Abstract
The MYC oncogene, which is commonly mutated/amplified in tumors, represents an important regulator of cell growth because of its ability to induce both proliferation and apoptosis. Recent evidence links MYC to altered miRNA expression, thereby suggesting that MYC-regulated miRNAs might contribute to tumorigenesis. To further analyze the impact of MYC-regulated miRNAs, we investigated a murine lymphoma model harboring the MYC transgene in a Tet-off system to control its expression. Microarray-based miRNA expression profiling revealed both known and novel MYC targets. Among the miRNAs repressed by MYC, we identified the potential tumor suppressor miR-26a, which possessed the ability to attenuate proliferation in MYC-dependent cells. Interestingly, miR-26a was also found to be deregulated in primary human Burkitt lymphoma samples, thereby probably being of clinical relevance. Although today only few miRNA targets have been identified in human disease, we could show that ectopic expression of miR-26a influenced cell cycle progression by targeting the bona fide oncogene EZH2, a Polycomb protein and global regulator of gene expression yet unknown to be regulated by miRNAs. Thus, in addition to directly targeting protein-coding genes, MYC modulates genes important to oncogenesis via deregulation of miRNAs, thereby vitally contributing to MYC-induced lymphomagenesis.
Introduction
Approximately 70% of human tumors overexpress c-MYC (hereafter referred to as MYC) because of alterations in signal transduction pathways or secondary to genomic aberrations including amplifications and translocations involving the MYC gene.1 For example, Burkitt lymphoma (BL), an aggressive variant of non-Hodgkin B-cell lymphoma, represents a prime example for MYC translocations involved in tumorigenesis.2 Although its sporadic form accounts for only 1% to 2% of adult lymphoma in the Western world, 40% to 50% of childhood non-Hodgkin lymphoma (NHL) belong to this tumor entity in nonendemic areas.3 With the introduction of novel treatment options (eg, monoclonal antibodies) based on the growing knowledge of the molecular biology of BL, the prognosis of the disease has changed significantly over the past years.4
MYC encodes a basic helix-loop-helix-zipper transcription factor, which heterodimerizes with MAX and binds to E-Box sequences in the promoter regions of target genes, such as cyclin D2 and E2F1, whose expression then mediates downstream effects of MYC on cell biology.5,6 MYC can act both as a transcriptional activator and a transcriptional repressor. In a complex with MAX and MIZ1 or SP1, it binds the initiator elements of several genes, such as p21 and p27, and helps to recruit the DNA methyltransferase DNMT3a.6 Furthermore, recent findings support a potential nontranscriptional function for MYC on chromatin structure.7 The molecular mechanisms by which MYC promotes tumorigenesis have not been comprehensively elaborated. Although large efforts aiming to define MYC target genes have been made by microarray expression studies and chromatin binding approaches, a dominant “gene-specific” model of MYC as a classic transcription factor could not be proposed.5,6
With the recent discovery of microRNAs (miRNAs), the network of MYC functions is getting even more complex. miRNAs represent a family of small (∼ 20-23 nt), nonprotein coding RNAs that function predominantly as posttranscriptional regulators of gene expression. They induce messenger RNA (mRNA) cleavage in case of perfect complementarity to the target mRNA. Alternatively, in case of imperfect base pairing, miRNAs can either initiate a translational block or accelerate mRNA deadenylation and decay.8,9 Deregulated miRNA expression has already been implicated in human malignancies,10,11 and several miRNAs were shown to be located at fragile sites commonly altered in cancer.12,13 However, only limited functional data are available on miRNA-mediated tumorigenesis, but for selected miRNAs their pathogenic role in this process either as tumor suppressors (eg, miR-15a/miR-16 and their target BCL2 in B-cell chronic lymphocytic leukemia (B-CLL)) or oncogenes (eg, miR-17-92 cluster and E2F1 in lymphoma) has been demonstrated.14,15 To better understand BL biology, it is indispensable to identify deregulated miRNA pathways involved in MYC-driven lymphomagenesis, for which various targets have recently been identified.16,17 For this purpose, mouse models, which allow the specific manipulation of single genes, represent attractive experimental setups. Recently, we have established a murine MYC-driven lymphoma model harboring the MYC transgene in a Tet-off system to control its expression.18,19 This model allows investigation of the impact of MYC-deregulated miRNA expression in lymphoma pathogenesis.
miRNA expression using DNA microarray technology was analyzed in a large series of MYC-driven lymphoma cell lines (n = 26) derived from our transgenic mice. Here we report the identification of deregulated miRNA expression patterns and highlight miRNAs and target genes that are of potential pathogenic relevance. Furthermore, we functionally evaluated the MYC-repressed miRNA miR-26a in more detail to (1) better understand its impact in MYC-driven lymphomagenesis and to (2) get new insights into the molecular biology of MYC-induced lymphoma by identifying the potential miR-26a target gene EZH2. Ultimately, these findings might provide a basis for novel molecular targeted therapeutic approaches in MYC-induced tumors like BL.
Methods
Cell lines and cell culture
The murine MYC-induced lymphoma cell lines (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article) as well as the human BL cell lines were cultured in RPMI medium 1640 with 10% fetal calf serum, 1% penicillin/streptomycin, 1% L-glutamine, 1% nonessential amino acids, and 0.1% β-mercaptoethanol. HEK-293 cells were maintained in Dulbecco modified Eagle medium with 10% fetal calf serum and 1% penicillin/streptomycin. In murine lymphoma cell lines, MYC expression was abrogated by doxycycline (Dox) treatment (1 μg/mL). miRNA expression was induced in the transfected BL cell lines by Dox treatment (0.5 μg/mL).
Lymphoma samples
Samples of 11 adult BL (2 peripheral blood: B-cell acute lymphoblastic leukemia, (B-ALL) and 9 lymph node (BL) specimens; all cases were translocation t(8,14) positive) and 12 B-CLL (9 unselected B-CLL and 3 CD19+-enriched B-CLL samples derived from peripheral blood) patients were drawn from the tissue and blood banks of the Departments of Pathology and Internal Medicine III, University of Ulm (Ulm, Germany). All tissue samples were pseudonymized to comply with the German law for correct usage of archived tissues for clinical research.20 For cell preparations, patients' informed consent was obtained in accordance with the Declaration of Helsinki, and the use of human samples was approved by the ethics committee of the University of Ulm. Lymphoma histologic diagnosis was made according to the criteria of the WHO classification at the Department of Pathology, University of Ulm, a German reference pathology institution for lymphoma diagnosis. As controls, human peripheral blood derived mononuclear cells (PBMNCs) were obtained from 3 healthy human donors.
miRNA expression profiling
For miRNA expression profiling, we set up a DNA microarray platform using a commercially available oligonucleotide probe set based on version 6.0 of the Sanger miRNA database (mirVana miRNA Probe Set; Ambion, Austin, TX). Total RNA was isolated from our lymphoma cell lines as well as differentiated healthy mouse tissues, which served as a common reference, using the mirVana miRNA Isolation Kit (Ambion). Size-fractionated small RNA (flashPAGE Fractionator System; Ambion) was prepared for microarray analysis using an end-labeling strategy (mirVana miRNA Labeling Kit; Ambion). Cohybridization of lymphoma cell lines and common reference was performed for 14 hours onto the miRNA microarrays and washed according to the manufacturer's protocol (Ambion). Microarrays were imaged using an Axon GenePix 4000B laser scanner (Molecular Devices, Sunnyvale, CA). Fluorescence ratios were extracted (tumor/common reference) after subtracting the background using the GenePix Pro 6.0 software (Molecular Devices). To identify differential miRNA expression between samples, the fluorescent ratios were log2-transformed, normalized, and filtered in analogy to mRNA microarrays.21 The complete filtered dataset is provided as Table S2, and raw data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo; accession number GSE12400). For hierarchical clustering, we used average linkage clustering (distance measure, correlation uncentered) and visualized results using TreeView.22
Northern blot analysis of miRNAs
For Northern blot analyses, 20 μg total RNA was separated on denaturing polyacrylamide gels and transferred to positively charged nylon membranes. After overnight incubation with 32P-labeled probes designed and processed according to the mirVana miRNA probe construction kit (Ambion), the membranes were washed and subjected to autoradiography. As a loading control, tRNAs were detected by ethidium bromide staining of the gels and 5S RNAs using a radioactive labeled probe.
Immunoblot analysis
Total cell extracts were fractionated on 10% sodium dodecyl sulfate polyacrylamide gels, electroblotted to polyvinylidene difluoride membranes (Millipore, Billerica, MA), and reacted with anti-EZH2 (Cell Signaling Technology, Danvers, MA), anti-ARF (Abcam, Cambridge, MA), anti-pAKT (Thr308), anti-AKT (both Cell Signaling Technology), anti-BCL2, anti-CDK8 (both Santa Cruz Biotechnology, Santa Cruz, CA), anti-ALS2CR2 (Abcam), anti-cyclinE2 (Cell Signaling Technology), or anti-actin or anti–α-tubulin (both Sigma-Aldrich, St Louis, MO) antibodies. Immunoreactivity was determined using the enhanced chemiluminescence method (Pierce Chemical, Rockford, IL).
Expression constructs and cell proliferation assay
Approximately 100-nt long oligos containing mature miRNA sequences were cloned into the pMIRTOP and pRTS vectors as described.23 Different BL cell lines were transfected using cell line specific Nucleofection protocols (Amaxa Biosystems, Gaithersburg, MD) followed by puromycin (1 μg/mL) selection from day 2 after transfection. Ten days after transfection, miRNA expression was induced with Dox (0.5 μg/mL) and green fluorescent protein (GFP) expression was determined by fluorescence-activated cell sorter. Cell counts were either determined by Trypan blue staining or fluorescence-activated cell sorter. Cell proliferation was measured in triplicate using a colorimetric assay reagent, WST1 (Roche Diagnostics, Mannheim, Germany).
Quantitative reverse-transcribed polymerase chain reaction
Total RNA was extracted using TRIzol reagent (Invitrogen, Carlsbad, CA). cDNA synthesis using Superscript II reverse transcriptase (Invitrogen) was primed with random hexamer primers (Roche Diagnostics). Analyses were carried out using SYBR Green PCR master mix (Applied Biosystems, Foster City, CA) and ABI Prism 7900 (GE Healthcare, Little Chalfont, United Kingdom). Primers for human EZH2: Fwd:5′-TACTTGTGGAGCCGCTGAC, Rev:5′-CTGCCACGTCAGATGGTG; and human PBGD: Fwd:5′-TGCCAGAGAAGAGTGTGGTG, Rev:5′-GAGGTTTCCC-CGAATACTCC. Results were normalized with respect to PBGD expression. Ct values for gene expression were calculated according to the Ct method. TaqMan microRNA assays (Applied Biosystems) were used to quantify the expression of mature miR-26a (4373070), miR-16 (4373121), miR-20a (4373286), and RNU6B (4373381) as a housekeeping gene.24 The mean Ct was determined from triplicate polymerase chain reaction (PCR).
Gene expression profiling
Gene expression profiling was performed in 6 cell line samples using Affymetrix microarray technology according to the manufacturer's recommendations (Human Genome U133 Plus 2.0 Array; Affymetrix, Santa Clara, CA). Fluorescence ratios were normalized by applying the RMA algorithm using the BRB Array Tools software package. For subsequent analyses, we only included probe sets whose expression varied as previously determined.25 The complete microarray data are available at the Gene Expression Omnibus (http://www.ncbi.nlm.nih.gov/projects/geo; accession number GSE12400). For hierarchical clustering, we used average linkage clustering (distance measure: correlation uncentered) and visualized results using TreeView.22
Luciferase reporter assay
In the 3′UTR of the Luciferase-containing TK-pGL3 reporter construct (Promega, Madison, WI), we included the 3′-UTR of human EZH2 or a 3′-UTR version with a mutated miR-26a binding site. The 3′-UTR was amplified with the following primers: 5′-ATTCTAGACTTGACATCTGCTA CCTCCTC and 5′-GGTCTAGAACAAGTTCAAGTATTCTTTA. The predicted miR-26a binding site (see Figure 5E) was mutated by base pair changes using DpnI mediated site-directed mutagenesis and the primers: 5′-GAATAAAGAATGCGTGAACTTG and 5′-CAAGTT CACGCATTCTTTATTC. For the reporter assays, HEK-293 cells were cultured in 24-well plates and transfected with TK-pGL3-EZH2 wt or mutated plasmid, Ubiquitin-Renilla, and either the precursor miR-26a or negative control RNA (Ambion) using Lipofectamine 2000 (Invitrogen). Luciferase activity was measured 24 hours after transfection using the Dual-luciferase reporter assay system (Promega).
EZH2 knock-down
Two shRNA sequences targeting human EZH2 (shRNA#1 5′-AAGCTAAGGCAGCTGTTTCAG, shRNA#2 5′-ATCACTGTCTGTATCCTTTGA) were constructed using the BLOCK-iT RNAi Designer (Invitrogen; available at https://rnaidesigner.invitrogen.com/rnaiexpress/) and cloned into the pMIRTOP and pRTS vectors as described.23 Cells were transfected and analyzed as described in “Expression constructs and cell proliferation assay.”
Data analysis
We identified miRNAs that were differentially expressed by using the SAM (Significance Analysis of Microarrays) method,26 which uses a modified t test statistic, with sample-label permutations to evaluate statistical significance. Furthermore, we applied a “class comparison analysis” using the BRB software package. Gene ontology groups of genes whose expression was differentially regulated among the classes were identified by computing the number of genes represented on the microarray in the respective gene ontology group, and the statistical significance P value for each gene in the group. These P values reflect differential expression among classes and were computed based on random variance t tests as previously described.25 To identify groups of genes belonging to distinct BioCarta or KEGG pathways whose expression was differentially regulated among the classes, the same computational algorithm was used. The analyses were performed using BRB-Array Tools Version 3.3.0 Beta_3 developed by Dr Richard Simon and Amy Peng Lam and using R, version 2.2.1 (available at http://www.r-project.org). The statistical tests were performed using also R, version 2.2.1, and all tests reported were 2-sided with an effect being considered significant if the P value was 0.05 or less.
Results
Identification of MYC-regulated miRNAs
To screen for MYC-regulated miRNAs, we used a microarray platform (Ambion) allowing the simultaneous measurement of approximately 200 murine miRNAs.27-29 We analyzed 26 T- and B-cell lymphoma cell lines derived from a murine-inducible MYC lymphoma model harboring the transgene in a Tet-off system.18,19 Cell lines were investigated at a high versus low MYC expression state comparing untreated with 18-hour Dox-treated samples. By supervised analysis, we identified 13 miRNAs associated with the MYC-expression state, including 7 miRNAs down-regulated after Dox treatment and 6 miRNAs being significantly up-regulated and hence repressed by MYC (Table 1). MYC-regulated miRNAs comprised known candidates, such as the miR-17–92 cluster (miR-17–5p, miR-18, miR-19b, miR-20a, and miR-92),17 which were mainly up-regulated by MYC, as well as newly identified MYC-associated miRNAs (Table 1). Interestingly, we observed marked differences in the miRNA expression changes among the 26 analyzed cell lines (Figure 1A). Whereas a down-regulation of the miR-17–92 cluster was seen in the majority of the cell lines, several cases also showed an up-regulation of the respective miRNAs on MYC inactivation.
miRNAs associated with MYC-induced lymphoma
| Class comparison . | Down-regulated miRNAs . | Up-regulated miRNAs . |
|---|---|---|
| High vs low MYC state | ||
| miR-16 | miR-17-5p | |
| miR-26a | miR-18 | |
| miR-134 | miR-19b | |
| miR-207 | miR-20a | |
| miR-489 | miR-92 | |
| miR-494 | miR-106a | |
| miR-130b | ||
| B-cell phenotype (B- vs T-cell phenotype) | ||
| let-7b | miR-30a-3p | |
| let-7c | miR-130b | |
| miR-15b | miR-155 | |
| miR-30a-5p | miR-187 | |
| miR-30b | miR-192 | |
| miR-30d | miR-345 | |
| miR-107 | miR-519e | |
| miR-128a | miR-527 | |
| miR-150 |
| Class comparison . | Down-regulated miRNAs . | Up-regulated miRNAs . |
|---|---|---|
| High vs low MYC state | ||
| miR-16 | miR-17-5p | |
| miR-26a | miR-18 | |
| miR-134 | miR-19b | |
| miR-207 | miR-20a | |
| miR-489 | miR-92 | |
| miR-494 | miR-106a | |
| miR-130b | ||
| B-cell phenotype (B- vs T-cell phenotype) | ||
| let-7b | miR-30a-3p | |
| let-7c | miR-130b | |
| miR-15b | miR-155 | |
| miR-30a-5p | miR-187 | |
| miR-30b | miR-192 | |
| miR-30d | miR-345 | |
| miR-107 | miR-519e | |
| miR-128a | miR-527 | |
| miR-150 |
miRNA expression profiling in MYC-dependent lymphoma. (A) Supervised analysis for MYC-regulated miRNAs (class comparison; P < .01, paired t test with random variance model paired samples) based on miRNAs displayed in panel C. Expression levels have been set to 0 in the untreated samples to display Dox-related changes. MYC-induced miRNAs are color coded in red, whereas repressed miRNAs are depicted in green. (B) Validation of microarray data by Northern blot analysis (doxycycline treatment for 18 hours). (C) Unsupervised average linkage cluster of MYC-induced lymphoma cell lines (n = 26) based on 154 differentially expressed miRNAs. Two major cell line clusters were identified, mainly characterized by a distinct miRNA pattern (blue bar).
miRNA expression profiling in MYC-dependent lymphoma. (A) Supervised analysis for MYC-regulated miRNAs (class comparison; P < .01, paired t test with random variance model paired samples) based on miRNAs displayed in panel C. Expression levels have been set to 0 in the untreated samples to display Dox-related changes. MYC-induced miRNAs are color coded in red, whereas repressed miRNAs are depicted in green. (B) Validation of microarray data by Northern blot analysis (doxycycline treatment for 18 hours). (C) Unsupervised average linkage cluster of MYC-induced lymphoma cell lines (n = 26) based on 154 differentially expressed miRNAs. Two major cell line clusters were identified, mainly characterized by a distinct miRNA pattern (blue bar).
For validation of findings, we performed Northern blot analyses for selected miRNAs in at least 4 cell lines. Representative results are shown for a miRNA predominantly up-regulated by MYC (miR-17-5p; Figure 1B) and an MYC-repressed candidate (miR-16; Figure 1B). Although Northern blot results were in agreement with our microarray findings, we also were able to confirm cell line-associated heterogeneous responses to MYC knock-down for miR-16 and miR-20a by quantitative RT-PCR (data not shown). Importantly, MYC-induced miRNAs, such as miR-17–5p, also showed higher expression levels in human BL cell lines compared with healthy PBMNCs, whereas MYC-repressed miRNAs were expressed at lower levels in malignant cells (“miR-26a expression in BL-derived cell lines and primary lymphoma samples” and data not shown).
Analysis of the kinetics of miRNA regulation by MYC suggested that mature miRNAs might possess a longer half-life than their pre-miRNA precursors (Figure S1). Rapid down-regulation of the precursor was seen as early as 6 hours after MYC inactivation, with miR-17-5p and miR-20a precursors being virtually undetectable after 18 hours, low amounts of mature miR-17-5p and miR-20a were still present in Dox-treated cell lines after 60 hours.
Evaluation of lymphoma heterogeneity
Having shown interindividual differences in our murine lymphoma cell lines on Dox treatment, we performed a hierarchical cluster analysis in the untreated lymphoma cell lines (n = 26) to determine whether the Dox-induced heterogeneity is already reflected in the untreated MYC-expressing cells (Figure 1C). Indeed, we identified 2 major cell line clusters, which were mainly characterized by differential expression of one prominent miRNA pattern containing among others the members of the miR-17-92 cluster (blue bar in Figure 1C). Whereas unsupervised clustering did not reflect the cellular origin of the cancer cells, supervised analysis revealed a distinct miRNA pattern associated with B- and T-cell phenotype (Table 1). B-cell lymphoma tended to contain lower expression levels of let-7b, let-7c, miR-15b, and miR-150 as well as higher expression levels of miR-155 compared with T-cell lymphoma (Table 1).
Given the observed miRNA expression heterogeneity, we looked at differences in signaling pathways known to be commonly involved in lymphomagenesis. Thus, we screened for expression differences of ARF, BCL2, and the phosphorylation status of AKT. Although ARF expression did not vary significantly between the groups, marked differences were seen for BCL2, which was exclusively detectable in lymphoma cell lines of B-cell origin, and AKT, which was expressed in all cell lines but only activated in 2 of them (Figure S2). However, these findings did not show a correlation with the miRNA expression-based sample grouping.
Impact of MYC-repressed miRNAs in a human BL-derived cell line
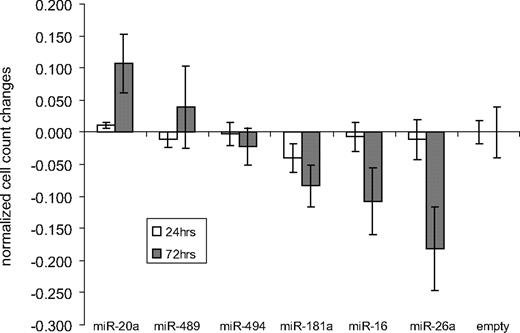
To address the biologic impact of the newly identified MYC-repressed miRNAs, we inducibly overexpressed selected miRNAs (miR-16, miR-26a, miR-181a, miR-489, and miR-494) in MYC-expressing lymphomas. As internal control, we overexpressed the known oncogenic miR-20a. We stably transfected the human BL cell line Namalwa with an episomal Tet-inducible expression system, allowing the expression of the respective miRNAs and GFP by Dox treatment. After puromycin selection, transfected cell lines were treated with Dox for 24 hours and 72 hours. Cell proliferation was determined using a colorimetric assay (Figure 2). No significant differences were seen after 24 hours, but changes were detectable after 72 hours of Dox treatment. Overexpression of miR-489 and miR-494 caused no significant cell count alterations, overexpression of miR-20a slightly increased cell counts, whereas miR-181a, miR-16, and most prominently miR-26a reduced cell proliferation.
Proliferation changes after inducible miRNA expression in Namalwa. Namalwa cells transfected with an episomal Tet-inducible expression system were treated with Dox for 24 hours and 72 hours; subsequently, cell proliferation was determined and normalized to the empty control. Overexpression of miR-20a slightly increased cell counts, whereas induction of miR-489 and miR-494 caused no significant cell count alterations. In contrast, miR-181a, miR-16, and most prominently miR-26a expression resulted in reduced cell proliferation.
Proliferation changes after inducible miRNA expression in Namalwa. Namalwa cells transfected with an episomal Tet-inducible expression system were treated with Dox for 24 hours and 72 hours; subsequently, cell proliferation was determined and normalized to the empty control. Overexpression of miR-20a slightly increased cell counts, whereas induction of miR-489 and miR-494 caused no significant cell count alterations. In contrast, miR-181a, miR-16, and most prominently miR-26a expression resulted in reduced cell proliferation.
miR-26a expression in BL-derived cell lines and primary human lymphoma samples
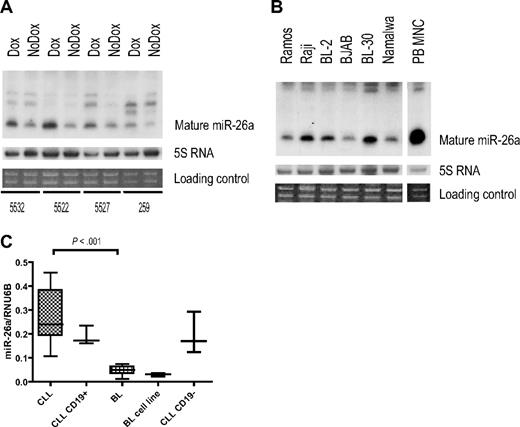
MYC-dependent repression of miR-26a was confirmed in murine lymphoma cell lines by Northern blot (Figure 3A). In addition, we found lower expression levels of miR-26a in human BL cell lines compared with normal PBMNCs (Figure 3B). However, we also observed a variable miR-26a expression among the BL cell lines, suggesting that, in accordance with our murine model, there also exists a certain level of molecular heterogeneity between individual human BL-derived cell lines.
miR-26a expression in MYC-induced lymphoma. (A,B) Northern blot analyses for miR-26a. (A) miR-26a expression in murine Myc-induced lymphoma cell lines after Dox treatment (18 hours). (B) miR-26a expression in human BL-derived cell lines and peripheral blood derived mononuclear cells (PBMNC). (C) Quantitative RT-PCR results evaluating miR-26a expression in primary human BL (n = 11), unselected B-CLL (n = 9), and CD19+-enriched B-CLL (n = 3) samples as well as in BL-derived cell lines (n = 3) and the CD19− fraction of the 3 enriched B-CLL cases. Normalized expression values are depicted (box plots indicate median expression and range of expression values). There are significant differences between BL and B-CLL samples with regard to miR-26a expression (P < .001, exact Wilcoxon rank sum test).
miR-26a expression in MYC-induced lymphoma. (A,B) Northern blot analyses for miR-26a. (A) miR-26a expression in murine Myc-induced lymphoma cell lines after Dox treatment (18 hours). (B) miR-26a expression in human BL-derived cell lines and peripheral blood derived mononuclear cells (PBMNC). (C) Quantitative RT-PCR results evaluating miR-26a expression in primary human BL (n = 11), unselected B-CLL (n = 9), and CD19+-enriched B-CLL (n = 3) samples as well as in BL-derived cell lines (n = 3) and the CD19− fraction of the 3 enriched B-CLL cases. Normalized expression values are depicted (box plots indicate median expression and range of expression values). There are significant differences between BL and B-CLL samples with regard to miR-26a expression (P < .001, exact Wilcoxon rank sum test).
To determine a possible clinical relevance, we investigated primary human BL samples. By quantitative RT-PCR, we determined miR-26a expression in 11 BL and for comparison in 12 B-CLL samples (9 unselected cases with high lymphocyte counts and 3 CD19+ selected cases), a lymphoma entity characterized by impaired apoptosis of slowly proliferating tumor cells without a prominent pathogenic role of MYC overexpression. There was a significant difference in miR-26a expression between BL and B-CLL samples (Figure 3C), supporting a potential role of miR-26a in MYC-dependent human lymphomagenesis.
miR-26a—a potential tumor-suppressor in MYC-induced lymphoma
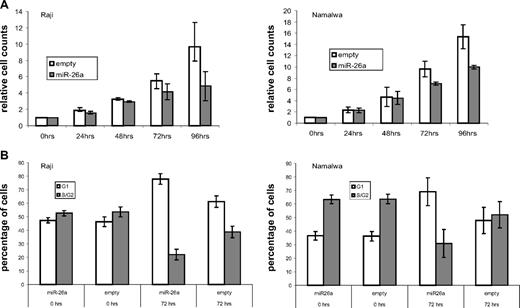
To elucidate tumorigenic properties of miR-26a deregulation, we inducibly overexpressed this miRNA in Raji and Namalwa using the episomal expression system. After puromycin selection, the proportion of GFP-positive cells was at least 95% after 24 hours of Dox treatment, and the expression of mature miR-26a was strongly elevated compared with the empty vector controls (Figure S3). Starting at 72 hours of miR-26a overexpression, cell numbers were reduced in transfected Raji and Namalwa cells with even more pronounced effects at 96 hours after miRNA induction (Figure 4A). Propidium iodide staining of Raji and Namalwa at 72 hours after Dox treatment revealed an increased percentage of cells in G1-phase within the miR-26a-expressing cell lines compared with the empty vector controls (78% vs 61% for Raji; 69% vs 48% for Namalwa) and less cells in the S/G2-phase, suggesting reduced cell cycle progression (Figure 4B). In contrast, elevated apoptosis analyzed by either propidium iodide staining (sub-G1 phase) or annexin V staining did not seem to contribute to the reduction in cell counts after miR-26a expression (data not shown).
miR-26a–induced phenotypic changes in a MYC expression background. (A) At indicated time points, cell count analysis was performed after induction of miR-26a expression in transfected Raji and Namalwa cells. After 72 hours of miR-26a overexpression, cell numbers were reduced in Raji and Namalwa cells (P = .031, exact Wilcoxon rank sum test). These effects were more pronounced after 96 hours of miRNA induction (P = .031, exact Wilcoxon rank sum test). As a control, empty vector- containing cells were treated with Dox for the same time periods. (B) Cell-cycle analysis in empty-vector and miR-26a-transfected Raji and Namalwa cells was determined by PI staining at indicated time points after Dox treatment. Compared with the empty-vector control, there was an increased percentage of cells in G1 phase within the miR-26a–expressing cell lines (P = .031, exact Wilcoxon rank sum test).
miR-26a–induced phenotypic changes in a MYC expression background. (A) At indicated time points, cell count analysis was performed after induction of miR-26a expression in transfected Raji and Namalwa cells. After 72 hours of miR-26a overexpression, cell numbers were reduced in Raji and Namalwa cells (P = .031, exact Wilcoxon rank sum test). These effects were more pronounced after 96 hours of miRNA induction (P = .031, exact Wilcoxon rank sum test). As a control, empty vector- containing cells were treated with Dox for the same time periods. (B) Cell-cycle analysis in empty-vector and miR-26a-transfected Raji and Namalwa cells was determined by PI staining at indicated time points after Dox treatment. Compared with the empty-vector control, there was an increased percentage of cells in G1 phase within the miR-26a–expressing cell lines (P = .031, exact Wilcoxon rank sum test).
In silico miR-26a target gene identification
Next, we wanted to investigate the molecular mechanisms by which miR-26a overexpression might affect growth in MYC-dependent lymphoma. Therefore, we screened different databases (PicTar, Target Scan, miRNAViewer, and miRBase as at June 2007), which predict potential miR-26a target genes using different computational algorithms. We focused on potential targets that were nominated in at least 2 different databases and might be involved in the observed G1 arrest by miR-26a overexpression. Within the group of putative candidates, we identified the Polycomb group protein Enhancer of Zeste Homolog 2 (EZH2), Cell Division Protein Kinase 8 (CDK8), CyclinE2 (CCNE2), Cell division cycle 6 homolog (CDC6), and Amyotrophic lateral sclerosis 2, juvenile, chromosome region 2 (ALS2CR2), also known as ILP-interacting protein (ILPIP).
Gene expression analysis after miR-26a overexpression
As imperfect miRNA binding to a certain mRNA does not only induce a translational block of the respective target but also negatively influences mRNA stability,30-33 we profiled gene expression in transfected BL cell lines 72 hours after miR-26a induction. Although we detected 1021 significantly differentially regulated genes by class comparison (P < .01, paired t test with random variance model [BRB-Array Tools]; Table S3), this signature displayed a significant enrichment of genes belonging to Gene Ontologies involved in cell cycle regulation, such as chromatin and nucleosome assembly, DNA replication factor C complex, cofactor binding, and DNA repair, as well as genes belonging to the “CDK Regulation of DNA Replication” pathway (P < .005, Fisher statistic and Kolmogorov-Smirnov permutation test [BRB-Array Tools]). In the group of genes down-regulated by miR-26a overexpression, we again identified the predicted target genes CDK8, CCNE2, CDC6, ALS2CR2, and EZH2, whose expression change was confirmed by quantitative RT-PCR (data not shown).
In accordance with our hypothesis that miR-26a might in part exert its action via inhibition of one of the potential target genes, we looked for an enrichment of genes known to be associated with respective candidates in our gene expression pattern. Interestingly, we observed a significant overrepresentation of EZH2 target genes in the gene expression pattern induced by miR-26a overexpression (P < .001, χ2 test). The significant overlap with an siRNA-mediated Polycomb gene (PcG) knock-down signature34 included deregulation of CCNE2, CCNB1 (cyclin B1), as well as genes involved in chromatin assembly and DNA replication, such as CHAF1A (chromatin assembly factor 1, subunit A) and RFC4 (replication factor C; Table 2).
EZH2 target gene enrichment in miR-26a-induced gene expression pattern
| Gene symbol . | Description . | Probe set . | Geometric mean of intensities (class empty/class miR-26a) . | Fold change after PcG knock-down* . |
|---|---|---|---|---|
| ARHGAP24 | Rho GTPase-activating protein 24 | 1566825_at | 0.7037292 | 1.5 |
| ATAD2 | ATPase family, AAA domain containing 2 | 222740_at | 1.3870516 | −1.9 |
| CCNB1 | Cyclin B1 | 214710_s_at | 1.3400383 | −1.5 |
| CCNE2 | Cyclin E2 | 205034_at | 1.5680341 | −2.5 |
| CDC25A | Cell division cycle 25 homolog A (S. cerevisiae) | 1555772_a_at | 1.3642922 | −1.9 |
| CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 203968_s_at | 1.4108894 | −3.1 |
| CDT1 | Chromatin licensing and DNA replication factor 1 | 228868_x_at | 1.5277866 | −2.2 |
| CHAF1A | Chromatin assembly factor 1, subunit A (p150) | 203976_s_at | 1.5635639 | −6.2 |
| CUGBP2 | CUG triplet repeat, RNA binding protein 2 | 242268_at | 0.7193129 | 1.7 |
| DCC1 | defective in sister chromatid cohesion homolog 1 (S. cerevisiae) | 219000_s_at | 1.3318879 | −1.6 |
| GMNN | Geminin, DNA replication inhibitor | 218350_s_at | 1.3320183 | −2 |
| GTSE1 | G2- and S-phase expressed 1 | 215942_s_at | 1.4042593 | −3.4 |
| HELLS | Helicase, lymphoid-specific | 220085_at | 1.3691602 | −2.3 |
| LRAP | Leukocyte-derived arginine aminopeptidase | 240338_at | 0.630956 | 1.4 |
| MCM10 | MCM10 minichromosome maintenance deficient 10 (S. cerevisiae) | 220651_s_at | 1.4244193 | −3.1 |
| MCM4 | MCM4 minichromosome maintenance deficient 4 (S. cerevisiae) | 222037_at | 1.2871303 | −4.8 |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 | 212022_s_at | 1.4520355 | −2.7 |
| MYBL1 | v-myb myeloblastosis viral oncogene homolog (avian)-like 1 | 213906_at | 1.5254951 | −1.7 |
| ORC1L | Origin recognition complex, subunit 1-like (yeast) | 205085_at | 1.511178 | −2.9 |
| PDE4DIP | Phosphodiesterase 4D interacting protein (myomegalin) | 232615_at | 0.7658641 | 1.1 |
| RFC4 | Replication factor C (activator 1) 4, 37kDa | 204023_at | 1.4484428 | −1.9 |
| SKP2 | S-phase kinase-associated protein 2 (p45) | 210567_s_at | 1.4732182 | −2.2 |
| SLC17A5 | Solute carrier family 17 (anion/sugar transporter), member 5 | 221041_s_at | 0.7774443 | 1.3 |
| SOX5 | SRY (sex determining region Y)-box 5 | 215768_at | 0.685878 | 1.7 |
| TncRNA | Trophoblast-derived noncoding RNA | 238320_at | 0.5457627 | 1.9 |
| ZBTB1 | Zinc finger and BTB domain containing 1 | 1557036_at | 0.7410104 | 4.9 |
| ZBTB10 | zinc finger and BTB domain containing 10 | 222863_at | 0.7321463 | 2.6 |
| Gene symbol . | Description . | Probe set . | Geometric mean of intensities (class empty/class miR-26a) . | Fold change after PcG knock-down* . |
|---|---|---|---|---|
| ARHGAP24 | Rho GTPase-activating protein 24 | 1566825_at | 0.7037292 | 1.5 |
| ATAD2 | ATPase family, AAA domain containing 2 | 222740_at | 1.3870516 | −1.9 |
| CCNB1 | Cyclin B1 | 214710_s_at | 1.3400383 | −1.5 |
| CCNE2 | Cyclin E2 | 205034_at | 1.5680341 | −2.5 |
| CDC25A | Cell division cycle 25 homolog A (S. cerevisiae) | 1555772_a_at | 1.3642922 | −1.9 |
| CDC6 | Cell division cycle 6 homolog (S. cerevisiae) | 203968_s_at | 1.4108894 | −3.1 |
| CDT1 | Chromatin licensing and DNA replication factor 1 | 228868_x_at | 1.5277866 | −2.2 |
| CHAF1A | Chromatin assembly factor 1, subunit A (p150) | 203976_s_at | 1.5635639 | −6.2 |
| CUGBP2 | CUG triplet repeat, RNA binding protein 2 | 242268_at | 0.7193129 | 1.7 |
| DCC1 | defective in sister chromatid cohesion homolog 1 (S. cerevisiae) | 219000_s_at | 1.3318879 | −1.6 |
| GMNN | Geminin, DNA replication inhibitor | 218350_s_at | 1.3320183 | −2 |
| GTSE1 | G2- and S-phase expressed 1 | 215942_s_at | 1.4042593 | −3.4 |
| HELLS | Helicase, lymphoid-specific | 220085_at | 1.3691602 | −2.3 |
| LRAP | Leukocyte-derived arginine aminopeptidase | 240338_at | 0.630956 | 1.4 |
| MCM10 | MCM10 minichromosome maintenance deficient 10 (S. cerevisiae) | 220651_s_at | 1.4244193 | −3.1 |
| MCM4 | MCM4 minichromosome maintenance deficient 4 (S. cerevisiae) | 222037_at | 1.2871303 | −4.8 |
| MKI67 | Antigen identified by monoclonal antibody Ki-67 | 212022_s_at | 1.4520355 | −2.7 |
| MYBL1 | v-myb myeloblastosis viral oncogene homolog (avian)-like 1 | 213906_at | 1.5254951 | −1.7 |
| ORC1L | Origin recognition complex, subunit 1-like (yeast) | 205085_at | 1.511178 | −2.9 |
| PDE4DIP | Phosphodiesterase 4D interacting protein (myomegalin) | 232615_at | 0.7658641 | 1.1 |
| RFC4 | Replication factor C (activator 1) 4, 37kDa | 204023_at | 1.4484428 | −1.9 |
| SKP2 | S-phase kinase-associated protein 2 (p45) | 210567_s_at | 1.4732182 | −2.2 |
| SLC17A5 | Solute carrier family 17 (anion/sugar transporter), member 5 | 221041_s_at | 0.7774443 | 1.3 |
| SOX5 | SRY (sex determining region Y)-box 5 | 215768_at | 0.685878 | 1.7 |
| TncRNA | Trophoblast-derived noncoding RNA | 238320_at | 0.5457627 | 1.9 |
| ZBTB1 | Zinc finger and BTB domain containing 1 | 1557036_at | 0.7410104 | 4.9 |
| ZBTB10 | zinc finger and BTB domain containing 10 | 222863_at | 0.7321463 | 2.6 |
Bracken et al.34
Evaluation of potential miR-26a target genes on miR-26a overexpression
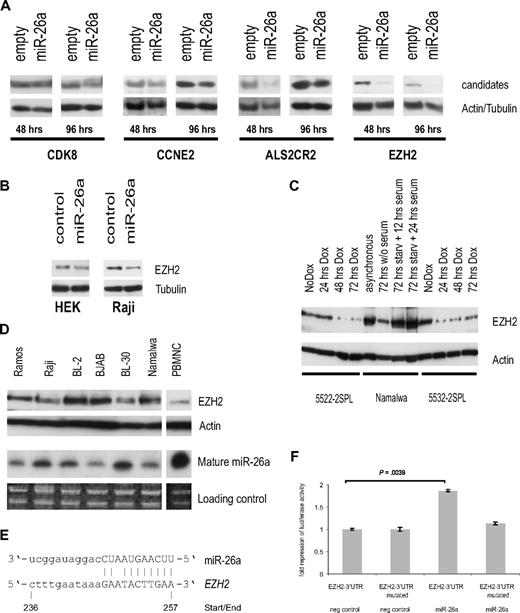
To determine whether the miR-26a–induced mRNA changes affect the protein expression of the aforementioned candidates, we investigated their protein levels on inducible miR-26a overexpression in stably transfected Raji cells. Whereas CDC6 was not detectable by Western blot (data not shown), for CDK8 we did not observe a considerable knock-down and only minor changes for CCNE2 and ALS2CR2. However, EZH2 expression was markedly reduced 48 hours and 96 hours after miR-26a induction (Figure 5A).
EZH2 as a potential miR-26a target. (A) Protein expression analysis of selected miR-26a candidate targets in stably transfected Raji cells at indicated time points after miR-26a induction compared with an empty vector control. Actin/tubulin served as a loading control. (B) Transient transfection of miR-26a precursor and scrambled RNA into HEK-293 and Raji cells. Western blot results are shown for EZH2 and tubulin 48 hours after transfection. (C) Protein expression analysis of EZH2 after Dox treatment in 2 MYC-induced murine lymphoma cell lines. Starvation of Namalwa cells served as a positive control. (D) EZH2 protein expression in several human BL cell lines compared with endogenous miR-26a expression. (E) Sequence homology between miR-26a and human EZH2, site of miR-26a binding in the 3′-UTR of EZH2 is indicated. (F) Fold repression of luciferase activity as determined by reporter assay in HEK-293 cells. Luciferase expression was determined 24 hours after cotransfection of either the precursor miR-26a or a scrambled RNA. There was a significant reduction in luciferase activity in the miR-26a transfected cells compared with the controls (P = .004, exact Wilcoxon rank sum test). Error bars for 3 independent experiments (measured each in triplicate) are depicted.
EZH2 as a potential miR-26a target. (A) Protein expression analysis of selected miR-26a candidate targets in stably transfected Raji cells at indicated time points after miR-26a induction compared with an empty vector control. Actin/tubulin served as a loading control. (B) Transient transfection of miR-26a precursor and scrambled RNA into HEK-293 and Raji cells. Western blot results are shown for EZH2 and tubulin 48 hours after transfection. (C) Protein expression analysis of EZH2 after Dox treatment in 2 MYC-induced murine lymphoma cell lines. Starvation of Namalwa cells served as a positive control. (D) EZH2 protein expression in several human BL cell lines compared with endogenous miR-26a expression. (E) Sequence homology between miR-26a and human EZH2, site of miR-26a binding in the 3′-UTR of EZH2 is indicated. (F) Fold repression of luciferase activity as determined by reporter assay in HEK-293 cells. Luciferase expression was determined 24 hours after cotransfection of either the precursor miR-26a or a scrambled RNA. There was a significant reduction in luciferase activity in the miR-26a transfected cells compared with the controls (P = .004, exact Wilcoxon rank sum test). Error bars for 3 independent experiments (measured each in triplicate) are depicted.
We also transiently transfected the miR-26a precursor or control RNA in HEK-293 cells as well as Raji. Although in HEK cells there was only a slight down-regulation of EZH2 on miR-26a overexpression, these effects were more pronounced in Raji cells, which express active MYC (Figure 5B).
We then went back to our murine lymphoma cell lines and analyzed EZH2 protein expression on turning off MYC expression. Western blot analyses revealed a down-regulation of EZH2 expression after 48 hours and 72 hours of Dox treatment (Figure 5C). Finally, we analyzed EZH2 and miR-26a expression in human BL cell lines, and we found the expected correlation: cell lines showing low miR-26a expression such as Namalwa and BJAB displayed elevated EZH2 protein levels, whereas miR-26a highly expressing cells such as BL-30 as well as PBMNCs showed low EZH2 expression (Figure 5D).
EZH2 as a potential miR-26a target gene
To determine whether EZH2 might be directly regulated by miR-26a, we used luciferase reporter assays. The full-length 3′UTR of human EZH2 containing the putative miR-26a responsive element (Figure 5E) was cloned into the 3′UTR region of the TK-pGL3 vector. As a control, 2 point mutations were introduced into the putative miR-26a binding site. Each construct was cotransfected in HEK-293 cells together with either the precursor of miR-26a or scrambled RNA as a negative control. Luciferase expression was significantly reduced in the miR-26a transfected cells compared with the controls (Figure 5F). Importantly, the mutant miR-26a responsive element was able to mitigate repression (Figure 5F), therefore suggesting a direct regulation of EZH2 via miR-26a.
EZH2 knock-down
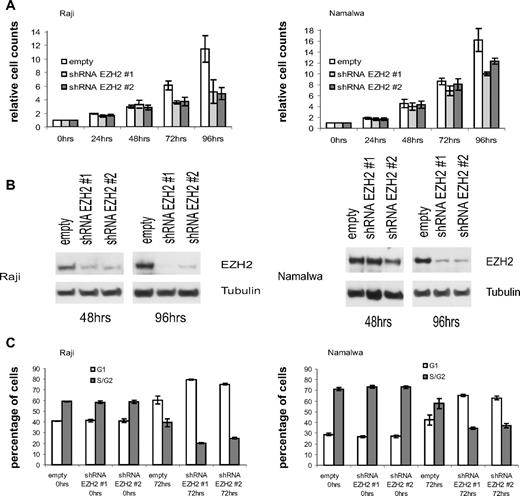
Based on our hypothesis that miR-26a effects might be in part mediated via the deregulation of EZH2, we used shRNAs to knock down the expression of EZH2 in BL cell lines. In agreement with the miR-26a findings, we observed a significant decrease in cell numbers in transfected Raji and Namalwa cells with a pronounced effect at 96 hours after shRNA induction and EZH2 down-regulation (Figure 6A,B). Effects on EZH2 protein levels and cell proliferation were delayed in Namalwa compared with Raji cells. We also observed an increased percentage of cells in G1-phase within the shRNA-expressing cell lines compared with the empty vector controls (80%/75% vs 61% for Raji; 65%/63% vs 42% for Namalwa; Figure 6C), suggesting reduced cell-cycle progression similar to the effects seen in miR-26a overexpressing cells. Thus, these findings further confirm that EZH2 deregulation might indeed play a role in MYC-dependent tumorigenesis.
EZH2 knock-down in MYC-dependent BL cell lines. (A) At indicated time points, cell count analysis was performed after induction of EZH2 shRNA#1 or EZH2 shRNA #2 expression in transfected Raji and Namalwa cells. After 96 hours of shRNA expression, cell numbers were significantly reduced in Raji and Namalwa cells (P = .031, exact Wilcoxon rank sum test). As a control, empty vector containing cells were treated with Dox for the same time periods. (B) Protein expression analysis of EZH2 after Dox induced shRNA expression at indicated time points. (C) Cell cycle analysis in empty vector and shRNA-transfected Raji and Namalwa cells was determined by PI staining at indicated time points after Dox treatment. Within the shRNA-expressing cell lines, there was an increased percentage of cells in G1 phase compared with the empty vector control (P = .031, exact Wilcoxon rank sum test).
EZH2 knock-down in MYC-dependent BL cell lines. (A) At indicated time points, cell count analysis was performed after induction of EZH2 shRNA#1 or EZH2 shRNA #2 expression in transfected Raji and Namalwa cells. After 96 hours of shRNA expression, cell numbers were significantly reduced in Raji and Namalwa cells (P = .031, exact Wilcoxon rank sum test). As a control, empty vector containing cells were treated with Dox for the same time periods. (B) Protein expression analysis of EZH2 after Dox induced shRNA expression at indicated time points. (C) Cell cycle analysis in empty vector and shRNA-transfected Raji and Namalwa cells was determined by PI staining at indicated time points after Dox treatment. Within the shRNA-expressing cell lines, there was an increased percentage of cells in G1 phase compared with the empty vector control (P = .031, exact Wilcoxon rank sum test).
Discussion
Many studies have shown that transgenic mice represent reliable model systems and strategies to conditionally regulate oncogene expression were used to investigate the oncogene dependence of a neoplastic phenotype.35 Accordingly, our lymphoma model induced by overexpression of the MYC oncogene offers the advantage to examine subsequent biologic effects after turning off MYC expression.18,19 Furthermore, we had established a large set of tumor cell lines derived from different MYC-induced lymphomas, which are still dependent on the oncogene expression. Therefore, our model provides an excellent tool to study the kinetics of MYC-induced lymphomagenesis, especially with regard to MYC-regulated miRNA expression and its impact on lymphoma pathogenesis.
Corresponding to several studies describing deregulated miRNA expression in tumorigenesis,11,13 we observed distinct MYC-associated miRNA expression changes. Although there is no human T-cell-BL like disease, the consistent finding of deregulated miRNA patterns in both B- and T-cell lines makes a strong argument for (1) the immediate involvement of MYC in regulating their expression and (2) the relevance of altered miRNA expression for MYC-induced lymphomagenesis. Although we detected an induction of the miR-17-92 cluster as previously reported,15,17 we also found several miRNAs to be significantly repressed on MYC expression and thus identified a larger set of MYC-regulated miRNAs than initially anticipated. In accordance with our findings, a recent work has also demonstrated a widespread MYC-induced repression of microRNAs, which tend to be direct MYC targets as determined by chromatin immunoprecipitation.16 As expected, there is a considerable overlap of findings, especially with regard to the expression of the miR-17-92 cluster and the MYC-repressed miRNAs, miR-16 and miR-26a (Table 1).
However, there were also distinct differences among the 2 tumor models. In our study, activation of MYC expression was the primary event in a probably multistep process of lymphoma development as determined by the phenotypic differences among the cell lines. On the other hand, in the Chang et al model, Myc was overexpressed in an Ebstein-Barr virus (EBV) immortalized B-cell background or in bone marrow cells derived from p53-null mice (Trp53−/−).16 Furthermore, the analysis of Chang et al was based on the comparison of only 2 tumors with high and 2 tumors with low Myc expression, thereby not reflecting potential influences of the underlying molecular heterogeneity. Although our unsupervised analysis of untreated lymphoma cell lines clearly demonstrated marked differences among a large cohort of 26 MYC-induced lymphoma-derived cell lines (Figure 1C), this molecular heterogeneity was also in part reflected by different responses to MYC repression (Figure 1A).
The molecular heterogeneity in our murine lymphoma model could not be explained by differences in ARF and BCL2 expression or altered phosphorylation status of AKT, pathways known to be commonly involved in MYC-induced tumorigenesis.5,6 However, part of the tumor heterogeneity might be explained by deregulated p53 signaling.16 Although expression changes in p53-induced miR-3436 were not observed between the clustering groups, additional genomic alterations, gene mutations, and/or epigenetic changes might underlie the different lymphoma phenotypes.
Despite methodologic differences to the Chang et al study and the marked lymphoma heterogeneity in our experimental setup, miR-26a was found to be a MYC-repressed miRNA in almost all examined MYC-dependent tumors, thereby representing one of the most interesting MYC-repressed miRNAs. In agreement, in vitro overexpression of miR-26a in MYC-dependent cells resulted in a strong antiproliferative effect, which was also seen by Chang et al in vivo.16 Finally, our findings established in a murine lymphoma model seem to be also relevant in primary human tumors as miR-26a expression is repressed in primary human BL samples compared with B-CLL.
To gain further insights in the molecular mechanisms underlying miR-26a–repressed proliferation, we screened several databases for predicted miR-26a target genes and correlated these findings with gene expression changes after prolonged miR-26a expression. Recently, this strategy has already been successfully used to identify miRNA targets.32,37 EZH2 turned out as a promising miR-26a target gene, which is known as a global regulator of gene expression and “bona fide” oncogene. EZH2 belongs to the family of PcG proteins, which are epigenetic gene silencers implicated in neoplastic development. As part of the polycomb repressive complex 2 (PRC2), it possesses methyltransferase activity directed toward lysine 27 of histone H3 (H3K27).38 Although trimethylated-H3K27 (H3K27me3) represents a quite stable and dominant mark for silenced gene expression, EZH2 especially targets genes involved in differentiation, such as homeobox genes and thereby promotes the late-stage development of cancer.39
Recently, EZH2 expression has been shown to be deregulated in human cancer including NHL,40 and its prognostic impact has been demonstrated in breast and prostate cancer.41,42 Thus, in MYC-induced lymphoma, there might also be an important role for EZH2. Our gene expression analyses after prolonged miR-26a expression showed a significant enrichment of EZH2 target genes as defined by Bracken et al in the miR-26a associated signature.34 Interestingly, we observed both an enrichment of up- and down-regulated EZH2 target genes. This finding further demonstrates that EZH2 does not only exert its biologic activity as a transcriptional repressor but also possesses transactivation activity, thereby being a dual function transcription regulator as recently suggested.43
By using a reporter assay, we were able to demonstrate direct regulation of EZH2 via miR-26a and by shRNA-mediated EZH2 knock-down we observed a phenotype corresponding to the miR-26 overexpression results. Recent data suggested that EZH2 can induce MYC expression.43 MYC might contribute to the up-regulation of EZH2 via down-regulation of its targeting miRNA, thereby generating a positive feedback loop that could be involved in maintaining the malignant phenotype. This oncogenic pathway, which is based on the repression of an oncogene targeting miRNA, might be as important as the induction of the miR-17-92 cluster in MYC-induced lymphomagenesis. Interestingly, there seems to be a potential connection between both signaling pathways. Recently, up-regulation of the miR-17-92 cluster has been shown to promote MYC-induced tumorigenesis.15,44 However, overexpression of the miR-17-92 cluster also led to repression of E2F1, a target directly induced by MYC.17 Thus, mechanisms exist through which MYC simultaneously activates E2F1 transcription and limits its translation to allow tightly controlled proliferative signaling. Interestingly, E2F1 was also shown to induce EZH2 expression.45 Therefore, by up-regulation of the direct MYC target E2F146 and simultaneous repression of miR-26a, MYC possesses 2 distinct pathways to up-regulate EZH2 expression thereby having the ability to enhance proliferation and maintain “cancer stem cell” properties. This suggests that MYC might exert different functions by deregulating miRNA expression, some of which negatively regulate direct MYC action to ensure fine-tuning, whereas others function synergistically thereby contributing to lymphomagenesis.
Thus, miRNAs apparently play an important role in the complex network of MYC-induced tumorigenesis. We identified a novel link toward the control of the PcG. It is currently unknown how exactly Polycomb complex protein activity is regulated. Recent data suggest that even subtle changes in Polycomb complex composition and temporary association of auxiliary factors play an important regulatory role.40 These subtle changes might well be transmitted via deregulated miRNA expression. Ultimately, future research will continue to unravel the specific contributions of miRNA expression to cell fate decisions and shed light into the molecular mechanisms underlying their involvement in neoplastic development, a prerequisite for improved lymphoma classification and individualized patient management.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank the staff of the Microarray Facility of the University of Ulm for providing high-quality DNA microarrays, K. Holzmann for his help with data analyses, G.W. Bornkamm and D. Eick for kindly providing the inducible miRNA expression vector system, and all Wirth laboratory members for fruitful discussion of this project.
This study was supported in part by the Deutsche José Carreras Stiftung e.V. (DJCLS R 06/41v; L.B.) and the Deutsche Forschungsgemeinschaft (DFG-SFB497 TP C5; T.W.). S. Sander is supported by the International Graduate School in Molecular Medicine Ulm.
Authorship
Contribution: S. Sander and L.B. designed and performed research, analyzed/interpreted data, and wrote the paper; K.K. and K.F. performed research and analyzed data; H.A.K. analyzed/interpreted data and wrote the paper; T.F.E.B., P.M., and S. Stilgenbauer contributed vital reagents and analyzed/interpreted data; J.R.P. analyzed/interpreted data and wrote the paper; and T.W. designed research, analyzed/interpreted data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Thomas Wirth, Institute of Physiological Chemistry, University of Ulm, Albert-Einstein-Allee 11, 89081 Ulm, Germany; e-mail: thomas.wirth@uni-ulm.de.


This feature is available to Subscribers Only
Sign In or Create an Account Close Modal