Abstract
CD38 and ZAP-70 are both useful prognostic markers for B-cell chronic lymphocytic leukemia (CLL), but are variably discordant with IGHV mutation status. A total of 5 human Fc receptor–like molecules (FCRL1-5) have tyrosine-based immunoregulatory potential and are expressed by B-lineage subpopulations. To determine their prognostic potential in CLL, FCRL expression was compared with IGHV mutation status, CD38 and ZAP-70 expression, and clinical features from 107 patients. FCRL1, FCRL2, FCRL3, and FCRL5 were found at markedly higher levels on CLL cells bearing mutated IGHV genes than on unmutated CLL cells or CD19+ polyclonal B lymphocytes. Univariate comparisons found that similar to CD38 and ZAP-70, FCRL expression was strongly associated with IGHV mutation status; however, only FCRL2 maintained independent predictive value by multivariate logistic analysis. Strikingly, FCRL2 demonstrated 94.4% concordance with IGHV mutation compared with 76.6% for CD38 and 80.4% for ZAP-70. Compared with other indicators, FCRL2 was also superior at predicting the time to first therapy; the median treatment-free interval was 15.5 years for patients with high FCRL2 expression compared with 3.75 years for FCRL2-low patients. Our studies indicate that FCRL2 has robust predictive value for determining IGHV gene mutation status and clinical progression and thus may further improve prognostic definition in CLL.
Introduction
The Rai and Binet classical staging systems have been useful for determining survival in chronic lymphocytic leukemia (CLL)1,2 ; however, they are unable to predict an indolent or aggressive clinical course at early stages in this heterogeneous disease. In terms of global gene transcription and protein phenotype, CLL cells resemble activated memory-like B cells,3-5 but can be divided into 2 groups based on the mutation status of their IGHV gene.6,7 This distinction has provided important clinical and biological insight into the significant differences in disease course,8 clinical outcome, and survival between these 2 subgroups.9,10 Thus, accurately distinguishing patients with mutated CLL (MT-CLL) who usually experience a more indolent disease course from patients with unmutated CLL (UM-CLL) who typically experience a more aggressive course is of increasing importance as treatment protocols evolve. Unfortunately, DNA sequencing is not a practical diagnostic test in most clinical laboratories. Therefore, a search for reliable markers of IGHV mutation status has been intensely investigated
Although CD38 overexpression was initially thought to correlate with UM-CLL,9 analysis of a larger series of samples uncovered up to 30% discordance and found that CD38 is an independent prognostic marker of aggressiveness and clinical progression.11,12 Profiling transcripts from UM-CLL and MT-CLL samples by microarray identified the ZAP-70 Syk family tyrosine kinase gene as the best indicator of UM-CLL,13 and follow-up studies by several groups confirmed that overexpression of its encoded product also correlates with an aggressive disease course.14-17 Unfortunately, the variable staining intensity and cytoplasmic localization of the ZAP-70 protein has created a technical hurdle for assigning a cutoff threshold value that has resulted in diagnostic inconsistency among clinical laboratories.18-25 Although great effort is being expended to standardize ZAP-70 staining assays internationally,20 other distinguishing markers would be helpful for easier prognostic confirmation to subgroup patients with CLL and perhaps better understand the biology of this malignancy.
A family of 5 Ig superfamily molecules termed Fc receptor–like (FCRL) possess tyrosine-based immunoregulatory potential and are differentially expressed by subpopulations of B-lineage cells.26-32 Although their ligands remain unknown, these characteristics suggest that FCRL family members influence B-cell signaling in a positive and/or negative manner, and thus may function to regulate innate and adaptive humoral immunity. The clinical significance of these receptors is suggested by their expression patterns in transformed B lymphocytes. According to microarray analysis, FCRL1 to FCRL4 (FCRL1-4) are differentially overexpressed by diffuse large B-cell lymphoma, mantle cell lymphoma, and CLL.33,34 Subsequent expression profiling of a large series of CLL samples revealed that, other than ZAP-70, FCRL2 and FCRL3 were among a small group of statistically significant genes overexpressed by MT-CLL cells.3,13 Several groups have also now detected the expression of different FCRL representatives by CLL cells.31,35,36 For example, using a panel of FCRL1-5 monoclonal antibodies (mAbs), Polson et al identified FCRL expression on CLL cells,31 and by microarray analysis, Huttmann et al found that FCRL2 was differentially up-regulated by indolent CD38−ZAP-70− CLL samples and confirmed FCRL2 protein expression with polyclonal Abs.35 In an attempt to explore the immunotherapeutic potential of FCRL molecules in B-cell malignancies, Du et al observed significant cytotoxicity upon targeting FCRL1+ malignancies such as CLL, hairy-cell leukemia, and other B-cell non-Hodgkin lymphomas, with immunotoxin-labeled anti-FCRL1 mAbs.36
These initial reports indicate the importance of these molecules in normal and pathologic B-cell biology. To determine if the FCRL proteins could be useful clinical and biologic markers in CLL, we examined samples from 107 patients and correlated FCRL expression with IGHV mutation status, CD38 surface expression, ZAP-70 cytoplasmic expression, and clinical features. Our data show that similar to ZAP-70 and CD38, FCRL1, FCRL2, FCRL3, and FCRL5 are able to stratify CLL samples according to the mutation status of the IGHV gene expressed by the leukemic clone. However in contrast to CD38 and ZAP-70, which are preferentially expressed by UM-CLL samples, the 4 FCRL representatives identified on CLL cells are distinctly overexpressed by patients with MT-CLL. Although all 4 FCRL proteins could distinguish MT-CLL from UM-CLL samples and were predictive of the need for treatment, our analysis indicates that FCRL2 is a superior prognostic marker in CLL because of its very strong correlation with mutated IGHV status and ability to predict clinical progression as defined by the time to first therapy.
Methods
Recruitment of patients with CLL
The study protocol was approved by the University of Alabama at Birmingham (UAB) institutional review board (IRB). CLL samples obtained from the North Shore University Hospital (NSUH) Long Island Jewish Medical Center and the Moores Cancer Center at the University of California, San Diego, met National Cancer Institute (NCI) criteria for the diagnosis of CLL,1 and were provided through material transfer agreements. Written informed consent was obtained from all participants before enrollment in the study in accordance with the Declaration of Helsinki, and all donor samples were anonymized to maintain health information confidentiality. Clinical characteristics, including sex, diagnosis age, diagnosis date, Rai stage, treatment history, and the date of initial therapy, were also reviewed for comparison. Although samples were derived from 3 different locations, no significant heterogeneity in the time from diagnosis to initial therapy was found among centers according to the product limit method of Kaplan-Meier and the log-rank test (data not shown).
Production of mAbs
FCRL-specific mAbs were generated by immunizing BALB/c mice with BW5147 mouse T-cell line transductants expressing individual hemagglutinin (HA)–tagged FCRL surface proteins,37 and hybridomas were produced according to standard methodology.38 Before subcloning, hybridoma supernatants were screened by staining FCRL1-6 stable transductants and analyzing reactivity by flow cytometry to confirm FCRL specificity and potential crossreactivity. FCRL mAb isotypes were determined by indirect capture enzyme-linked immunosorbent assay (ELISA; Zymed, San Francisco, CA). The anti-FCRL1 (clone 5A3; γ2bκ) mAb has been described previously.29 The anti-FCRL2 (clone 7F2; γ1κ), FCRL3 (clone 3D2; γ1κ), FCRL4 (clone 10E4; γ2bκ), and FCRL5 (clone 2B4; γ1κ) mAbs were newly generated for these studies. Purified mAbs were biotinylated using the EZ-Link Sulfo-NHS-LC-Biotin kit (Pierce, Rockford, IL).
Multiparameteric flow cytometry
After collection in anticoagulant containing tubes, peripheral blood mononuclear cells (PBMCs) were isolated by Ficoll-Hypaque (Mediatech, Herndon, VA) density gradient centrifugation and either analyzed immediately or cryopreserved for future use. Mononuclear cells were stained with anti-CD5 (clone UCHT2) fluorescein isothiocyanate (FITC) and CD19-allophycocyanin (APC; clone HIB19) to identify the expanded CLL population, costained with biotinylated anti-FCRL1–5 mAbs, and incubated with streptavidin-phycoerythrin (SA-PE) prior to 3-color flow cytometric analysis (BD Biosciences, San Diego, CA).29 Fluorochrome-conjugated isotype-matched negative control mAbs were used to detect nonspecific staining. Necrotic cells were excluded with propidium iodide (PI; Fluka, Buchs, Switzerland). After gating the lymphoid population according to typical light scatter characteristics, a total of 105 events were acquired for analysis using a FACSCalibur flow cytometer equipped with CellQuest (BD Biosciences) and WinMDI software (Scripps Institute, La Jolla, CA). To calculate relative expression levels, mean fluorescence intensity (MFI) ratios were determined by dividing the MFI of the antigen-specific fluorochrome-conjugated mAb by the MFI of the irrelevant fluorochrome-conjugated isotype-matched negative control mAb. The MFI ratio calculation assists by eliminating arbitrary assignment of the negative control mAb threshold.
CD38 surface measurements were determined with anti-CD38–PE (clone HB-7; BD Biosciences) according to published criteria.9,12 A cut-off value of greater than 30% was used to determine a positive result. ZAP-70 cytoplasmic staining was performed according to comparative studies with minor modifications.19,21,39,40 Briefly, 106 purified mononuclear cells were stained with anti-CD19–APC and CD5-FITC mAbs and incubated for 20 minutes at room temperature in the dark. Cells were then fixed and permeabilized using the Fix and Perm kit (Invitrogen, Burlingame, CA), counterstained with anti–ZAP-70–PE (clone 1E7.2; Invitrogen) or an isotype-matched control mAb, incubated in the dark for 20 minutes, washed twice, and analyzed with a FACSCalibur flow cytometer. A threshold of greater than 20% relative to the isotype control was considered a positive result. A second internal control-based method was also used by comparing the CD19−CD5+ T-cell population with the CD19+CD5+ B-CLL population. A T-cell /B-CLL cell MFI ratio less than 4 was considered a positive result.40
IGHV gene sequencing analysis
Statistical analysis
The Kruskal-Wallis test followed by the Dunn multiple comparison analysis was applied to assess FCRL expression differences among UM-CLL, MT-CLL, and normal B cells. Maximum Youden index values were used to determine optimal threshold cut-off values for prognostic factors as continuous variables based on the receiver operating characteristic (ROC).42 This allowed for optimizing the cut points for outcome of CLL subtypes based on IGHV mutation status. Recursive partitioning also referred to as classification and regression tree (CART) analysis was additionally used as an alternative means for determining cutoff values.43 Comparisons between IGHV mutation status and the different prognostic factors were performed using univariate and multivariate logistic regression models. The C-Statistic, which is a generalization of the area under the ROC curve (AUC), was used to assess the overall predictive accuracy of these models.44 The Kendall tau-b was used to quantify the strength of association between 2 variables, with values of plus or minus 0.50 indicating a strong relationship.45 After optimizing cut points, data assessing FCRL expression and clinical progression as defined by the time to initial therapy was estimated by constructing Kaplan-Meier survival plots and using the log-rank test. Comparisons between potential prognostic factors and time from diagnosis to initial therapy were analyzed using the Cox regression model. All statistical analysis was performed using SAS 9.1 (SAS Institute, Cary, NC) or the RPART Package - R version 2.5.1 (The R Foundation for Statistical Computing, Vienna, Austria).
Results
Generation of FCRL1-5-specific mAbs
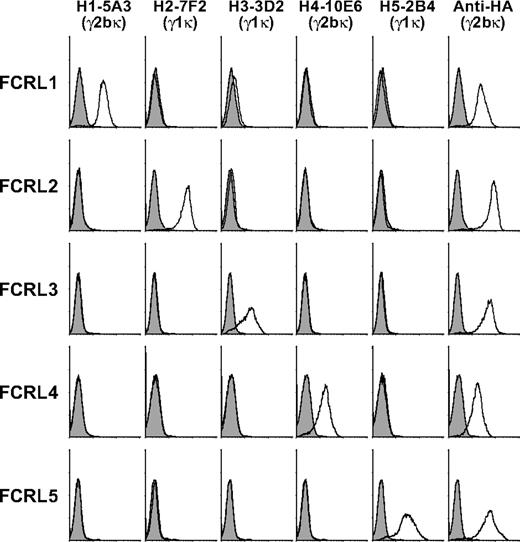
A panel of FCRL-specific mAbs was generated by immunizing mice with HA-tagged retroviral transductants stably expressing human FCRL1-5 in the BW5147 mouse thymoma cell line. Reactive hybridoma subclones were screened for cross-reactivity with all 5 transductant cell lines as well as FCRL6 (data not shown). These mAbs were found to be receptor specific and were capable of detecting their respective proteins by immunofluorescence (Figure 1) and immunoprecipitation (data not shown).
Generation of a panel of human FCRL1-5-specific mAbs. FCRL1-5 mAbs were screened for cross-reactivity with BW5147 cells retrovirally transduced with plasmids expressing individual HA-tagged FCRLs. Cells were stained with the indicated biotin-coupled mouse IgG isotype FCRL mAbs (histogram, black line) or isotype matched controls (histogram, gray shade) followed by SA-PE, and analyzed by flow cytometry. Positive control staining was performed with the 12CA5 (anti-HA) mAb. FCRL1-5 mAbs also did not react with FCRL6 transductants (not shown).
Generation of a panel of human FCRL1-5-specific mAbs. FCRL1-5 mAbs were screened for cross-reactivity with BW5147 cells retrovirally transduced with plasmids expressing individual HA-tagged FCRLs. Cells were stained with the indicated biotin-coupled mouse IgG isotype FCRL mAbs (histogram, black line) or isotype matched controls (histogram, gray shade) followed by SA-PE, and analyzed by flow cytometry. Positive control staining was performed with the 12CA5 (anti-HA) mAb. FCRL1-5 mAbs also did not react with FCRL6 transductants (not shown).
FCRL expression differs between polyclonal CD19+ B cells and CLL cells
Except for FCRL3, which is also identified on subsets of natural killer (NK) and T cells (Polson et al31 ; unpublished data), FCRL1-5 are all expressed by subpopulations of human B cells (reviewed in Davis32 ). These findings, as well as the intriguing microarray data from the studies described in the Introduction,3,13,33,35 led to an investigation of FCRL1-5 protein expression in CLL.
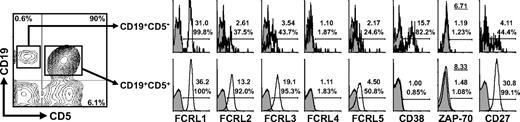
To assess whether FCRL1-5 are expressed by CLL cells, mononuclear cells prepared from the blood of a single patient were first analyzed. Staining for CD5 and CD19 identified both polyclonal CD19+CD5− B cells (0.6%) and a large population of expanded CD19+CD5+ CLL B cells (90%). Consistent with previous data, FCRL1, FCRL2, FCRL3, and FCRL5 were detected on both the polyclonal B cells and transformed CLL cells, but FCRL4 was not (Figure 2).31 In particular, FCRL1 was expressed by all blood B cells, whereas FCRL2, FCRL3, and FCRL5 were expressed by subsets of CD19+ cells. Interestingly, FCRL2, FCRL3, FCRL5, and CD27, but not FCRL1, were clearly overexpressed on the malignant CD19+CD5+ B-cell population compared with polyclonal CD19+CD5− B cells. In this representative sample, CD38 expression was identified among the CD19+CD5− B cells, but was not detected on the CLL clone. ZAP-70 staining was also negative in the CD19+ B cells and CLL cells from this donor.
Comparison of FCRL1-5 surface expression on polyclonal B cells and malignant CLL cells. Ficoll-purified mononuclear cells isolated from the blood of a patient with CLL (CLL37; Table S1) were stained with discriminating mAbs to detect polyclonal CD19+CD5− B cells and the CD19+CD5+ leukemic CLL population in order to assess FCRL1-5, CD38, ZAP-70, and CD27 expression by multiparametric flow cytometry. Histograms reflect the staining of specific mAbs (black line) versus isotype-matched controls (gray shade) within the respective gated populations. The numbers indicated within histograms specify the MFI ratio (top values) and the percent of the population staining positive (bottom values). The MFI ratio is derived from the MFI of the test mAb divided by that of the isotype-matched control. Note that the underlined numbers in the ZAP-70 panel indicate the T/B-CLL MFI ratio.
Comparison of FCRL1-5 surface expression on polyclonal B cells and malignant CLL cells. Ficoll-purified mononuclear cells isolated from the blood of a patient with CLL (CLL37; Table S1) were stained with discriminating mAbs to detect polyclonal CD19+CD5− B cells and the CD19+CD5+ leukemic CLL population in order to assess FCRL1-5, CD38, ZAP-70, and CD27 expression by multiparametric flow cytometry. Histograms reflect the staining of specific mAbs (black line) versus isotype-matched controls (gray shade) within the respective gated populations. The numbers indicated within histograms specify the MFI ratio (top values) and the percent of the population staining positive (bottom values). The MFI ratio is derived from the MFI of the test mAb divided by that of the isotype-matched control. Note that the underlined numbers in the ZAP-70 panel indicate the T/B-CLL MFI ratio.
An expanded analysis of 107 CLL samples and 10 age-matched healthy volunteers was then performed to determine the relative abundance of FCRL expression on malignant CLL cells versus polyclonal CD19+ B cells. In general, FCRL1 had the highest MFI ratios regardless of the population analyzed; however, no significant difference was found for FCRL1 expression between the entire CLL cohort and healthy donor samples or for the 3 other FCRLs by nonparametric Kruskal-Wallis test comparisons. In agreement with a prior report,31 no significant difference in FCRL expression was found among polyclonal CD19+, CD19+CD5−, or CD19+CD5+ B cells from healthy volunteers (data not shown). This data indicates that FCRL1, FCRL2, FCRL3, and FCRL5 are expressed by CLL cells, but FCRL4 is not. Furthermore, FCRL expression does not significantly differ between this sample cohort of malignant CLL cells as a whole compared with normal polyclonal B cells.
FCRL surface expression correlates with IGHV mutation status
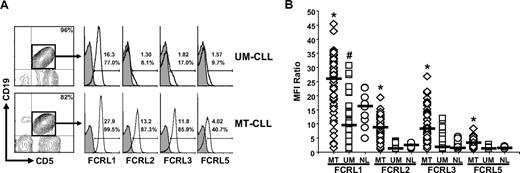
CLL samples were next compared to determine whether FCRL surface expression varies between patients. An analysis of 2 representative patients with UM-CLL and MT-CLL that have marked differences in the surface density of FCRL proteins is depicted in Figure 3A. While FCRL1 was expressed at relatively high levels on the leukemic expansion in both patients, FCRL5 staining was somewhat lower. In contrast, an even greater degree of difference in MFI was evident for FCRL2 and FCRL3 staining between these samples. Nevertheless, all 4 of these FCRL representatives demonstrated differences in staining between these samples, with higher levels on the MT-CLL donor relative to the UM-CLL donor.
Heterogeneous FCRL protein expression on CLL cells correlates with IGHV mutation status. (A) FCRL expression varies between CLL samples. Purified mononuclear cells from the blood of 2 patients with CLL, representing the UM-CLL (CLL08) and MT-CLL (CLL32) subtypes, were stained with discriminating mAbs (black line) to define FCRL1, FCRL2, FCRL3, and FCRL5 expression on the expanded CD19+CD5+ CLL population. Isotype-matched control staining is indicated by histograms with a gray shade. Numerical values indicate the MFI ratio (top number) and percentage of the population staining positive (bottom number). (B) FCRL surface expression correlates with IGHV gene mutation status. MFI ratio comparisons determined for FCRL1, FCRL2, FCRL3, and FCRL5 expression on CD19+CD5+ cells from 55 MT-CLL samples (◇), 52 UM-CLL samples (□), and CD19+ B cells from 10 healthy volunteers (NL; ○). *P < .05 compared with UM and NL; #P < .05 compared with NL. The characteristics of the patient samples are described in Table S1.
Heterogeneous FCRL protein expression on CLL cells correlates with IGHV mutation status. (A) FCRL expression varies between CLL samples. Purified mononuclear cells from the blood of 2 patients with CLL, representing the UM-CLL (CLL08) and MT-CLL (CLL32) subtypes, were stained with discriminating mAbs (black line) to define FCRL1, FCRL2, FCRL3, and FCRL5 expression on the expanded CD19+CD5+ CLL population. Isotype-matched control staining is indicated by histograms with a gray shade. Numerical values indicate the MFI ratio (top number) and percentage of the population staining positive (bottom number). (B) FCRL surface expression correlates with IGHV gene mutation status. MFI ratio comparisons determined for FCRL1, FCRL2, FCRL3, and FCRL5 expression on CD19+CD5+ cells from 55 MT-CLL samples (◇), 52 UM-CLL samples (□), and CD19+ B cells from 10 healthy volunteers (NL; ○). *P < .05 compared with UM and NL; #P < .05 compared with NL. The characteristics of the patient samples are described in Table S1.
To determine whether FCRL expression consistently correlates with the mutation status of the IGHV gene expressed by leukemic CLL cells, FCRL1, FCRL2, FCRL3, and FCRL5 surface staining on the CD19+CD5+ population from 107 IGHV genotyped CLL samples was compared with CD19+ polyclonal B cells from 10 healthy donors by flow cytometry. This analysis, which included 52 UM-CLL and 55 MT-CLL donors (Table S1), found that FCRL1, FCRL2, FCRL3, and FCRL5 are all expressed at significantly higher levels on MT-CLL cells compared with UM-CLL cells or polyclonal B cells from healthy volunteers (Figure 3B). While FCRL1 was expressed at higher levels on MT-CLL, the magnitude and range of the MFI ratios were much greater than those of other family members on the 3 different populations. Compared with normal CD19+ B cells, however, FCRL1 expression was significantly lower on UM-CLL samples. These data indicate that FCRL proteins have a heterogeneous pattern of expression in CLL that correlates with IGHV mutation status.
Elevated FCRL2 expression strongly correlates with the MT-CLL subtype
The prognostic value of FCRL1, FCRL2, FCRL3, and FCRL5 was then evaluated in the same cohort of 107 patients with CLL. The clinical characteristics of these samples are summarized in Tables 1 and S1. This cohort of patients was typical for individuals with CLL. Among the 107 samples, the ratio of men to women was 1.5:1, with a median age of 57 years. At the time of diagnosis, 42% of patients were Rai stage 0, 27% were stage I, 18% were stage II, and 13% were stage III or IV. A total of 52% of the samples were from patients who were treated. The median time to first therapy was 6.4 years. This analysis identified 52 patients with UM-CLL characterized by IGHV sequences 98% or more identical to the germline and 55 donors of the MT-CLL subtype defined by IGHV sequences less than 98% germline identical. The previously reported overrepresentation of the 1-69, 3-23, 4-34, and 3-07 IGHV genes frequently found among patients with CLL,6,7,46,47 and the VH 3-21 gene that has a distinct association with poor prognosis,48 were also observed in this analysis (Table S1).
Clinical characteristics of CLL samples
| Parameters . | All patients . | UM, ≥ 98% . | MT, < 98% . |
|---|---|---|---|
| No. of patients (%) | 107 (100) | 52 (49) | 55 (51) |
| Sex, no. (%) | |||
| Male | 64 (60) | 34 | 30 |
| Female | 43 (40) | 18 | 25 |
| Age at diagnosis, y | |||
| Median | 57.0 | 58.0 | 56.0 |
| Range | 33.0-83.0 | 33.0-83.0 | 41.0-82.0 |
| Rai stage, no. (%) | |||
| 0 | 45 (42) | 10 | 35 |
| I | 29 (27) | 17 | 12 |
| II | 19 (18) | 14 | 5 |
| III-IV | 14 (13) | 11 | 3 |
| Time to treatment | |||
| Median time to first treatment, y | 6.4 | 3.5 | 13.5 |
| No. treated (%) | 56 (52) | 37 | 19 |
| No. censored events (%) | 51 (48) | 15 | 36 |
| Parameters . | All patients . | UM, ≥ 98% . | MT, < 98% . |
|---|---|---|---|
| No. of patients (%) | 107 (100) | 52 (49) | 55 (51) |
| Sex, no. (%) | |||
| Male | 64 (60) | 34 | 30 |
| Female | 43 (40) | 18 | 25 |
| Age at diagnosis, y | |||
| Median | 57.0 | 58.0 | 56.0 |
| Range | 33.0-83.0 | 33.0-83.0 | 41.0-82.0 |
| Rai stage, no. (%) | |||
| 0 | 45 (42) | 10 | 35 |
| I | 29 (27) | 17 | 12 |
| II | 19 (18) | 14 | 5 |
| III-IV | 14 (13) | 11 | 3 |
| Time to treatment | |||
| Median time to first treatment, y | 6.4 | 3.5 | 13.5 |
| No. treated (%) | 56 (52) | 37 | 19 |
| No. censored events (%) | 51 (48) | 15 | 36 |
The relationship between IGHV mutation status and FCRL expression, CD38 surface expression, and ZAP-70 cytoplasmic expression were first compared (Table 2). In agreement with prior reports,9,10,49 elevated CD38 surface expression (≥ 30%) was significantly associated with UM-CLL samples (P < .001), and 25 (23%) of 107 patients were similarly found to be discordant.12
Correlation of IGHV mutation status with FCRL, CD38, and ZAP-70 expression
| . | FCRL1 . | FCRL2 . | FCRL3 . | FCRL5 . | CD38 . | ZAP-70 (T/B) . | ZAP-70 (%) . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UM | ||||||||||||||
| Median (range) | 9.53 (1.01-30.7) | 1.79 (1.00-3.90) | 2.04 (1.02-12.1) | 1.43 (1.00-2.72) | 49.8 (0.68-99.4) | 2.79 (0.85-6.28) | 26.9 (0.58-94.7) | |||||||
| Mean ± SD | 10.0 ± 7.05 | 1.93 ± 0.89 | 2.89 ± 2.36 | 1.52 ± 0.40 | 48.5 ± 36.6 | 2.82 ± 1.11 | 29.5 ± 19.1 | |||||||
| MT | ||||||||||||||
| Median (range) | 26.0 (1.58-45.5) | 8.89 (1.05-19.4) | 8.15 (1.22-26.9) | 3.45 (1.05-8.35) | 1.63 (0.71-99.9) | 4.80 (1.40-13.0) | 10.1 (1.00-59.9) | |||||||
| Mean ± SD | 24.2 ± 9.86 | 9.07 ± 3.73 | 9.49 ± 6.03 | 3.57 ± 1.31 | 11.3 ± 26.0 | 5.24 ± 2.17 | 14.6 ± 13.1 | |||||||
| Cutoff value | ≥ 15 | < 15 | ≥ 4.2 | < 4.2 | ≥ 4.0 | < 4.0 | ≥ 2.3 | < 2.3 | < 30% | ≥ 30% | ≥ 4.0 | < 4.0 | < 20% | ≥ 20% |
| UM (n = 52) | 7 | 45 | 0 | 52 | 7 | 45 | 2 | 50 | 19 | 33 | 5 | 47 | 14 | 38 |
| MT (n = 55) | 45 | 10 | 49 | 6 | 46 | 9 | 49 | 6 | 49 | 6 | 39 | 16 | 42 | 13 |
| Sensitivity, % | 81.8 | 89.1 | 83.6 | 89.1 | 89.1 | 70.9 | 76.4 | |||||||
| Specificity, % | 86.5 | 100 | 86.5 | 96.2 | 63.5 | 90.4 | 73.1 | |||||||
| PPV, % | 86.5 | 100 | 86.8 | 96.1 | 72.1 | 88.6 | 75.0 | |||||||
| NPV, % | 81.8 | 89.7 | 83.3 | 89.3 | 84.6 | 74.6 | 74.5 | |||||||
| Concordance, % | 84.1 | 94.4 | 85.0 | 92.5 | 76.6 | 80.4 | 74.8 | |||||||
| C-statistic | 0.861 | 0.964 | 0.877 | 0.946 | 0.803 | 0.864 | 0.760 | |||||||
| . | FCRL1 . | FCRL2 . | FCRL3 . | FCRL5 . | CD38 . | ZAP-70 (T/B) . | ZAP-70 (%) . | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| UM | ||||||||||||||
| Median (range) | 9.53 (1.01-30.7) | 1.79 (1.00-3.90) | 2.04 (1.02-12.1) | 1.43 (1.00-2.72) | 49.8 (0.68-99.4) | 2.79 (0.85-6.28) | 26.9 (0.58-94.7) | |||||||
| Mean ± SD | 10.0 ± 7.05 | 1.93 ± 0.89 | 2.89 ± 2.36 | 1.52 ± 0.40 | 48.5 ± 36.6 | 2.82 ± 1.11 | 29.5 ± 19.1 | |||||||
| MT | ||||||||||||||
| Median (range) | 26.0 (1.58-45.5) | 8.89 (1.05-19.4) | 8.15 (1.22-26.9) | 3.45 (1.05-8.35) | 1.63 (0.71-99.9) | 4.80 (1.40-13.0) | 10.1 (1.00-59.9) | |||||||
| Mean ± SD | 24.2 ± 9.86 | 9.07 ± 3.73 | 9.49 ± 6.03 | 3.57 ± 1.31 | 11.3 ± 26.0 | 5.24 ± 2.17 | 14.6 ± 13.1 | |||||||
| Cutoff value | ≥ 15 | < 15 | ≥ 4.2 | < 4.2 | ≥ 4.0 | < 4.0 | ≥ 2.3 | < 2.3 | < 30% | ≥ 30% | ≥ 4.0 | < 4.0 | < 20% | ≥ 20% |
| UM (n = 52) | 7 | 45 | 0 | 52 | 7 | 45 | 2 | 50 | 19 | 33 | 5 | 47 | 14 | 38 |
| MT (n = 55) | 45 | 10 | 49 | 6 | 46 | 9 | 49 | 6 | 49 | 6 | 39 | 16 | 42 | 13 |
| Sensitivity, % | 81.8 | 89.1 | 83.6 | 89.1 | 89.1 | 70.9 | 76.4 | |||||||
| Specificity, % | 86.5 | 100 | 86.5 | 96.2 | 63.5 | 90.4 | 73.1 | |||||||
| PPV, % | 86.5 | 100 | 86.8 | 96.1 | 72.1 | 88.6 | 75.0 | |||||||
| NPV, % | 81.8 | 89.7 | 83.3 | 89.3 | 84.6 | 74.6 | 74.5 | |||||||
| Concordance, % | 84.1 | 94.4 | 85.0 | 92.5 | 76.6 | 80.4 | 74.8 | |||||||
| C-statistic | 0.861 | 0.964 | 0.877 | 0.946 | 0.803 | 0.864 | 0.760 | |||||||
FCRL cutoff values were determined according to the ROC and CART methods. The indexes including sensitivity, specificity, PPV, NPV, concordance (concordance with IGHV mutation status), or C-statistic (predictive of model accuracy) were evaluated. A C-statistic of 1.0 indicates perfect predictive discrimination, while values greater than 0.9 indicate outstanding, 0.8 to 0.9 indicate excellent, and 0.7 to 0.8 indicate fair discrimination.44
Methods for assessing ZAP-70 protein expression and the use and interpretation of controls vary among laboratories internationally.18,19,21,25,39 Based on these studies, 2 methods for measuring ZAP-70 expression were performed. In this study, the concordance rates for ZAP-70 expression and IGHV mutation status were 74.8% using the 20% and greater cutoff and 80.4% using a T/B-CLL ratio of less than 4; both results are consistent with previous findings.16,17,20,23,39,50
After defining the clinical and prognostic characteristics of the samples, these parameters were compared with FCRL1, FCRL2, FCRL3, and FCRL5 staining results obtained by flow cytometric analysis (Table 2). MFI ratio cutoff values for the different FCRLs were determined by ROC- and CART-based analyses (“Methods”). A significant correlation among the expression patterns of the 4 FCRL proteins was identified. As indicated by their mean and median values, and from the results obtained in Figure 3B, in general, FCRL proteins were expressed at higher levels on MT-CLL samples and lower levels on UM-CLL samples. The concordance in expression patterns among the 4 FCRL molecules was 77% (ie, all FCRL representatives were either up-regulated or down-regulated on the expanded CLL population regardless of IGHV subtype). Furthermore, among the 23% of patients that differed in FCRL1, FCRL2, FCRL3, and FCRL5 expression, 18% were discordant in the expression of only a single FCRL molecule. Importantly, this study found that the 4 FCRL proteins had concordance values of 84.1% to 94.4% with IGHV gene mutation status. This exceeded the association observed for CD38 and ZAP-70 expression, regardless of which staining method was used.
To evaluate the statistical significance of these different variables, univariate logistic regression was initially performed. By this analysis, 8 significant prognostic factors, including FCRL1, FCRL2, FCRL3, and FCRL5; CD38; ZAP-70 T/B-CLL ratio; ZAP-70 percentage; and Rai stage, were identified that could distinguish patients with MT-CLL from those with UM-CLL (all were P < .001); however, both age and sex did not segregate these subgroups. Note that with this particular cohort of 107 patients with CLL, using both ROC- and CART-based analyses, we found that a cutoff value of 13% for CD38 was slightly better in predicting IGHV mutation status, resulting in 81% concordance. Interestingly, these studies verified the utility of the 20% cut-off by both statistical methods using the ZAP-70 percentage technique; however, a value of 3.6 was slightly better for predicting IGHV mutation status according to the ZAP-70 T/B-CLL ratio scheme, yielding 83% concordance. When these same 10 parameters were considered by multivariate logistic analysis with stepwise selection, only FCRL2 maintained a significant association with IGHV mutation status (odds ratio = 3.73; 95% confidence interval [CI], 1.98-7.04; P < .001) regardless of which cutoff value was selected for CD38 and ZAP-70.
In comparison with IGHV mutation status, FCRL2 had the highest sensitivity, specificity, positive predictive value (PPV), and negative predictive value (NPV) among FCRL family members (Table 2). Strikingly, FCRL2 demonstrated 94.4% concordance with IGHV mutation status compared with 76.6% for CD38 and 80.4% for ZAP-70 by T/B-CLL ratio analysis. Significant correlations between FCRL2 expression and ZAP-70 T/B-CLL ratio, CD38, and ZAP-70 percentage were also found. Using the Kendall tau-b to determine the degree of association, we found values of 0.64, −0.61, and −0.52, respectively, indicating that FCRL2 expression correlates with these markers as well. We also confirmed the value of CD38 (sensitivity for MT-CLL was 89.1%) and ZAP-70 T/B-CLL ratio (specificity for UM-CLL was 90.4%) for predicting IGHV mutation status. These findings indicate that the preferential overexpression of FCRL proteins by MT-CLL correlates with IGHV mutation status, and remarkably, FCRL2 has the strongest potential for stratifying UM-CLL and MT-CLL.
FCRL2 expression is stable and can predict mutation status and time to first therapy
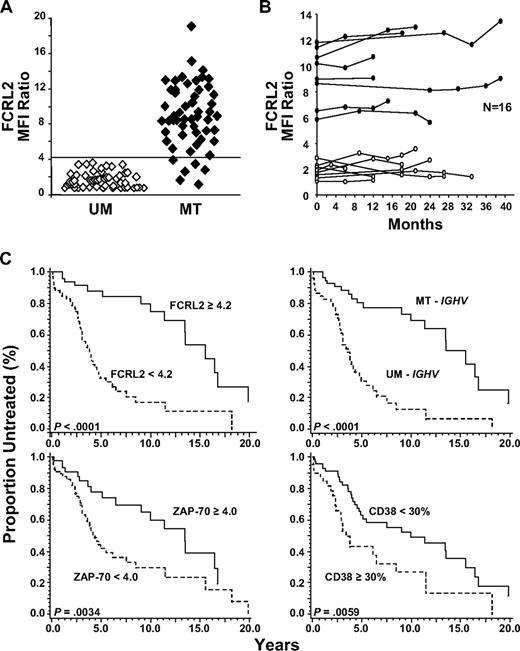
FCRL1, FCRL2, FCRL3, and FCRL5 exhibited higher surface levels of staining on MT-CLL samples compared with UM-CLL samples. Among these proteins, however, FCRL2 was distinguished as the strongest indicator of IGHV mutation status and by the magnitude of difference in its expression levels between the 2 subtypes (Table 2). The mean and median of FCRL2 staining was significantly higher among patients with MT-CLL than patients with UM-CLL at 4.7 times higher and 5.0 times higher, respectively. Using this particular anti-FCRL2 subclone (7F2) in a 2-step staining procedure, we found by ROC- and CART-based analyses that a cutoff MFI ratio value of 4.2 could clearly segregate patients with MT-CLL from patients with UM-CLL (Figure 4A). Among the 107 samples however, 6 donors with MT-CLL demonstrated decreased levels of FCRL2 surface expression (MFI ratio less than 4.2) despite having IGHV regions that were less than 98% germline. Furthermore, in 4 of 6 of these patients, the expression patterns of all 4 FCRL proteins were concordant. Mutation status of the IGHV regions of these 6 samples was also reconfirmed by sequencing. Remarkably, however, among the 52 UM-CLL samples, no discordant results were observed for FCRL2 expression.
FCRL2 is a stable indicator of IGHV mutation status and can predict time from diagnosis to initial treatment. (A) FCRL2 expression strongly correlates with the IGHV mutation status of 107 CLL samples. Cells were stained as previously with the biotinylated 7F2 mAb or an isotype-matched control followed by SA-PE, and the FCRL2 MFI ratio was derived from the gated CD19+CD5+ population. The horizontal line indicates the 4.2 MFI ratio cutoff value. (B) FCRL2 surface expression on CLL cells is stable over time. MFI ratios were determined for 16 CLL samples (8 MT, ●; 8 UM, ○) at multiple time points (2-5) over a 39-month interval. The stability of FCRL2 expression was evaluated using the overall concordance correlation coefficient (CCC).51 Values greater than 0.60 suggest satisfactory stability; those greater than 0.80 indicate excellent stability based on criteria established for the K coefficient.52 (C) FCRL2, IGHV, ZAP-70 T/B-CLL ratio, and CD38 can predict time from diagnosis to initial treatment. The median time to initial therapy for patients with high FCRL2 expression (MFI ratio ≥ 4.2) was 15.5 years compared with 3.75 years for patients with low FCRL2 expression (MFI ratio < 4.2). The median time to first therapy for patients with MT-CLL was 13.5 years versus 3.51 years for patients with UM-CLL; for ZAP-70 T/B-CLL ratio of 4.0 or greater it was 13.4 years versus 4.25 years for ZAP-70 T/B-CLL ratio less than 4.0; and for CD38 less than 30%, it was 9.92 years versus 3.34 years for CD38 of 30% or greater. P values are indicated in each panel.
FCRL2 is a stable indicator of IGHV mutation status and can predict time from diagnosis to initial treatment. (A) FCRL2 expression strongly correlates with the IGHV mutation status of 107 CLL samples. Cells were stained as previously with the biotinylated 7F2 mAb or an isotype-matched control followed by SA-PE, and the FCRL2 MFI ratio was derived from the gated CD19+CD5+ population. The horizontal line indicates the 4.2 MFI ratio cutoff value. (B) FCRL2 surface expression on CLL cells is stable over time. MFI ratios were determined for 16 CLL samples (8 MT, ●; 8 UM, ○) at multiple time points (2-5) over a 39-month interval. The stability of FCRL2 expression was evaluated using the overall concordance correlation coefficient (CCC).51 Values greater than 0.60 suggest satisfactory stability; those greater than 0.80 indicate excellent stability based on criteria established for the K coefficient.52 (C) FCRL2, IGHV, ZAP-70 T/B-CLL ratio, and CD38 can predict time from diagnosis to initial treatment. The median time to initial therapy for patients with high FCRL2 expression (MFI ratio ≥ 4.2) was 15.5 years compared with 3.75 years for patients with low FCRL2 expression (MFI ratio < 4.2). The median time to first therapy for patients with MT-CLL was 13.5 years versus 3.51 years for patients with UM-CLL; for ZAP-70 T/B-CLL ratio of 4.0 or greater it was 13.4 years versus 4.25 years for ZAP-70 T/B-CLL ratio less than 4.0; and for CD38 less than 30%, it was 9.92 years versus 3.34 years for CD38 of 30% or greater. P values are indicated in each panel.
Confirming the stability and reliability of biomarkers is essential for validating their diagnostic utility as prognostic markers and clinical tools. Among the 107 donors, 16 patients (8 with MT-CLL and 8 with UM-CLL) were analyzed at various time points over the disease course (Figure 4B). To determine the stability of FCRL2 expression levels as a function of time, the overall concordance correlation coefficient (CCC) was calculated.51 Based on these 16 donors, a value of 0.931 (95% CI, 0.864-0.966) was obtained, indicating excellent stability for FCRL2.52 Sample preservation also did not appear to affect FCRL2 expression when cells were stained fresh, kept overnight at room temperature or 4°C (as whole blood or after Ficoll), or thawed following cryopreservation (data not shown). These data indicate that FCRL2 surface expression is stable over time.
To determine the clinical value of FCRL2 expression, the treatment-free interval from the time of diagnosis to initial therapy was calculated. The Cox proportional model with univariate analysis found that, similar to IGHV mutation status (hazard ratio [HR] = 4.45; 95% CI, 2.47–8.00; P < .001), FCRL2 could also predict time to first therapy (HR = 4.83; 95% CI, 2.43-8.40; P < .001). ZAP-70 T/B-CLL ratio, ZAP-70 percentage, and CD38 were also predictive of the time to first therapy with HR of 2.32 (95% CI, 1.30-4.15; P = .004), 1.98 (95% CI, 1.16-3.38; P = .012), and 2.11 (95% CI, 1.23-3.62; P = .006), respectively. The log-rank test was used to calculate the median treatment-free interval for the entire cohort of 107 patients. In agreement with prior reports,9,10,12,14-17 this analysis confirmed that IGHV mutation status (P < .001), CD38 (P = .005), and ZAP-70 percentage (P = .011; data not shown) can all predict the time to first therapy with statistical significance (Figure 4C). This study also found that determining ZAP-70 expression according to the T/B-CLL ratio within a sample is also predictive of the initial treatment-free interval (P = .003; Figure 4C). Patients with UM-CLL required treatment at a median interval of 3.51 years compared with patients with MT-CLL who first received therapy at a median interval of 13.5 years. Interestingly, all 4 FCRL proteins could predict the time to first treatment with statistical significance; however, FCRL2+ (MFI ratio of 4.2 or greater) patients had a median time to first therapy of 15.5 years, whereas for FCRL2− donors, it was 3.75 years (P < .001; Figure 4C). The Cox model of multivariate analysis with stepwise selection found that both FCRL2 (HR = 4.44; P < .001) and clinical disease stage (HR = 1.33; P = .003) could predict clinical progression as defined by the time to first therapy, while the 9 other parameters considered in this study, including IGHV, FCRL1, FCRL3, FCRL5, CD38, ZAP-70 T/B-CLL ratio, ZAP-70 percentage, age, and sex, could not. Considering the interaction of these variables and the clinical significance of these markers, these data indicate that FCRL2 surface expression may exhibit greater prognostic power than other FCRL proteins and presently used markers, including IGHV mutation status, CD38 expression, and ZAP-70 expression.
Discussion
By comparing FCRL1-5 expression with IGHV mutation status, CD38 and ZAP-70 expression, and clinical features from a cohort of 107 CLL samples, we found that FCRL2 possesses significant prognostic value for predicting both IGHV mutation status and the time to first therapy. In general, 4 of the 5 FCRL proteins were overexpressed by MT-CLL samples, and each demonstrated overall higher concordance with IGHV mutation status than CD38 or ZAP-70. According to this analysis, FCRL2 had the most applicable and statistically significant association for predicting clinical progression according to the time to first therapy in CLL.
Although FCRL1 exhibited higher overall levels of surface expression on CLL cells than other FCRLs, its relationship to IGHV mutation status was the most discordant (84.1%). Rather, its discrete expression by B-lineage cells29,31 and moderate to high level of staining on most CLL samples analyzed indicates that FCRL1 could be a potential immunotherapy target candidate. This is in accord with a recent study by Du et al in which immunotoxin-labeled anti-FCRL1 mAbs were used for cytotoxicity studies in CLL and other B-cell malignancies.36 FCRL3 expression was slightly more concordant with mutation status (85%); however, a single nucleotide polymorphism in an NF-κB consensus binding site within its promoter region significantly alters its expression.53 The resulting variability in FCRL3 expression among patients could be problematic for its reliability and diagnostic utility in the clinical setting, but ultimately might still be a useful indicator. Nevertheless, these data verify the transcript profile identified by Wiestner et al in which both FCRL2 and FCRL3 were among a small number of statistically significant genes overexpressed by MT-CLL samples.13 Similar to FCRL2, FCRL5 was also highly concordant with CLL according to IGHV subtype (92.5%), but by our analysis had a dim and narrow range of expression, making its possible use a prognostic tool to separate patients with UM-CLL from those with MT-CLL difficult. As more than one FCRL5 isoform has been identified,26 and discrepancy in its expression pattern among blood B cells has been observed by other groups,31,54 it is possible that a different FCRL5 mAb could provide a brighter and more discriminating profile for phenotyping patients with UM-CLL and MT-CLL. Although FCRL5 could still be a useful marker, the analysis reported here, and 2 independent microarray studies using different strategies,3,13,35 have found that FCRL2 expression at the transcript and protein level is strongly concordant with MT-CLL.
The initial goal of this analysis was to assess the correlation between FCRL surface expression and IGHV mutation status in CLL. This study identified FCRL2 as the most concordant with this hallmark indicator compared with other FCRL family members and the 2 currently used prognostic markers, CD38 and ZAP-70. Despite the modest incongruity evident for CD38 and ZAP-70 with IGHV subtype by this analysis and others,12,16,17,23,24,49,50 the expression patterns of both proteins have value as independent markers of aggressive disease course in CLL. Importantly, when stained in combination or compared with mutation status, these markers may also distinguish more distinct subgroups of patients with an intermediate prognosis.12,16,17 Based on a cutoff value of 4.2, defined by 2 different statistical methods (ROC and CART), it was surprising to find complete agreement between unmutated IGHV mutation status and low FCRL2 expression. In contrast, 6 donors with MT-CLL demonstrated decreased levels of FCRL2 surface expression (MFI ratio less than 4.2) despite having IGHV regions that were less than 98% germline. It is intriguing that among these discordant cases, 4 patients (C545, C853, K0008, and K0107) were both ZAP-70+ and CD38+ with IGHV sequence identities of 94.6%, 97.9%, 97.3%, and 97.6%, respectively (Table S1). These 4 samples were also negative for the 3 other FCRLs. Interestingly, 2 of these individuals (C853 and K0008) had been previously treated, and one was VH 3–21 restricted (K0008). Considering that the IGHV sequence identity of 3 of these 4 patients was between 97% and 98%, perhaps low FCRL2 expression levels also have significance for identifying patients with limited IGHV gene diversity near the 98% cutoff and/or possessing aggressive disease characteristics. Notably, patients with CLL who have borderline IGHV mutation (97%-97.9%) may have inferior clinical outcomes.55 Thus, according to their FCRL2, ZAP-70, and CD38 status, these 4 patients would appear more likely to have an aggressive disease course regardless of their mutation status. Of the 2 additional MT-CLL outliers with an FCRL2− profile, both were CD38− and ZAP-70−, but one had undergone treatment and was FCRL1−, FCRL3−, and FCRL5−. Although this number of discordant patients is small, these data suggest that FCRL2 may have additional prognostic potential independent of mutation status and could be helpful for further subgrouping patients with aggressive disease.
The biological role of the FCRL family of proteins presently remains unclear. While these molecules share many similar features with the classical Fc receptors for IgG and IgE, such as related extracellular Ig domain composition and tyrosine-based signaling function, the FCRL ligands are currently unknown. Among normal B cells, the FCRLs preferentially mark mature subpopulations where they may exert immunomodulatory function.29,37,56 Since CLL is a disease of progressive immunodeficiency and altered immune regulation with autoimmune features, it seems logical that as modulating proteins, the FCRLs could possibly influence disease pathogenesis. Assuming that FCRL function is intact in CLL cells, and depending on the nature of their physiologic ligands, the inhibitory characteristics of several of these receptors and overexpression by MT-CLL, as also pointed out by Huttmann et al, could also be clinically advantageous by perhaps downmodulating cellular activation and proliferation or possibly affecting anergy that is typical of this CLL subtype.35 For example, FCRL2 has 2 consensus ITIM motifs and a noncanonical ITAM that appear to assist in the recruitment of the SH-2 domain containing phosphatase, SHP-1.57 In contrast, the decreased expression of FCRL family members by UM-CLL might instead contribute to this subtype's more aggressive features. Although several different scenarios can be envisioned for the potentially altered biology of these molecules in CLL, more fundamental information is needed about their normal function and ligand(s) to better define their roles in this context.
Our results may also have implications for defining the normal cellular counterpart of the 2 CLL subtypes. In particular, FCRL2 expression peaks on memory B cells and plasma cells in the tonsils and spleen and appears to be enriched among CD27+ memory B cells in the blood.31 While the normal cellular counterpart(s) of the 2 CLL subtypes remain unknown, it is apparent that both UM-CLL and MT-CLL express CD27 as well as other markers of activation.5 Furthermore, regardless of their IGHV mutation status, both subtypes have the gene expression signature of antigen-experienced B cells.3,4 Based on these findings, marginal zone (MZ) B cells are perhaps the most frequently postulated pretransformed CLL counterpart8,58,59 ; however, the recent characterization of CD5+ subepithelial B cells, a possible equivalent subpopulation in tonsils, may also be a legitimate candidate.60 With regard to the distinct pattern of FCRL expression in CLL, it is quite notable that 4 of the 5 FCRL molecules are up-regulated on MT-CLL cells. Support for the possibility that this lymphoproliferative disorder represents a transformed MZ-like B cell is also suggested by microarray analysis that identified FCRL2 and FCRL3 among a small number of statistically significant overexpressed genes that distinguish splenic and circulating blood MZ B cells.61 FCRL expression is also primarily concentrated in areas that harbor MZ-type B cells, such as the interfollicular and subepithelial regions in tonsils, and of note for FCRL2 and FCRL3, in the marginal zone of the spleen (see Davis32 for review). Although only 2 of the 5 human FCRL representatives are evident in mice (FCRL1 and FCRL5), FCRL5, which has similar features to human FCRL3 and FCRL5, is distinctly expressed by MZ and B1 B cells.62 These data suggest that the expression patterns of FCRL representatives may also be useful for future studies designed at further defining the normal CLL counterpart.
In a comprehensive comparison of established biologic and clinical prognostic characteristics in CLL, this study has identified remarkable prognostic potential for FCRL2. Although the 3 other FCRL proteins expressed by CLL cells demonstrate significant biological and clinical promise, FCRL2 was statistically superior in stratifying CLL samples according to IGHV gene mutation status and through its capacity to predict time from diagnosis to initial treatment. The localization of FCRL2 on the cell surface and its stability over time makes its detection by conventional staining and flow cytometry simple to perform and control for, and thus easy to implement in the clinical diagnostic laboratory. As a protein distinctly overexpressed by MT-CLL samples, FCRL2 could help distinguish patients more likely to experience an indolent disease course, in contrast to CD38 and ZAP-70, which are preferentially expressed by UM-CLL. Importantly, these 3 markers in combination might not only provide useful diagnostic verification, but could also identify patients with aggressive features independent of mutation status and/or with an intermediate prognosis.25 Future studies using optimized reagents and expanded patient sample numbers should be helpful for validating the clinical utility of FCRL2 in CLL and hopefully in improving biologic understanding and the care of people with other B-cell malignancies as well.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank members of the Davis and Cooper laboratories for their advice, critique, and insightful comments. We are grateful to Peter Burrows and Harry Schroeder for comments on the manuscript and Kanti Rai, Thomas Kipps, and Laura Rassenti for their generosity in providing patient samples. We would also like to thank the clinicians who enrolled patients and the volunteers and donors who participated in this study.
This work was supported in part by National Institutes of Health/National Institute of Allergy and Infection Diseases K08 award AI055638, the Dana Foundation Human Immunology Program, and the V Foundation for Cancer Research (R.S.D).
National Institutes of Health
Authorship
Contribution: F.J.L. performed research, did statistical analysis, and wrote the paper; S.D. and Y.L. performed statistical analysis; J.P., M.A.S., E.K., and J.W. performed research; S.-j.S. supervised the statistical analysis; N.C. provided clinical samples and helpful discussion, and reviewed the manuscript; and R.S.D. conceived the project, directed research, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Randall S. Davis, Division of Hematology/Oncology, Departments of Medicine, Microbiology, and Biochemistry and Molecular Genetics, University of Alabama at Birmingham, 1825 University Blvd SHEL 412, Birmingham, AL 35294-2182; e-mail: rsdavis@uab.edu.