Abstract
Three-dimensional (3D) reconstruction of organs and tissues is a powerful tool to establish anatomical and functional relationships of microscopic structures. We developed whole-mount tissue processing methods for 3D in situ visualization of murine and human bone marrow; our methods are compatible with fluorescent labeling of different cell types and other structures of interest in the tissue microenvironment. The major technical problems addressed were the conditions for tissue fixation in the absence of permeabilization and sectioning; antibody penetration and binding; and the acquisition of high quality images by adequate laser scanning confocal microscope. For murine bone marrow, the sternum was bisected sagitally; for human tissue, 2–3 mm fragments of core biopsies were utilized. Bone marrow tissue and cells were exposed to fluorescence labeled nucleic acid dyes and antibodies, with or without prior chemical fixation. Single and double labeling of cells was feasible with combinations of various antibodies and direct and indirect immunofluorescent techniques. In some experiments, cells were visualized from transgenic mice with cell populations expressing green fluorescence protein (GFP). Series of two dimensional (xy) images 600 μm × 600 μm were collected along the z-axis at 5 μm z-intervals to depths of 60–100 μm using a Zeiss LSM 510 confocal microscope. Two dimensional images were assembled to reconstruct 3-dimensional volumes by Bitplane’s Imaris 3D computer software. Antigenicity was preserved, allowing simultaneous labeling of cell types and structures by immunohistochemistry or nuclear dyes. Different hematopoietic cell types as well as blood vessels, adipose cells, and extracellular matrix were visualized in complex 3-dimensional organization of intact bone marrow tissue revealing unknown features of multicellular architecture.
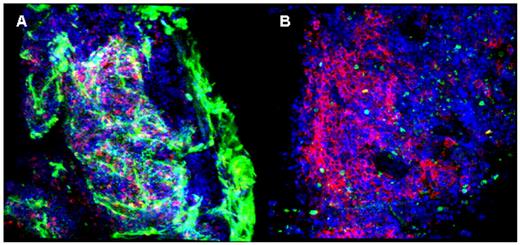
Normal murine bone marrow, after brief fixation formaldehyde, is shown in Figure A. Rat anti-mouse basement-membrane monoclonal antibody (MAb) and fluorescent isothiocyanate (FITC)-labeled donkey anti-rat monoclonal antibody were used to visualize the extracellular matrix and micro-vessels (appearing green). Allophycocyanin (APC)-labeled rat anti-mouse CD45R cells permitted visualization of B lymphocytes (red). 4’,6-diamidino-2-phenylindole(DAPI) stained all nuclei (blue). Nests of lymphocytes appeared encased by extracellular matrix, fed by microvessels running from the bone edge. An example of the architecture of a human hematologic malignancy is shown in figure B, from a marrow biopsy of a patient with multiple myeloma prior to therapy. Mouse anti-human CD20 MAb and FITC-labeled donkey anti-mouse IgG were used to visualize mature B cells (green). APC-conjugated mouse anti-human CD38 MAb identified plasma cells (red). DAPI stained nuclei (blue). The large tumor cells appeared in unevenly distributed cell clumps. In mouse experiments, (not illustrated), marrow cells were easily observed in animals in which GFP was driven by the ubiquitin-C promoter. In humans (also not illustrated), we observed malignant cell populations stained with appropriate lineage-specific antibodies in patients with leukemia and compared CD34 cell numbers in normal with aplastic bone marrow. Confocal laser scanning microscopy, a powerful technique to generate serial sections of whole-mount tissue and their digital reassembly into virtual 3-dimensional structures, has been readily adapted to examination of murine and human bone marrow. The wide variety of MAbs available for specific antigens in combination with this imaging method should aid in conceptualizing microanatomical relationships among hematopoietic cells, stroma, blood vessels, and extracellular matrix in normal and diseased bone marrow.
Disclosures: No relevant conflicts of interest to declare.
Author notes
Corresponding author

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal