma cell disorder, where numerous CAs have been recognized as pathogenetic factors. Meanwhile, molecular testing has become a standard in diagnostic evaluation to identify patient subgroups regarding prognosis. Based on ploidy data, hyperdiploid and nonhyperdiploid forms of MM have been delineated as 2 major pathogenetic pathways.10-13 Hyperdiploid MM is characterized by the accumulation of extra copies of chromosomes. Multiple trisomies most frequently involve chromosomes 3, 5, 7, 9, 15, 19, and 21.14 By a clustering of CAs, we recently found nonhyperdiploid MM to separate in 3 branches: first, the translocation t(11;14)(q13;q23), which juxtaposes the immunoglobulin heavy chain locus (IgH) to the oncogene cyclin D1; second, deletion of 13q14, which is frequently in association with translocation t(4;14)(p16;q23) and involves the oncogenes MMSET and FGFR3; and third, gain of 1q21.15
Monoclonal gammopathy of undetermined significance (MGUS) is a premalignant disorder characterized by an expansion of monoclonal plasma cells, which can progress to frankly malignant MM.16 Though the interpretation of CA results in MGUS has been complicated by an inferior plasma cell purity due to a lesser degree of bone marrow plasmocytosis and a lower proliferative index,17 chromosome aberrations have been consistently detected by interphase–fluorescence in situ hybridization (FISH) in a high proportion of patients.17-19 Markedly, CAs in MGUS were similar to those in symptomatic MM, with IgH translocations and deletions of 13q14 observed at comparable frequencies.
In this study, we address the distribution pattern of CAs in AL compared with those in MGUS by interphase FISH analysis.
Methods
Patients
We evaluated a series of 231 consecutive patients with plasma cell disorders from a single institution who were tested for cytogenetic (FISH) abnormalities from May 2003 to February 2007. The AL cohort consisted of 85 patients, all of them with a histologically confirmed diagnosis. Of these, 76 were untreated and only 9 had received prior therapy without achieving a remission of their underlying plasma cell disorder. A series of 146 patients with MGUS or MM stage I (MM I) not requiring therapy were used as control groups. Patients with MM stage II or III were not considered in this analysis, also in case of a concomitant AL. As recommended by a consensus workshop report,19 patients with an IgM heavy chain subtype were also not eligible for this study. For the classification of patients into the MGUS and MM I groups we applied standard diagnostic criteria,20 except for bone marrow plasmocytosis of 30% instead of 10% as the cutoff value between MGUS and MM I, according to the Boston group and Mayo Clinic criteria for AL21,22 and our own practice.23 Thus, we obtained 4 patient groups: group 1 (n = 75) included patients with AL without concomitant MM; group 2 (n = 10) included patients with AL with concomitant MM I; group 3 (n = 127) included patients with MGUS; and group 4 (n = 19) included patients with MM I. Groups 1 and 2 were combined for the diagnosis of AL versus groups 3 and 4. Groups 1 and 3 were combined from the point of view of monoclonal gammopathy versus MM I (groups 2 and 4). The characteristics of the patient groups are given in detail in Table 1. As expected, lack of an intact immunoglobulin and lambda light chain restriction prevailed among patients with AL. However, age and sex were not statistically differently distributed among patient groups.
Patient characteristics
| . | Group 1: AL without MM I, n = 75 . | Group 2: AL plus MM I, n = 10 . | Group 3: MGUS, n = 127* . | Group 4: MM I, n = 19 . |
|---|---|---|---|---|
| Median age, y (range) | 62 (39-81) | 52 (45-70) | 59 (30-84) | 65 (41-77) |
| Sex, no. male/female | 44/31 | 2/8 | 58/69 | 10/9 |
| Light chain, no. kappa/lambda | 18/57 | 3/7 | 80/46 | 8/11 |
| Intact immunoglobulin, no. yes/no | 34/41 | 7/3 | 122/5 | 19/0 |
| Median plasma cell content, % (range) | 10 (2-26) | 17 (5-42) | 9 (1-28) | 28 (3-50) |
| Median LC urine, mg/d (range)† | 99 (< 5-850) | 1368 (< 5-2457) | < 5 (< 5-871) | 37 (< 5-1960) |
| No. patients with MM stage I as defined by the value of serum M protein | 0 | 0 | 0 | 11 |
| . | Group 1: AL without MM I, n = 75 . | Group 2: AL plus MM I, n = 10 . | Group 3: MGUS, n = 127* . | Group 4: MM I, n = 19 . |
|---|---|---|---|---|
| Median age, y (range) | 62 (39-81) | 52 (45-70) | 59 (30-84) | 65 (41-77) |
| Sex, no. male/female | 44/31 | 2/8 | 58/69 | 10/9 |
| Light chain, no. kappa/lambda | 18/57 | 3/7 | 80/46 | 8/11 |
| Intact immunoglobulin, no. yes/no | 34/41 | 7/3 | 122/5 | 19/0 |
| Median plasma cell content, % (range) | 10 (2-26) | 17 (5-42) | 9 (1-28) | 28 (3-50) |
| Median LC urine, mg/d (range)† | 99 (< 5-850) | 1368 (< 5-2457) | < 5 (< 5-871) | 37 (< 5-1960) |
| No. patients with MM stage I as defined by the value of serum M protein | 0 | 0 | 0 | 11 |
Age and sex were statistically equally distributed between AL and MGUS, whereas lambda light chain restriction and intact immunoglobulin deficiency prevailed among patients with AL.
This group includes one patient with a gammopathy biclonal for kappa and lambda.
LC indicates light chain.
Informed consent was obtained from the patients in accordance with the Declaration of Helsinki. The study was approved by the Ethics Committee of the University of Heidelberg.
Cytogenetic testing
Density gradient centrifugation of bone marrow aspirates over Ficoll Hypaque (Biochrom, Berlin, Germany) was performed to separate mononuclear cells by standard protocol. CD138+ plasma cells were isolated by magnetic-activated cell sorting using anti CD138 immunobeads and an auto–magnetic-activated cell sorter (MACS) separation system (Miltenyi Biotec, Bergisch Gladbach, Germany) according to the manufacturer's protocol. Purity was confirmed by the CD38+ and CD138+ phenotypes in flow cytometric analysis.
FISH was carried out using a panel of commercial 2-color probe sets for the detection of numerical chromosome changes for the following loci: 1q21/8p21, 11q23/13q14, and 17p13/19q13; and t(11;14), t(4;14), and t(14;16) for the identification of translocations. From May 2003 to November 2005, the first 94 patients were assessed for t(11;14) and t(4;14) only. Starting with the 95th patient of this study in November 2005, the strategy was switched to a determination of t(11;14) and the IgH breakapart probe first. In the event that t(11;14) turned out to be negative and the IgH breakapart probe yielded a positive result, the search of the translocation partner was pursued with probes for t(4;14) and t(14;16). Hybridization was performed according to the manufacturer's instructions (Kreated, Amsterdam, The Netherlands and Vysis, Downers Grove, IL). A total of 100 interphase nuclei per probe were evaluated using a DM RXA fluorescence microscope (Leica, Wetzlar, Germany). Hybridization efficiency was validated on metaphase spreads and interphase nuclei obtained from the peripheral blood and bone marrow of a healthy donor. The thresholds for gains, deletions, and translocations were set at 10%.
Statistical analysis
The frequencies of CAs of these patient groups were compared by the Fisher exact test. Of note, for each parameter, P values resulting from all tests were adjusted by the Holm method to control the family-wise error rate at .05.24
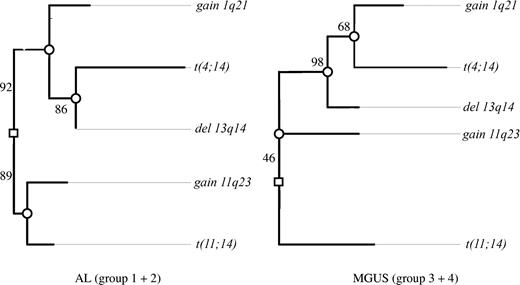
Oncogenetic tree models derived by maximum likelihood estimation were described by von Heydebreck et al.25 The root of the tree represents the state of normal cells. The marginal probabilities of the observed events are encoded by the length of the paths between the root and the corresponding nodes representing the following CAs: gain of 1q21 and 11q23, deletion of 13q14, and translocations t(11;14) and t(4;14). The length (horizontal distance) from the event (CA) to the root or the next inner node is the negative logarithm of the conditional probability that this aberration occurs at this node, given that the hidden events corresponding to this node happened. Independent events are represented by nonoverlapping paths to the root. To assess the uncertainty of the obtained tree models, a nonparametric bootstrap by Felsenstein et al26 is used. In Figure 1, confidence values for the internal edges of the maximum likelihood trees (based on 500 bootstrap data sets) are given. Therefore, the proposed tree structure has to be interpreted with caution, still allowing us to formulate hypotheses about the association between the CAs under discussion.
To analyze the association between CAs and hematologic factors, the Fisher exact test and exact Wilcoxon rank-sum test were performed, respectively, with Bonferroni-Holm correction for resulting P values.
Clinical factors and patient characteristics were compared by the Fisher exact test for categorical variables and by the exact Wilcoxon rank-sum test for quantitative variables, respectively. All statistical tests were 2-sided.
Statistical analysis was carried out using the software package R, version 2.5.1 together with the R package on comodel, version 0.7 (http://www.r-project.org/; Vienna, Austria).27
Results
Frequencies of chromosomal aberrations
In the AL without concomitant MM cohort (group 1), a CA could be detected in 95% of patients. Translocation involving the IgH heavy chain locus on chromosome 14q32 was found in 73% of patients. The most frequent translocation partner was chromosome 11. The resulting translocation t(11;14) was detected in 47% of patients, whereas other known translocations of the IgH heavy chain locus, t(4;14) and t(14;16), were rarely found in AL, accounting only for 3% and 4% of patients, respectively. Other frequent aberrations in AL included gain of 11q23 (33%), deletion of 13q14 (31%), gain of 1q21 (19%), and gain of 19q13 (15%), whereas deletion of 8p21 was a rare event and deletion of 17p13 could not be detected. The frequencies of all CAs tested are specified according to the 4 patient subgroups in Table 2. The frequency of 47% for t(11;14) in AL was higher than the incidence of 26% in MGUS (patient group 3; P = .03).
Frequencies of the respective chromosomal aberrations
| . | Group 1: AL without MM I, n = 75 . | Group 2: AL plus MM I, n = 10 . | Group 3: MGUS, n = 127 . | Group 4: MM I, n = 19 . |
|---|---|---|---|---|
| IgH translocations | ||||
| IgH breakapart | 37/51 (73) | 4/7 (57) | 39/66 (59) | 10/12 (83) |
| t(11;14) | 35/75 (47) | 3/10 (30) | 33/127 (26) | 5/19 (26) |
| t(4;14) | 2/75 (3) | 1/10 (10) | 12/127 (9) | 5/19 (26) |
| t(14;16) | 2/54 (4) | 0/7 (0) | 2/74 (3) | 0/12 (0) |
| IgH rearrangement with unknown translocation partner | 11/51 (22) | 2/7 (29) | 13/66 (20) | 1/11 (9) |
| Gains | ||||
| 1q21 | 14/74 (19) | 5/10 (50) | 16/109 (15) | 9/17 (53) |
| 11q23 | 25/75 (33) | 4/10 (40) | 42/126 (33) | 6/19 (32) |
| 19q13 | 8/55 (15) | 3/8 (38) | 20/79 (25) | 4/13 (31) |
| Deletions | ||||
| 8p21 | 4/55 (7) | 1/8 (13) | 5/77 (6) | 3/12 (25) |
| 13q14 | 23/75 (31) | 4/10 (40) | 39/127 (31) | 10/19 (53) |
| 17p13 | 0/74 (0) | 1/10 (10) | 5/124 (4) | 0/19 (0) |
| . | Group 1: AL without MM I, n = 75 . | Group 2: AL plus MM I, n = 10 . | Group 3: MGUS, n = 127 . | Group 4: MM I, n = 19 . |
|---|---|---|---|---|
| IgH translocations | ||||
| IgH breakapart | 37/51 (73) | 4/7 (57) | 39/66 (59) | 10/12 (83) |
| t(11;14) | 35/75 (47) | 3/10 (30) | 33/127 (26) | 5/19 (26) |
| t(4;14) | 2/75 (3) | 1/10 (10) | 12/127 (9) | 5/19 (26) |
| t(14;16) | 2/54 (4) | 0/7 (0) | 2/74 (3) | 0/12 (0) |
| IgH rearrangement with unknown translocation partner | 11/51 (22) | 2/7 (29) | 13/66 (20) | 1/11 (9) |
| Gains | ||||
| 1q21 | 14/74 (19) | 5/10 (50) | 16/109 (15) | 9/17 (53) |
| 11q23 | 25/75 (33) | 4/10 (40) | 42/126 (33) | 6/19 (32) |
| 19q13 | 8/55 (15) | 3/8 (38) | 20/79 (25) | 4/13 (31) |
| Deletions | ||||
| 8p21 | 4/55 (7) | 1/8 (13) | 5/77 (6) | 3/12 (25) |
| 13q14 | 23/75 (31) | 4/10 (40) | 39/127 (31) | 10/19 (53) |
| 17p13 | 0/74 (0) | 1/10 (10) | 5/124 (4) | 0/19 (0) |
Frequencies [no./N(%)]of the respective CA specified according to the four patient groups are given. Whereas the AL amyloidosis group displays the highest frequency of t(11;14), the MM I group is characterized by a higher frequency of gain 1q21. The determination of the IgH breakapart frequency is based on 136 patients, whereas the subgroups t(11;14), t(4;14), and t(14;16) are based on a larger cohort of 231, 231, and 147 patients, respectively. Due to the different sizes of the patient groups, the sum of the single translocations does not correspond exactly to the overall IgH breakapart frequency.
In AL and MGUS, disease progression to MM I was associated with an increased frequency of gain of 1q21 (16% vs 52%; P = .001) and t(4;14) (7% vs 21%); however, statistical significance for t(4;14) was lost after correction for multiple testing (P = .23).
Clustering of cytogenetic aberrations
In order to detect clustering of CAs we applied an oncogenetic tree model15,25 that was based on the whole study population, also including patients with MM I. Translocations of t(11;14) and t(4;14) and gains of 11q23 and 1q21, as well as deletion of 13q14, were chosen for this model. For these 5 major CAs, the data set was complete in 210 (91%) of 231 patients. A total of 72 of (86%) 84 patients with AL and 97 (77%) of 126 patients with MGUS displayed at least one of the 5 aberrations and could thus be included into the cluster analysis.
In the AL group we could discern 2 major independent branches (Figure 1). The first branch was characterized by t(11;14) together with gain of 11q23, and the second was characterized by deletion of 13q14, gain of 1q21, and t(4;14). In the first branch, t(11;14) and gain of 11q23 separated early from each other. In the second branch, t(4;14) was placed at the node farthest from the root, whereas gain of 1q21 separated earlier from this pathway.
Clustering of chromosomal aberrations in the oncogenetic tree model. Maximum likelihood tree models for the AL group (groups 1 and 2) and MGUS group (groups 3 and 4) based on the CAs observed in 210 of the patients. The length of each horizontal edge “e” is proportional to −log(pe) (eg, in groups 1 and 2, the distance of deletion 13q14 to the next inner node is 0, which means that this aberration has always occurred at this node and precedes the translocation t(4;14)). Bootstrap confidence values (percentage) for the inner edges are given. In groups 1 and 2, the translocation t(11;14) was often observed together with gain of 11q23 (88.6%) in contrast to only 16% in groups 3 and 4. Also, in 46.4% of bootstrap samples, t(11;14) was individually separated from the other 4 aberrations in groups 3 and 4.
Clustering of chromosomal aberrations in the oncogenetic tree model. Maximum likelihood tree models for the AL group (groups 1 and 2) and MGUS group (groups 3 and 4) based on the CAs observed in 210 of the patients. The length of each horizontal edge “e” is proportional to −log(pe) (eg, in groups 1 and 2, the distance of deletion 13q14 to the next inner node is 0, which means that this aberration has always occurred at this node and precedes the translocation t(4;14)). Bootstrap confidence values (percentage) for the inner edges are given. In groups 1 and 2, the translocation t(11;14) was often observed together with gain of 11q23 (88.6%) in contrast to only 16% in groups 3 and 4. Also, in 46.4% of bootstrap samples, t(11;14) was individually separated from the other 4 aberrations in groups 3 and 4.
The major difference between the AL and the MGUS groups was the grouping of gain of 11q23. Whereas it was linked with the t(11;14) branch in AL, it was placed on the opposite side of the root in the MGUS group. The significance of this finding in cluster analysis was confirmed by the Fisher exact test, where the combination of t(11;14) and gain of 11q23 proved more frequent in AL than in MGUS (20% vs 7%; P = .005). As a minor difference between the respective AL and MGUS trees, gain of 1q21 separated earlier from the deletion of 13q14 branch in AL.
In spite of these 2 differences, the trees for AL and MGUS shared common features: they both showed a t(11;14) branch independent of deletion of 13q14 pathway. Also, in both tree models, t(4;14) and gain of 1q21 were grouped together with deletion of 13q14.
Association of CAs with hematologic parameters
We analyzed the association of CAs with hematologic parameters, including the detection of an intact immunoglobulin and—if detected—its subtype, kappa versus lambda light chain restriction, and the bone marrow plasma cell content.
In the overall study population, the detection of t(11;14) was associated with a lack of intact immunoglobulin in immunofixation (P < .001; Table 3). Gain of 19q13 was associated with the detection of an intact immunoglobulin (94% vs 72%; P = .05). The immunoglobulin heavy chain subtype retained no statistical significance following P value adjustment for multiple testing (data not shown) for all analyzed CAs. Gain of 1q21 turned out to be the only CA which was associated with a lambda light chain restriction (Table 4). Gain of 1q21 and 11q23 as well as deletion of 13q14 were associated with a higher plasma cell content (median of 11% vs 9% [P = .002], 12% vs 9% [P = .03], and 12% vs 9% [P = .04], respectively).
Association of t(11;14) with hematologic parameters
| . | Overall study population, patient groups 1-4, n = 231 . | AL amyloidosis series, patient groups 1-2, n = 85 . | ||||
|---|---|---|---|---|---|---|
| t(11;14) . | No t(11;14) . | P . | t(11;14) . | No t(11;14) . | P . | |
| Intact immunoglobulin by immunofixation, no. patients (%) | 45/76 (59) | 137/155 (88) | < .001 | 7/38 (18) | 34/47 (72) | < .001 |
| Lambda light chain restriction, no. patients (%) | 43/75 (57) | 78/155 (50) | 1.0 | 26/38 (68) | 38/47 (81) | 1.0 |
| Median involved FLC concentration, mg/L | ND | ND | ND | 257 | 193 | 1.0 |
| Median plasma cell content, % | 10 | 10 | 1.0 | 10 | 10 | 1.0 |
| . | Overall study population, patient groups 1-4, n = 231 . | AL amyloidosis series, patient groups 1-2, n = 85 . | ||||
|---|---|---|---|---|---|---|
| t(11;14) . | No t(11;14) . | P . | t(11;14) . | No t(11;14) . | P . | |
| Intact immunoglobulin by immunofixation, no. patients (%) | 45/76 (59) | 137/155 (88) | < .001 | 7/38 (18) | 34/47 (72) | < .001 |
| Lambda light chain restriction, no. patients (%) | 43/75 (57) | 78/155 (50) | 1.0 | 26/38 (68) | 38/47 (81) | 1.0 |
| Median involved FLC concentration, mg/L | ND | ND | ND | 257 | 193 | 1.0 |
| Median plasma cell content, % | 10 | 10 | 1.0 | 10 | 10 | 1.0 |
Table shows the association of the CA t(11;14) with hematologic parameters [n/N(%)]. he strongest association was detected between the detection of t(11;14) and the lack of an intact immunoglobulin.
FLC indicates full light chain; ND, not determined.
Association of gain of 1q21 with hematologic parameters
| . | Overall study population, patient groups 1-4, n = 231 . | AL amyloidosis series, patient groups 1-2, n = 85 . | ||||
|---|---|---|---|---|---|---|
| Gain 1q21 . | No gain 1q21 . | P . | Gain 1q21 . | No gain 1q21 . | P . | |
| Intact immunoglobulin by immunofixation, no. patients (%) | 40/44 (91) | 122/166 (73) | .12 | 15/19 (79) | 26/65 (40) | .03 |
| Lambda light chain restriction, no. patients (%) | 34/44 (77) | 81/165 (49) | .01 | 18/19 (95) | 45/65 (69) | .29 |
| Median involved FLC concentration, mg/L | ND | ND | ND | 231 | 204 | 1.0 |
| Median plasma cell content, % | 11 | 9 | .002 | 11 | 9 | .55 |
| . | Overall study population, patient groups 1-4, n = 231 . | AL amyloidosis series, patient groups 1-2, n = 85 . | ||||
|---|---|---|---|---|---|---|
| Gain 1q21 . | No gain 1q21 . | P . | Gain 1q21 . | No gain 1q21 . | P . | |
| Intact immunoglobulin by immunofixation, no. patients (%) | 40/44 (91) | 122/166 (73) | .12 | 15/19 (79) | 26/65 (40) | .03 |
| Lambda light chain restriction, no. patients (%) | 34/44 (77) | 81/165 (49) | .01 | 18/19 (95) | 45/65 (69) | .29 |
| Median involved FLC concentration, mg/L | ND | ND | ND | 231 | 204 | 1.0 |
| Median plasma cell content, % | 11 | 9 | .002 | 11 | 9 | .55 |
Table shows the association of the CA t(11;14) and gain 1q21 with hematologic parameters [n/N(%)]. Effects could be shown for the detection of an intact immunoglobulin, light chain restriction, and bone marrow plasmocytosis.
FLC indicates free light chain; ND, not determined.
In AL (groups 1 and 2), the association of t(11;14) with lacking intact immunoglobulin could also be shown (P < .001). Gain of 1q21 was more frequent in patients with AL who displayed an intact immunoglobulin (P = .03). Detection of gain of 11q23 was associated with kappa light chain restriction (P = .03). CAs were not associated with serum-free light chain levels.
Delineation of characteristic features of AL by multivariate analysis
A multivariate analysis was performed in order to identify the parameters that are characteristic for AL from a genetics and monoclonal gammopathy standpoint and those that set it apart from other plasma cell dyscrasias. The following variables were included as possible explanatory factors in the multivariate logistic regression model for development of an AL phenotype: the 5 major CAs, namely gain of 11q23 and 1q21, deletion of 13q14, and t(11;14) and t(4;14), and the hematologic parameters bone marrow plasmocytosis, light chain restriction, and detection of intact immunoglobulin. The lack of an intact immunoglobulin and the preponderance of lambda light chain restriction maintained themselves as hallmarks of AL (P < .001 and P < .001, respectively). Neither the higher incidence of t(11;14) nor the lower frequency of t(4;14) in AL reached statistical significance in this model (P = .91 and P = .11, respectively). Thus, none of the CAs by itself could explain the phenotype of AL.
Lacking association of CAs with clinical parameters in the AL group
Next we tested the association of CAs of the AL cohort with clinical characteristics. The following clinical parameters were included: sex; age; number of affected organs; N-terminal pro–brain natriuretic peptide (NT-proBNP) values; involvement of heart, kidney, gastrointestinal (GI) tract, liver, and soft tissue; peripheral neuropathy; Karnofsky index; and eligibility for high-dose chemotherapy, the latter only being evaluated in patients younger than 70 years. Markedly, none of these clinical parameters showed a significant association with any of the CAs (data not shown). CAs therefore do not seem to influence the pattern of organ involvement in AL.
Discussion
Our analysis of CAs in AL revealed a strikingly high frequency of 47% for the translocation t(11;14), which was significantly higher than the 26% for our MGUS group and also exceeded by far the frequencies reported for patients with symptomatic MM in previous studies, which range from 13% to 21%.28-32 This high t(11;14) positivity in AL in our analysis is in tune with previous reports, which—though based on a lower patient number—detected this translocation in 16 (55%) of 29 and 9 (38%) of 24 of patients with AL, respectively.7,8 The overrepresentation of the t(11;14) translocation among patients with AL suggests that cyclin D1 up-regulation33,34 and disruption of the heavy chain locus might also be important pathogenetic mechanisms in AL. Indeed, 82% of patients with a t(11;14) lacked an intact immunoglobulin in serum immunofixation. This finding in our study is reminiscent of the higher t(11;14) frequencies of 31%, 79%, and 88% reported for light chain only, nonsecretory, and IgM myelomas, respectively, compared with the lower frequencies in myelomas with an intact rearranged immunoglobulin, namely 15%, 10%, and 22% for IgG, IgA, and IgD in studies by Avet-Loiseau et al.35,36 Whereas the high frequency of t(11;14) in AL accounts for the high rate of intact immunoglobulin deficiency, it is probably unrelated to the lambda light chain predilection in AL, which is another hallmark of AL from the standpoint of monoclonal gammopathy. In AL, the kappa patients even had a slightly higher rate of t(11;14) than the lambda patients (57% vs 41%).
Next, we evaluated the association of t(11;14) with other CAs. As shown in the tree model and Fisher testing, t(11;14) in AL was frequently detected in combination with a gain of 11q23, which has been identified as a critical genomic region for CAs in MM29 and other B-cell disorders.37 Since this could only rarely be detected in the MGUS group, the combination of t(11;14) and gain of 11q23 appears to be a specific feature of AL.
Translocation t(4;14), another IgH translocation on which conflicting frequencies have been reported in AL so far,7,9 could only be detected in 4% of patients with AL in our cohort. This was lower than that of our MGUS group with 12% and also inferior to frequencies reported for MM, which range from 11% to 17%.15,28,31,32,38
Like MGUS and MM, karyotypic instability was a recurring feature of AL. With our testing panel, 95% of patients with AL displayed at least one CA, and all major CAs identified in MGUS were also detected in our AL group. Aberrations like t(14;16); gains of chromosomes 1q21, 11q23, and 19q13; and deletions of chromosomes 8p21, 13q14, and 17p13 were found at comparable frequencies in the AL and the MGUS groups. Further similarities between the AL and the MGUS groups were revealed by the clustering of CAs based on the oncogenetic tree model.15 The association of t(11;14) with gain of 11q23 was more frequent in AL, but apart from this, we found that the tree models for the AL and the MGUS/MM I groups widely overlapped. In addition to the t(11;14) and 11q23 branches, deletion of 13q14, t(4;14), and gain of 1q21 clustered together as a common branch. This association of deletion of 13q14 with t(4;14) and gain of 1q21 previously described in MM15,39,40 could thus be confirmed in our AL and MGUS groups.
Markedly, gain of 1q21, which has been recognized as a prognostically unfavorable marker,39,40 was found at higher frequencies in the MM I groups both with and without AL (50% and 53%, respectively) compared with the monoclonal gammopathy groups (19% and 15%, respectively). Our data therefore suggest that the concept of gain of 1q21 as a progression marker characteristic of cytogenetic evolution during disease progression41 applies to AL and MGUS alike.
The similarities of cytogenetic patterns between AL and MGUS were also highlighted by our multivariate analysis, where none of the major CAs proved statistically significantly associated with the AL phenotype in competition with hematologic parameters. Furthermore, none of the CAs had an influence on the tissue affinity of amyloid fibrils as also shown by Bryce et al42 or determined the pattern and the severity of organ involvement. This also suggests that the CAs do not directly mediate amyloidogenicity.
In conclusion, the concept of distinct pathogenetic CAs and karyotypic instability, which was developed for MM, is also applicable to AL. Accordingly, the clinical picture of AL is rather dictated by the amyloidogenic properties of the light chain than by karyotypic instability itself. This concept has also been referred to in AL as “MGUS with an unlucky protein.”8 In this context, the low frequencies of t(4;14) and deletion of 17p13, which are considered to bring about a rapid progression of the plasma cell disorder,31,32,35,38,43-45 fit the clinical course of patients, since complications in AL arise rather from the amyloidogeneic potential of the secreted light chains than from progression into symptomatic MM. Conversely, the high frequency of t(11;14) in AL suggests that this translocation—though less aggressive—nevertheless sustains the proliferation of the aberrant plasma cell clone so that amyloidosis-related symptoms prompt the detection of this “more benign” gammopathy that otherwise may not have become symptomatic so soon. In tune with this model, Fonseca et al18 —based on a higher frequency of t(11;14) in MGUS than in MM—have suggested that t(11;14) is negatively selected for progression from an early-stage plasma cell disorder to symptomatic MM.
It will be very interesting to evaluate in future studies whether CAs in AL are of prognostic value for achieving hematologic remission after melphalan-based chemotherapy regimens.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported by the Tumorzentrum Heidelberg/Mannheim (grant no. FSPI./6) and by the “Dietmar Hopp Stiftung.”
Authorship
Contribution: T.B., S.O.S., U.H., F.C., A.J., and H.G. designed research; A.J., D.H., F.C., and J.B. performed research; U.H., S.O.S., T.B., A.J., and F.C. collected data; U.H., S.O.S., T.B., F.C., A.J., M.M., C.B., A.D.H., and H.G. analyzed and interpreted data; C.H. and A.B. performed statistical analysis; U.H., T.B., S.O.S., and C.H. drafted the manuscript; and all coauthors revised the manuscript.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Stefan Schönland, Medizinische Klinik V, Universitätsklinikum Heidelberg, Im Neuenheimer Feld 410, 69120 Heidelberg, Germany; e-mail: stefan.schoenland@med.uni-heidelberg.de.
References
Author notes
*T.B. and U.H. contributed equally to this paper.
†A.J. and S.O.S. are to be considered equal last authors.