Abstract
HIV-1 infection is associated with B-cell abnormalities, such as hypergammaglobulinemia, poor immunization responses, and loss of serologic memory. To determine whether altered expression of chemokine receptors and their ligands may play a role in B-cell dysfunctions during HIV-1 infection, the expression of CXC chemokine receptor 4 (CXCR4), CXCR5, and CC chemokine receptor 7 (CCR7) and their respective ligands on CD19+ B cells were examined in HIV-1–infected patients and controls. We report a decreased CXCR5 expression on B cells from patients (P < .05), a phenomenon associated with a low CD4 T-cell count (< 350 cells/μL). Interestingly, an increased expression of CXC chemokine ligand 13 (CXCL13), the ligand for CXCR5, was found in peripheral B cells from HIV-1–infected patients. Moreover, on B-cell activation in vitro, CXCL13 was secreted in culture. CXCL13+ B cells were also found in the lymph nodes of HIV-1–infected patients, but not in control tissue. B-cell migration toward CXCL13, CXCL12, and CC chemokine ligand 21 (CCL21), ligands for CXCR5, CXCR4, and CCR7 was also evaluated. In patients with a low CD4 T-cell count, migration toward all ligands was increased. Our findings indicate that altered expression of the chemokine receptor-ligand pair, CXCR5/CXCL13, may participate in the establishment of B-cell dysfunctions during HIV-1 infection.
Introduction
HIV-1 infection is associated with extensive B-cell abnormalities, manifested by phenotypic alterations and polyclonal B-cell activation, increased frequencies of B-cell malignancies, hypergammaglobulinemia, as well as poor antigen-specific immune responses to recall and de novo antigens.1-5 In secondary lymphoid tissue, HIV-1 infection induces follicular hyperplasia and alterations in the architecture of germinal center (GC)6 and splenic marginal zones.7 Defects in the B-cell compartment become overt already during primary HIV-1 infection8 as measured by a decline of B-cell number, increased expression of activation, and apoptosis markers.9 The mechanisms by which HIV-1 impairs humoral immunity may be the result of intrinsic B-cell defects and/or a lack of functional dialogue between B and T cells in secondary lymphoid organs.
Lymphocyte migration and recirculation between the periphery and lymphoid tissue are critical for effective immunity and are in part regulated by chemokine receptors on lymphocytes together with the expression of their respective ligands (chemokines) in different tissue compartments.10,11 In recent years, increasing attention has been given to the potential role of viruses to interfere with chemokine receptor expression, binding, and signaling.12-14 In this respect, HIV-1 has been extensively studied because the virus uses CXC chemokine receptor 4 (CXCR4) and CC chemokine receptor 5 (CCR5) as coreceptors for entry into target cells, in addition to the main receptor, the CD4 molecule.15 However, the expression of chemokine receptors in the context of B-cell trafficking is still poorly studied in chronic HIV-1 infection.
The chemokine receptor CXCR4 is broadly expressed on a majority of B cells in the bone marrow (BM), as well as in the periphery, and plays an important role for early B-cell development16,17 and plasma cell homing to the BM.18,19 On the other hand, CXCR5 is expressed by mature B cells and contributes to the recruitment of naive B cells into the lymph nodes20 where the GC reaction occurs with class switch, somatic hypermutation, and affinity maturation. The microanatomic organization of GCs into light and dark zones has been attributed to the expression of CXCR4 and CXCR5.21,22 B cells also express a moderate amount of CCR7, which contributes to the migration within the lymph node.23
In the present study, we examined the cell surface expression of chemokine receptors CXCR4, CXCR5, and CCR7 on B cells isolated from the blood of HIV-1–infected patients because these receptors mediate important events of B-cell homing to lymphoid tissue.20,23-26 Using gene expression profiling of chemokine receptors and chemokines, we found a high level of CXC chemokine ligand 13 (CXCL13) mRNA in B cells from HIV-1–infected patients compared with controls; in addition, these cells secreted a high level of the CXCL13 protein after in vitro activation. Histopathology studies performed in lymphoid tissues revealed the presence of CXCL13+ B cells in lymph nodes from HIV-1 patients but not in controls. Taken together, our results suggest that an impaired expression of CXCR5/CXCL13 may contribute to B-cell immunopathology during HIV-1 infection.
Methods
Subjects
For this study, blood specimens from 60 HIV-1–infected persons (40 males; 20 females) and 60 healthy, age- and sex-matched controls were collected. Patients had a median age of 41 years (range, 21-65 years). In the HIV-1 cohort, the median CD4+ T-cell count was 375 cells/μL (range, 50-1230 cells/μL) and viral load ranged from less than 20 to 250 000 copies/mL blood. The viral load was determined using the NASBA system (Organon Teknika, Boxtel, The Netherlands). Fifty-two patients were undergoing highly active antiretroviral therapy (HAART), with a median time on HAART of 7 years (range, 4-15 years), and 8 patients were untreated. Because of sample limitations, all patients were not included in all analyses. Informed consent was obtained from all subjects in accordance with the Declaration of Helsinki, and the ethical committees of the Karolinska University Hospital and Karolinska Institutet approved the study.
Lymph nodes obtained from 10 HIV-1–infected patients with CD4+ counts more than 250 were examined at the Bernhard-Nochts Institute for Tropical Diseases (Hamburg, Germany). The patients had received no or minimal treatment consisting of zidovudine. From the diagnostic file of the department, 5 HIV-negative lymph nodes exhibiting follicular hyperplasia of unknown genesis were also selected. The ethical committee at the Bernhard-Nochts Institute for Tropical Diseases approved the study on lymph node tissue.
Cell preparation and cell cultures
Peripheral blood mononuclear cells (PBMCs) were prepared from blood by standard Ficoll-Hypaque gradient centrifugation (Lymphoprep; Medinor, Oslo, Norway). B cells were separated using magnetic cell sorting (B-Cell Isolation Kit II; Miltenyi Biotec, Bergisch Gladbach, Germany). The purity of the separated B cells was more than 95% as verified by anti-CD19 staining. Cells were cultured in serum-free X-Vivo 15 culture medium (Cambrex, Walkersville, MD) overnight before the migration assay was performed.
For the detection of CXCL13 in the culture medium, B cells were activated in vitro with monoclonal antibody to CD40 (1 μg/mL; Nordic Bio-site, Stockholm, Sweden), interleukin-2 (IL-2; 100 ng/mL), and IL-10 (100 ng/mL; Tebu-Bio, Roskilde, Denmark), cytosine guanine dinucleotide (CpG) oligodeoxynucleotide (ODN) 2006 (CpG type B; 10 μg/mL; Microsynth, Balgach, Switzerland), and for BCR ligation with Affini Pure F(ab′)2 fragment goat antihuman IgA plus IgG plus IgM (H + L; Jackson ImmunoResearch Laboratories, Suffolk, United Kingdom). After activation, cells were kept in culture (106 cells/mL) for 6 days before CXCL13 measurements were performed using the Quantikine kit (R&D Systems Europe, Abingdon, United Kingdom). For neutralization of the CXCL13 bioactivity in vitro, the antihuman CXCL13/BLC/BCA-1 antibody (100 ng/mL; R&D Systems Europe) was used according to the manufacturer's instructions.
Flow cytometry
Briefly, 0.5 × 106 PBMCs were stained for 30 minutes at room temperature in phosphate-buffered saline containing 1% bovine serum albumin with the following antibodies (Abs): Cy-Chrome–conjugated anti-CD19, fluorescein isothiocyanate–conjugated anti-CD27 and anti-CD38, phycoerythrin (PE)–conjugated anti-CXCR4 (clone 12G5), anti-CCR7 (clone 3D12; BD Biosciences, Stockholm, Sweden), PE-conjugated anti-CXCR5 (FAB190P; clone 51 501.11; R&D Systems), and the negative fluorescein isothiocyanate- and PE-conjugated mouse IgG Ab. Cells were acquired on a BD Biosciences FACScan and analyzed using CellQuest software.
cDNA expression array
Purified B cells from patients (n = 4) and controls (n = 4) were collected in RNeasy lysis buffer for total RNA extraction using RNeasy Mini Kit (QIAGEN, Stockholm, Sweden). Approximately 10 to 20 mg of RNA was purified from each individual sample, and cDNA expression microarray analysis was performed using the GEArray Q series Human Chemokines and Receptors Gene Array (SABiosciences, Frederick, MD).
A total of 0.5 mg of total RNA was reverse-transcribed using Ampolabelling (LPR) Kit (SABiosciences) with biotin-16-dUTP (Enzo Life Sciences, Farmingdale, NY), and amplification of cDNA was performed during 30 cycles. The biotinylated cDNA probes were denatured and added to the hybridization solution. GEArray Q Series membranes were prehybridized at 60°C overnight and thereafter hybridized with the cDNA probes. Membranes were then washed, blocked, and incubated with alkaline phosphatase–conjugated streptavidin, and the labeled biotin was detected by chemoluminescence using the GEArray Chemoluminescence Detection Kit (D-01; SABiosciences). All microarray data have been submitted to Gene Expression Omnibus under accession number GSE12597.27
The luminescence intensities of hybridized cDNA probes were analyzed by GEArray Expression Analysis Suite software (Superarray Bioscience). Local background was subtracted for each point, and obtained values were adjusted to a common mean of 100.0 and minimal positive value was set to 10.0% of mean.
Real-time quantitative polymerase chain reaction
PBMCs (2.5 × 105 cells/mL) were incubated with 250 ng/mL CXCL13 for 24 hours. The CXCR5 surface expression was examined by flow cytometry, and gene expression level was investigated by quantitative polymerase chain reaction (qPCR) (TaqMan Gene Expression Assays, Hs 00173527_m1, lot 414094; Applied Biosystems, Stockholm, Sweden). The minor groove binder probes were labeled with 6-carboxyfluorescein (FAM). The PCR contained 10 μL master mix (TaqMan 2× PCR Master Mix, 58003365-01, lot HB3858; Applied Biosystems), 1-μL reverse-transcribed reaction product including 100 ng of each of the primers containing minor groove binder probe, cDNA, and water to a final volume of 20 μL. The reaction was performed in ABI Prism 7900 (Applied Biosystems), and the reaction conditions were 50°C for 2 minutes, 95°C for 10 minutes, and then 40 cycles of 95°C for 15 seconds and 60°C for 1 minute. The final results were analyzed using ABI Prism 7900 SDS software (Applied Biosystems).
Immunohistochemistry
Paraffin-embedded tissue were prepared and dewaxed as 5-μm sections and placed in a domestic pressure cooker containing 2 mM ethylenediaminetetraacetic acid (pH 8), boiled for 5 minutes, and thereafter cooled to room temperature. Tissue samples were incubated with a biotinylated antihuman BLA/BCA1/CXCL13 Ab (code BAF801; R&D Systems; dilution 1:50) overnight followed by incubation with StreptABComplex/HRP (code K0391; Dako, Hamburg, Germany) using 3-amino-9-ethylcarbazole (Sigma-Aldrich, Deisenhofen, Germany) as the substrate. The sections were counterstained with hematoxylin and mounted. For detection of CXCR5, frozen cryostat sections, fixed in 2% paraformaldehyde for 10 minutes, were incubated with an unconjugated mouse antihuman CXCR5 monoclonal Ab (clone 51505, R&D Systems; dilution 1:50) followed by the alkaline phosphatase antialkaline phosphatase (APAAP method) using Fast Red as chromogen.
Double labeling for lineage characterization of CXCL13+ cells was performed after the initial CXCL13 labeling by heat treatment of the sections for 5 minutes with 0.01 M buffered sodium citrate solution (pH 6.0), followed by overnight incubation with the following Abs: immature dendritic cells (DCs) (CD1a; NeoMarkers, Freemont, CA; clone 010) (dilution 1:50), mature DCs (CD83; Novocastra Laboratories, Newcastle-upon-Tyne, United Kingdom; clone 1H4b 1.30), B cells (CD79a; Dako), macrophages (CD68; Dako; clone KP1; dilution 1:20), or CD8 cells (Dako; clone C8/144B). For detection of CD4+ cells (Novocastra Laboratories; clone 1F6; dilution 1:20), the high temperature antigen retrieval was performed in 50 mM Tris and 2 mM ethylenediaminetetraacetic acid (pH 9.0) for 3 minutes. Antibody binding was detected with the APAAP method using Fast Blue as chromogen. No counterstain was performed.
Semiquantitative analysis of the CXCL13+ cells
Using a 40× objective, a standard area (unit area) was set and a photomicrograph was taken with a Zeiss AxioImager M1 microscope equipped with AxioCam MRc5 camera (Carl Zeiss, Jena, Germany). Ten nonoverlapping unit areas of the extrafollicular lymphoid tissue were selected. Using AxioVision (Release 4.6) software (Carl Zeiss), positive cells within the unit area were determined by manual counting. The individual values were pooled, and the mean number of positive cells per unit area was calculated.
Transmigration assay
Transwell culture system with a 5-μm–diameter pore filter (Transwell, 24-well plate; Corning Life Sciences, Lowell, MA) was used as previously described.28 Different concentrations of the chemokines CXCL12, CXCL13, and CC chemokine ligand 21 (CCL21; R&D Systems) were included in the migration assay (results not shown), and 250 ng/mL was evaluated as the optimal amount for B-cell migration. Purified B cells (2.5 × 105) were resuspended in medium, loaded into the upper chamber of the Transwell, and thereafter, 600 μL medium containing 250 ng/mL of the respective chemokine was added to the lower well. After a 4-hour incubation at 37°C, migrated cells were collected and counted by flow cytometry for 1 minute. Migration is presented as an index of migrated cells toward the individual chemokine divided by spontaneous cell migration as previously described.29
Statistical analysis
Statistical analyses were performed using GraphPad Prism (GraphPad Software, San Diego, CA). Differences between patients and controls were analyzed by parametric (t test) or nonparametric tests (Mann-Whitney U test) when appropriate. Correlation between clinical parameters and expression of chemokine receptors and chemokines were performed by linear regression.
Results
Reduced expression of the chemokine receptor CXCR5 on B cells from HIV-1–infected patients
The expression (median fluorescence intensity [MFI]) of the chemokine receptors CXCR4, CXCR5, and CCR7 was studied in the CD19+ B-cell compartment in the blood of HIV-1–infected patients and noninfected control subjects (Figure 1A). The majority of mature B cells expressed CXCR4, CXCR5, and, to a lesser extent, CCR7 on the cell surface. There were no significant differences in the expression of CD19 between patients and controls, and the expression of chemokine receptors on cells from the healthy controls was similar to previous findings.30
Chemokine receptor expression on B cells. Representative staining histograms for chemokine receptor expression on total CD19+ B cells, from one control (top panel) and one HIV-1 patient (bottom panel). An acquisition forward/side scatter dot plot was used to gate live lymphocytes, and chemokine receptor expression was analyzed on B cells (A). (B-E) The expression of chemokine receptors on CD19+ B cells is shown as median fluorescence intensity (MFI); in the left panels, the expression of the CXCR5 (B), CXCR4 (C), CXCR3 (D), and CCR7 (E) in patients (n = 30) and controls (n = 30) is shown. The receptor expression in groups of patients divided according to their CD4+ T-cell count (< 350 cells/μL or > 350 cells/μL) is shown in the right panels. CXCR5 expression (B) on B cells from HIV-1–infected is decreased (P = .012) compared with controls, and CXCR5 expression was also significantly lower in patients with low CD4+ T-cell count (P = .04). There were no significant differences in the expression of CXCR4, CXCR3, and CCR7 between patients and controls. The expression of CCR7 (E) was increased in patients with low CD4+ T-cell counts (P = .004).
Chemokine receptor expression on B cells. Representative staining histograms for chemokine receptor expression on total CD19+ B cells, from one control (top panel) and one HIV-1 patient (bottom panel). An acquisition forward/side scatter dot plot was used to gate live lymphocytes, and chemokine receptor expression was analyzed on B cells (A). (B-E) The expression of chemokine receptors on CD19+ B cells is shown as median fluorescence intensity (MFI); in the left panels, the expression of the CXCR5 (B), CXCR4 (C), CXCR3 (D), and CCR7 (E) in patients (n = 30) and controls (n = 30) is shown. The receptor expression in groups of patients divided according to their CD4+ T-cell count (< 350 cells/μL or > 350 cells/μL) is shown in the right panels. CXCR5 expression (B) on B cells from HIV-1–infected is decreased (P = .012) compared with controls, and CXCR5 expression was also significantly lower in patients with low CD4+ T-cell count (P = .04). There were no significant differences in the expression of CXCR4, CXCR3, and CCR7 between patients and controls. The expression of CCR7 (E) was increased in patients with low CD4+ T-cell counts (P = .004).
The CXCR5 expression on B cells was significantly reduced comparing HIV-1–infected patients with healthy controls (MFI 111 [range, 26-276] vs 169 [range, 14-286]; P = .01; Figure 1B left panel). CXCR4 expression on B cells was similar in HIV-1–infected subjects compared with healthy controls (MFI 64 [range, 17-250] vs 70 [range, 14-220]; P > .05; Figure 1C left panel). We also studied the expression of CXCR3 because it may be an important marker for B-cell activation, and we found that CXCR3 expression was similar in patients and controls (MFI 29 [range, 10-60] vs 29 [range, 11-64]; P = .29; Figure 1D left panel). In addition, the CCR7 expression on B cells showed no differences (MFI 13 [range, 7-34] vs 13 [range, 7-30]) comparing patients and controls (Figure 1E left panel).
To clarify whether CD4+ T-cell count correlates to the expression of chemokine receptors, patients were divided into 2 groups depending on the CD4+ T-cell counts. The expression of CXCR5 in HIV-1–infected patients (n = 18) with a CD4+ T-cell count less than 350 cells/μL was decreased compared with patients (n = 12) with a CD4+ T-cell count more than 350 cells/μL (P = .04; Figure 1B right panel). The decreased expression of CXCR5 was also confirmed by the use of calibrated fluorescent beads (data not shown). When performing regression analysis, a low CXCR5 expression also correlated to low CD4+ T-cell counts (P = .04). There was no correlation between viral load and CXCR5 expression. In addition, HAART treatment did not affect the expression of CXCR5 on B cells (P > .05). The CXCR4 and CXCR3 receptors were similarly expressed on B cells of all patients independently of CD4+ T-cell counts (Figure 1C,D right panels). On the contrary, an increased expression of CCR7 was detected in patients with low CD4+ T-cell count compared with patients with high CD4+ T-cell count (Figure 1E right panel).
CXCR5 expression on subpopulations of B cells
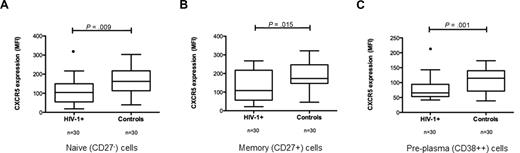
To further characterize the CXCR5 expression in relation to subpopulations of B cells, additional stainings were performed. The expression of CXCR5 on naive (CD27−) B cells from pa-tients were significantly reduced compared with controls (MFI 104 [range, 19-217] vs 162 [range, 39-303]; P = .009; Figure 2A), and this was also found on memory (CD27+) B cells (MFI 108 [range, 22-268] vs 173 [range, 46-322]; P = .015; Figure 2B). Pre-plasma B cells (CD38++CD19+) from HIV-1 patients also showed a decreased CXCR5 expression compared with controls (MFI 66 [range, 42-215] vs 115 [range, 39-173]; P = .001; Figure 2C). The expression of CXCR3, CXCR4, and CCR7 on the different subpopulations of B cells did not differ between patients and controls (results not shown).
CXCR5 expression on subpopulations of B cells. An acquisition forward/side scatter dot plot was used to gate live lymphocytes, and CXCR5 expression was analyzed on different subpopulations of B cells based on the expression of CD19, CD27, and CD38. (A) CXCR5 expression on naive (CD27−CD19+) B cells in HIV-1+ and control samples. CXCR5 expression on memory (CD27+CD19+) B cells (B) and early pre-plasma B cells (CD38++CD19+) (C) is also shown. CXCR5 expression was significantly lower in all B-cell subpopulations in HIV-1 subjects compared with controls.
CXCR5 expression on subpopulations of B cells. An acquisition forward/side scatter dot plot was used to gate live lymphocytes, and CXCR5 expression was analyzed on different subpopulations of B cells based on the expression of CD19, CD27, and CD38. (A) CXCR5 expression on naive (CD27−CD19+) B cells in HIV-1+ and control samples. CXCR5 expression on memory (CD27+CD19+) B cells (B) and early pre-plasma B cells (CD38++CD19+) (C) is also shown. CXCR5 expression was significantly lower in all B-cell subpopulations in HIV-1 subjects compared with controls.
Gene expression of chemokine receptors and chemokines in B cells
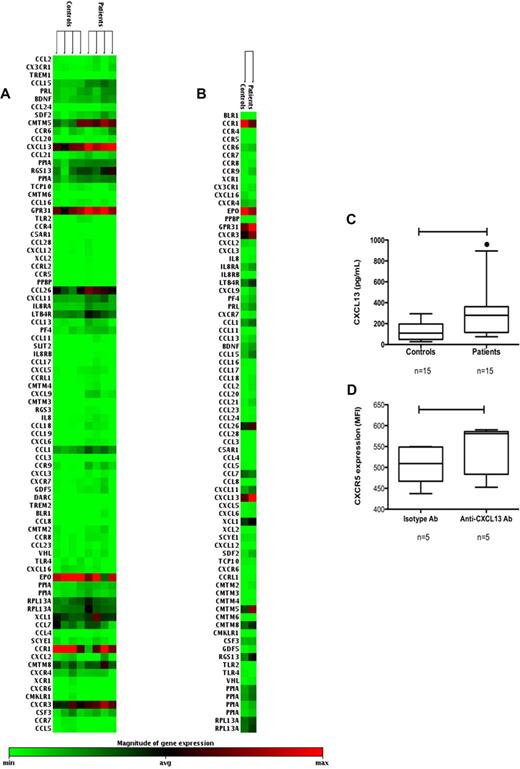
To further characterize the expression of chemokine and chemokine receptors in B cells from HIV-1–infected subjects and controls, gene expression profiling was used. The mRNA expression of the chemokine receptors CXCR4, CXCR5 (BLR1), and CCR7 did not show any changes between the patient and control samples (Figure 3A). In addition, the mRNA levels of the ligands CXCL12 (CXCR4), CCL19, and CCL21 (CCR7) were not considerably changed in patients compared with controls. However, the mRNA levels of CXCL13, the ligand for CXCR5 (BLR1), were consistently up-regulated in all patient samples compared with controls (Figure 3A,B). Increased CXCL13 expression was detected in all individual patient samples (Figure 3A) but also when grouping patients or controls together (Figure 3B).
Microarray analysis of mRNA of chemokines and chemokine receptors. (A) mRNA clustergrams of genes and relative expression of mRNAs are shown for individual samples from purified B cells from controls (n = 4) and patients (n = 4). Color intensity changes from light green (low or absent expression) to black (average expression) and from black to intense red (maximum expression level). (B) mRNA expression clustergrams are shown after grouping the data from patients and controls. CXCL13 expression was up-regulated in all patient samples compared with controls. The mRNA levels for the CXCR4, CXCR5, CCR7, and the ligands CXCL12 and CCL21 showed no major differences. (C) CXCL13 is secreted from purified B cells after in vitro activation for 6 days. The levels found in patients (n = 15) were higher compared with controls (n = 15; P < .005). (D) Purified B cells from HIV-1 patients (n = 5) were cultured after in vitro activation (as above) with or without a neutralizing anti-CXCL13 Ab and CXCR5 expression (MFI) was measured. B cells cultured with the neutralizing CXCL13 Ab showed a higher CXCR5 expression compared with cells cultured with the isotype control Ab (P = .02).
Microarray analysis of mRNA of chemokines and chemokine receptors. (A) mRNA clustergrams of genes and relative expression of mRNAs are shown for individual samples from purified B cells from controls (n = 4) and patients (n = 4). Color intensity changes from light green (low or absent expression) to black (average expression) and from black to intense red (maximum expression level). (B) mRNA expression clustergrams are shown after grouping the data from patients and controls. CXCL13 expression was up-regulated in all patient samples compared with controls. The mRNA levels for the CXCR4, CXCR5, CCR7, and the ligands CXCL12 and CCL21 showed no major differences. (C) CXCL13 is secreted from purified B cells after in vitro activation for 6 days. The levels found in patients (n = 15) were higher compared with controls (n = 15; P < .005). (D) Purified B cells from HIV-1 patients (n = 5) were cultured after in vitro activation (as above) with or without a neutralizing anti-CXCL13 Ab and CXCR5 expression (MFI) was measured. B cells cultured with the neutralizing CXCL13 Ab showed a higher CXCR5 expression compared with cells cultured with the isotype control Ab (P = .02).
The expression of CXCL13 mRNA in B cells was a new finding, and we asked whether the CXCL13 protein also could be secreted from B cells. Therefore, purified B cells from patients (n = 15) and controls (n = 15) were cultured in the presence or absence of different stimuli. When B cells were kept in culture without stimulation over a period of 36 hours, CXCL13 could not be detected in culture supernatant (data not shown). However, when B cells were activated by the presence of anti-CD40 monoclonal Ab, IL-2, IL-10, and CpG, the protein CXCL13 was detected in all culture supernatants. B cells purified from HIV-1 patients secreted higher amounts of CXCL13 compared with controls (226 pg/mL [range, 77-1095] vs 109 pg/mL [range, 27-295] P = .005; Figure 3C). There was a trend toward higher CXCL13 secretion from B cells purified from patients with a low CD4+ T-cell count (P = .12; data not shown). Cultures were also activated by B-cell receptor ligation, but this procedure did not induce secretion of CXCL13 (data not shown).
To test whether autocrine CXCL13 could affect the CXCR5 expression on B cells, purified B cells were activated in the presence of a neutralizing anti-CXCL13 Ab or isotype control Ab (Figure 3D). As shown, when neutralizing CXCL13 in cultures, the cell surface expression of CXCR5 increased (MFI 581 [range, 453-590] vs 509 [range, 437-547]; P = .02).
CXCL13 induces internalization of CXCR5 with a rapid and transient increase in mRNA expression
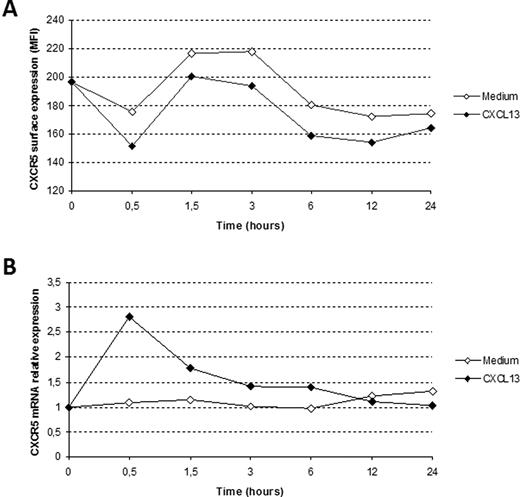
B cells from HIV-1–infected patients did not show any reduced mRNA expression for the CXCR5 molecule, despite the low cell surface expression of this receptor. To determine the kinetics of CXCR5 protein expression in relation to mRNA, we quantified CXCR5 protein and mRNA after incubation of PBMCs with the ligand CXCL13. After 30 minutes, a clear decrease in CXCR5 surface expression occurred on cells incubated with CXCL13 (Figure 4A), and this was accompanied by a rapid increase in CXCR5 mRNA expression (Figure 4B). At 12 and 24 hours, only minor differences in mRNA expression and surface expression could be detected between the different conditions.
CXCR5 receptor internalization resulting from CXCL13 ligation is followed by a rapid increase of CXCR5 mRNA levels. Surface expression (A) and mRNA levels (B) of CXCR5 on PBMCs incubated with or without CXCL13 over a 24-hour time-course experiment. The surface expression of CXCR5 decreased, despite a rapid increase of CXCR5 mRNA expression already after 30 minutes, in cells cultured with the chemokine CXCL13. At 12 and 24 hours, only minor differences could be detected between the cultures. One representative experiment of 3 is shown.
CXCR5 receptor internalization resulting from CXCL13 ligation is followed by a rapid increase of CXCR5 mRNA levels. Surface expression (A) and mRNA levels (B) of CXCR5 on PBMCs incubated with or without CXCL13 over a 24-hour time-course experiment. The surface expression of CXCR5 decreased, despite a rapid increase of CXCR5 mRNA expression already after 30 minutes, in cells cultured with the chemokine CXCL13. At 12 and 24 hours, only minor differences could be detected between the cultures. One representative experiment of 3 is shown.
CXCL13 expression in lymph nodes from HIV-1–infected patients and controls
We characterized the CXCL13 expression in lymphoid tissue from HIV-1 patients. The distribution of CXCL13 protein expression in HIV-1 follicular hyperplasia was associated with the GC light zone (Figure 5A) where it displayed an interwoven pattern resembling the DC network. The CXCL13 expression in GCs was similar in HIV-1 and noninfected samples with follicular hyperplasia. Interestingly, double-positive cells expressing both CXCL13 and the B-cell marker CD79a were detected in HIV-1 samples but not in uninfected samples (Figure 5B). These CXCL13+CD79a+ cells showed no preferential localization in the microarchitecture of the lymph node.
CXCL13 expression in HIV-1+ lymph nodes. Immunohistochemical detection of CXCL13 expression (brown) on paraffin sections and characterization of the positive cells (blue). Slides were viewed with a Zeiss AxioImager M1 microscope for brightfield and fluorescence application (Carl Zeiss, Jena, Germany) using Plan-NEOFLUAR lens at 20×/0.5 and 63×/1.3 oil objective. Images were acquired using AxioCam MRc5 camera (Zeiss) and Axio-Vision (release 4.6) software. Digitalized images were processed with the Module Imaging Plus (Zeiss) and Adobe Photoshop version CS3 Extended software (Adobe Systems, San Jose, CA). Original magnifications, panels A and D, ×80; panels B and C, ×160. (A) In the GC light zone, part of the follicular DC network stained positive for CXCL13. (B) In HIV-1+ tissue, double-positive cells (CXCL13+CD79a+), B cells could be detected. (C) The majority of CXCL13+ cells were found in the T-dependent zone, and double-staining showed that cells were CD68+ macrophages, (D) but that the majority of double-positive cells were CD1a+ immature DCs.
CXCL13 expression in HIV-1+ lymph nodes. Immunohistochemical detection of CXCL13 expression (brown) on paraffin sections and characterization of the positive cells (blue). Slides were viewed with a Zeiss AxioImager M1 microscope for brightfield and fluorescence application (Carl Zeiss, Jena, Germany) using Plan-NEOFLUAR lens at 20×/0.5 and 63×/1.3 oil objective. Images were acquired using AxioCam MRc5 camera (Zeiss) and Axio-Vision (release 4.6) software. Digitalized images were processed with the Module Imaging Plus (Zeiss) and Adobe Photoshop version CS3 Extended software (Adobe Systems, San Jose, CA). Original magnifications, panels A and D, ×80; panels B and C, ×160. (A) In the GC light zone, part of the follicular DC network stained positive for CXCL13. (B) In HIV-1+ tissue, double-positive cells (CXCL13+CD79a+), B cells could be detected. (C) The majority of CXCL13+ cells were found in the T-dependent zone, and double-staining showed that cells were CD68+ macrophages, (D) but that the majority of double-positive cells were CD1a+ immature DCs.
There was also a high amount of CXCL13+ cells in the T-dependent zone, and double-labeling showed an interesting pattern in both HIV-1 and control samples. Some cells were CD4+ T cells and macrophages (CD68+) (Figure 5C), but the majority of cells expressed CD1a, an antigen expressed by immature DCs (Figure 5D). The CD1a+CXCL13+ cells could also be found in the marginal sinuses. The number of double-positive CD1a+CXCL13+ cells was higher than CD83+CXCL13+ mature DCs in our samples. CXCR5 expression in tissue was found mainly in the mantle zone, and no major changes were found between HIV-1 and control samples (data not shown).
Altered B-cell migration in patients with low CD4+ T-cell count
The capacity of B cells isolated from HIV-1–infected patients to functionally respond to chemokines may be altered because of the changes in chemokines and chemokine receptors expression reported in Figure 1. To assess for the functional significance of the changes occurring at the level of chemokine receptor expression on B cells during HIV-1 infection, specific migration toward the ligands CXCL12, CXCL13, and CCL21 was studied in a transmigration assay with B cells isolated from the blood of HIV-1–infected patients and controls.
The specific migration toward CXCL13 showed a similar pattern in controls and HIV-1 patients with a high CD4+ T-cell count (> 350 cells/μL; Figure 6A). The migration index was 1.6 plus or minus 0.4 SD in both groups. However, in patients with low CD4+ T-cell count (< 350 cells /μL), B-cell migration index toward CXCL13 was higher (2.4 ± 1.0 SD; Figure 6A). Migration toward CXCL12 in patients with high CD4+ T cells and controls was in a similar range (3.7 ± 2.0 SD vs 3.5 ± 1.7 SD), whereas B cells isolated from patients with a low CD4+ T-cell count showed a great increase in CXCL12 migration with an index of 9.5 plus or minus 6.0 SD (Figure 6B). The degree of CCL21-induced migration of B cells was in a similar range in controls and patients with high CD4+ T-cell count (index 3.0 ± 1.4 SD vs 2.5 ± 1.1 SD; Figure 6C), whereas in patients with low CD4+ T-cell count, B cells showed an increase in migration to this cytokine (index 5.9 ± 3.4 SD). There was no increase in spontaneous migration in HIV-1 samples compared with controls (data not shown).
Transmigration assay of purified B cells. Migration of primary B cells from HIV-1–infected patients and controls toward CXCL13 (A), CXCL12 (B), and CCL21 (C) in a transmigration assay. Migration index is presented as migrated cells toward the specific chemokine divided by spontaneous migration. Migration of purified B cells toward CXCL13, CXCL12, and CCL21 is increased in HIV-1–infected patients with low CD4+ T-cell counts compared with healthy donors and patients with high CD4+ T cells.
Transmigration assay of purified B cells. Migration of primary B cells from HIV-1–infected patients and controls toward CXCL13 (A), CXCL12 (B), and CCL21 (C) in a transmigration assay. Migration index is presented as migrated cells toward the specific chemokine divided by spontaneous migration. Migration of purified B cells toward CXCL13, CXCL12, and CCL21 is increased in HIV-1–infected patients with low CD4+ T-cell counts compared with healthy donors and patients with high CD4+ T cells.
Discussion
HIV-1 has established a unique interaction with the chemokine receptors CXCR4 and CCR531,32 as they function as coreceptors for virus entry. Although B cells are not the major target of HIV-1 infection, alterations in the balance between chemokines and their respective receptors may impact B-cell biology during HIV-1 infection. The present study focused on characterizing the expression and function of chemokines and chemokine receptors pivotal for B-cell migration from blood to lymphoid tissue.
We report that the expression of CXCR5 is altered on B cells from HIV-1–infected subjects compared with controls; the naive and the memory B cells, and the pre-plasma cells, from the patients had a decreased expression of the CXCR5 receptor. B cells from patients with low CD4+ T-cell count had a significantly lower CXCR5 expression, indicating that disease progression may lead to dysregulation of chemokine receptor expression. In this group of patients, the CCR7 expression was slightly increased compared with controls, which may alter B-cell migration toward CCL21. Reduced expression of CXCR5 has been previously described on naive B cells during HIV-1 infection,33 and it has also been reported that CXCR5− B cells appear in the blood of HIV-1+ patients.34 The altered expression of this receptor is interesting because CXCR5 mediate migration of B cells into spleen and lymph nodes,35 and further GC organization is also dependent on CXCR5 and CXCR4.20,22 Moreover, it is known that the GC architecture is severely disturbed during HIV-1 or SIV infection, and this is only in part corrected by HAART.36,37
Modulation of the cell surface expression of CXCR4 and CXCR5 occurs after interaction with the respective ligands, leading to rapid internalization and endocytosis of the receptors.38 A possible mechanism for the reduced cell surface expression of CXCR5 on B cells could be the altered chemokine levels found in serum of HIV-1–infected patients, which may lead to increased receptor internalization. In support of this hypothesis is that serum levels of CXCL12 and CXCL13 are known to be elevated during HIV-1 infection and could thereby affect chemokine receptor surface expression. In these previous studies, the level of chemokines correlated to disease progression measured by CD4+ T-cell count and immune activation markers, such as soluble CD27 and inflammatory chemokine CXCL10/inducible protein-10.39,40 Similarly, in our cohort, the reduced CXCR5 expression on B cells was associated with low CD4+ T-cell counts.
To study whether infection with HIV-1 alters the expression of a larger set of chemokine and chemokine receptor genes, we performed microarray analysis on B cells from HIV-1–infected and noninfected subjects. Although the cell surface expression of the CXCR5 receptor was significantly decreased, we could not detect any decrease of the CXCR5 mRNA in patients compared with controls. Additional experiments comparing cell surface expression and gene expression of CXCR5 after incubation of cells with CXCL13 demonstrated a transient increase in CXCR5 mRNA expression, which suggests that CXCR5 expression is also in part regulated at the posttranscriptional level through endocytosis.
The most striking finding in the gene profile expression of B cells was the high CXCL13 mRNA level in HIV-1 patient samples. This is a novel finding in that CXCL13 expression has not previously been reported in primary B cells; thus, we investigated lymphoid tissue from HIV-1 patients and controls. Interestingly, CXCL13+ B cells could also be detected in tissue from HIV-1 patients but were not found in control samples.
Previous reports have indicated follicular dendritic cells (FDCs) and follicular stromal cells as a source of CXCL1341-43 ; and in the present study, CXCL13 protein was readily detected in a pattern resembling the FDC network, in the GCs of both HIV-1 samples and controls. It has also been shown that CXCL13 expression only partially colocalize with FDC markers in lymphoid tissues.44 In that perspective, it is interesting that the main expression of CXCL13 in our samples was found in immature CD1a+ DCs, both in HIV-1 and control samples. The expression was less prominent in mature DCs, which could suggest that the CXCL13 protein might be lost during DC maturation. In previous studies, in vitro–derived DCs have been shown to up-regulate the expression of CXCL13 mRNA after stimulation with endotoxin (lipopolysaccharide) from Escherichia coli,45 and infection in vitro with Bartonella henselae also induced secretion of CXCL13 from DCs.46 However, this is the first report of immature DCs expressing CXCL13 in human tissue. It is possible that also CXCL13 produced from immature DCs may participate in the down-regulation of the CXCR5 receptor on B cells.
CXCL13 expression has been previously reported in GC-derived human CD4+ T-helper cells where CXCL13 expression was regulated by TCR activation.47 We could not detect any spontaneous secretion of CXCL13 from purified B cells but on in vitro activation of B cells with CD40 ligation, CpG, IL-2, and IL-10 the CXCL13 protein was readily detected in cultures. Surprisingly, B-cell receptor ligation did not induce secretion of CXCL13. Therefore, it is possible that the high degree of unspecific immune activation occurring during HIV-1 infection,48 may be a contributing factor leading to up-regulation of mRNA expression and secretion of CXCL13 in B cells. As a consequence, the cell surface expression of CXCR5 on B cells may be decreased by autocrine or paracrine secretion of CXCL13 as shown by our in vitro data. In diseases characterized by a high degree of immune activation and inflammation, such as rheumatoid arthritis and neuroborreliosis, high levels of CXCL13 have been reported in association with lymphoid neogenesis and immunoglobulin production.49,50 CXCL13 has also been implicated in production of natural antibodies and CD5+ B-cell homing.43
The responsiveness of chemokine receptors for their respective ligands is tightly regulated during migration of different lymphoid subpopulations into different tissue compartments.29,51 A recent study by Moir et al described a tissue-like memory B-cell population (CD20hi/CD27−/CD21lo) in peripheral blood with altered chemokine receptor expression compared with classical memory B cells (CD27+) and suggested that these tissue-like B cells may have an altered homing capacity.52 To analyze whether the alterations in chemokine receptor expression found in our patient cohort could impact on migration of peripheral B lymphocytes, a transmigration assay of B cells toward the corresponding chemokines was performed. In our experiments, a difference in migration was found in cells from patients with low CD4+ T-cell count, where specific migration to all ligands was increased, compared with cells from controls and patients with high CD4+ T-cell count. It has been shown that B-cell activation via CD40 ligation increases CXCL12-mediated migration without changing the chemokine receptor expression14,51,53 ; furthermore, that B-cell activation with lipopolysaccharide also increases chemokine-induced migration.51 Type 1 interferons (IFNs) have also been shown to increase CXCL12- and CCL21-mediated migration,24 which may be relevant for HIV-1 pathogenesis as the levels of IFNs are elevated with disease progression.54 Our finding of an increase in migration toward CXCL12, CXCL13, and CCL21 in HIV-1 patients with CD4+ T-cell counts less than 350 cells/μL may thus reflect polyclonal stimulation of B cells, either by CD40-CD40L interactions, by IFNs or via TLR signaling during HIV-1. Our findings suggest that there may be an altered migration profile of B cells during HIV-1.
Further understanding of the role that impaired expression of chemokines play during HIV-1 infection may lead to intervention to correct impaired humoral immunity during HIV-1 infection. In this frame, the novel approach by Castelletti et al55 demonstrated that VSV-immunization in mice in presence of the chemokine MEC/CCL28 led to an increased presence of IgA-secreting cells on the gastrointestinal mucosa.
In conclusion, we show that HIV-1 infection induces changes in the expression of both CXCR5 and CXCL13, a phenomenon that may contribute to impaired humoral immunity during HIV-1 infection and hypergammaglobulinemia.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
The authors thank Anna Pasetto for helpful assistance with the microarray analysis; Mauro D'Amato, Gösta Winberg, and Daniel Palm for helping with the RT-PCR setup; Petra Meyer for the excellent histotechnical assistance; and Wendy Murillo for practical help.
This work was supported by grants received from the Swedish Research Council (MRC), the Swedish International Development Agency, the Regione Autonoma della Sardegna, Cagliari, Italy and the Istituto Italiano di Cultura C.M. Lerici Foundation, Stockholm, Sweden, and the German Ministry of Education and Research (Bundesministerium für Bildung und Forschung), Compentence Network for HIV/AIDS (FKZ 01KI0501). Financial support was also provided through Stockholm County Council and the Karolinska Institutet. F.C. is a member of the European Union (EU) Fp6 Network of Excellence Europrise. L.V.P.D. is supported through a fellowship from the EU Marie Curie Program (contract MEST/CT).
Authorship
Contribution: A.C. and F.M. designed and performed research, analyzed data, and wrote the paper; L.V.P.D. performed research; A.A. and S.G. contributed with analytical tools; K.T.-R. and P.R. performed research and contributed with analytical tools; and F.C. and A.N. designed and performed research, contributed with analytical tools, analyzed data, and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Anna Nilsson, Department of Microbiology, Tumor and Cell Biology, Karolinska Institutet, 171 77 Stockholm, Sweden; e-mail: anna.nilsson.1@ki.se.
References
Author notes
*A.C. and F.W. contributed equally to this work.