Abstract
Recent studies suggest the potential involvement of common antigenic stimuli on the ontogeny of monoclonal T-cell receptor (TCR)–αβ+/CD4+/NKa+/CD8−/+dim T-large granular lymphocyte (LGL) lymphocytosis. Because healthy persons show (oligo)clonal expansions of human cytomegalovirus (hCMV)–specific TCRVβ+/CD4+/cytotoxic/memory T cells, we investigate the potential involvement of hCMV in the origin and/or expansion of monoclonal CD4+ T-LGL. Peripheral blood samples from patients with monoclonal TCR-αβ+/CD4+ T-LGL lymphocytosis and other T-chronic lymphoproliferative disorders were evaluated for the specific functional response against hCMV and hEBV whole lysates as well as the “MQLIPDDYSNTHSTRYVTVK” hCMV peptide, which is specifically loaded in HLA-DRB1*0701 molecules. A detailed characterization of those genes that underwent changes in T-LGL cells responding to hCMV was performed by microarray gene expression profile analysis. Patients with TCR-αβ+/CD4+ T-LGL displayed a strong and characteristic hCMV-specific functional response, reproduced by the hCMV peptide in a subset of HLA-DRB1*0701+ patients bearing TCRVβ13.1+ clonal T cells. Gene expression profile showed that the hCMV-induced response affects genes involved in inflammatory and immune responses, cell cycle progression, resistance to apoptosis, and genetic instability. This is the first study providing evidence for the involvement of hCMV in the ontogeny of CD4+ T-LGL, emerging as a model disorder to determine the potential implications of quite a focused CD4+/cytotoxic immune response.
Introduction
Monoclonal T-cell receptor (TCR)-αβ+/CD4+/NKa+/CD8−/+dim T-large granular lymphocyte (LGL) lymphocytosis is a subgroup of T-LGL lymphoproliferative disorders characterized by (mono)clonal expansion of cytotoxic/Th1 CD4+ T cells.1 Although CD4+ T-LGL patients typically display an indolent clinical course, its outcome remains unknown and is frequently determined by its association with other neoplasias.1 In a recent study,2 we showed that monoclonal CD4+ T-LGL cells display a restricted TCR-Vβ repertoire with a predominance of TCR-Vβ13.1 cases, in clear association with an HLA-DRB1*0701 haplotype. In addition, these cases show a high degree of homology in the CDR3 region of the TCR-β chain, supporting the existence of a common chronic antigen-driven origin for this monoclonal CD4+ T-LGL lymphoproliferative disorder.2 However, in contrast to other T-LGL neoplasias (eg, TCR-γδ+ and CD8+ TCR-αβ+ T-LGL),3,4 CD4+ T-LGL cases did not show a clear association with autoimmune disorders, pointing out the potential involvement of non–self-antigens in this lymphoproliferative disorder.1,2
At present, it is well established that some viruses (eg, human T-cell lymphotropic virus type 1 (HTLV-1), Epstein-Barr virus (EBV), human herpesvirus 8 (HHV8), cytomegalovirus [CMV]) are capable of establishing long-term, persistent infections with chronic immune activation and inflammation.5 Such a latent virus could act as a source of peptides, which are presented to CD4+ T-LGL cells, potentially leading to its clonal expansion. Indeed, infection by HTLV-1, EBV, and HHV8 is often followed by lymphoproliferative disorders, which can even evolve into an overt neoplasia (leukemia/lymphoma) of mature lymphocytes.6-8 However, in such cases, clonal T-cell expansions are frequently the result of infection of the neoplastic cells by the virus, and they have not been directly related to the immune response against the virus.6,9,10
In contrast to other viruses, human CMV (hCMV) has not been proven to be involved in human cancer.11,12 However, recent studies from our and other groups in hCMV-seropositive healthy persons show the presence of TCR-Vβ (oligo)clonal expansions of hCMV-specific CD4+/cytotoxic/memory T cells in the peripheral blood (PB) of immunocompetent healthy adults.13-15 Interestingly, these hCMV-specific CD4+ T cells display a heterogeneous Th1 memory/effector antigen-experienced phenotype with a clear association between specific HLA haplotypes and unique TCRVβ expansions. Among others, these include oligoclonal expansions of TCRVβ13.1+ CD4+ T cells in HLA-DRB1*0701+ healthy adults.15 Altogether, these results point out the potential involvement of hCMV in the origin and/or expansion of monoclonal lymphocytes in CD4+ T-LGL patients.
In this study, we show, for the first time, that in most patients with monoclonal TCR-αβ+/CD4+ T-LGL lymphocytosis, the expanded T-cell clone displays a highly characteristic cellular and soluble response to hCMV, in contrast to other T-LGL and T-chronic lymphoproliferative disorders (T-CLPDs). Interestingly, in TCRVβ13.1+/CD4+ T-LGLs, the anti-hCMV response can be reproduced by a single peptide, “MQLIPDDYSNTHSTRYVTVK,” from the hCMV gB protein. Likewise, we provide a detailed characterization of the “in vitro” response to hCMV by the expanded monoclonal CD4+ T cells, which specifically affects genes involved in both immune and inflammatory responses, cell proliferation/cell-cycle progression, apoptosis, protein synthesis, G-protein–mediated intracellular signaling, and the DNA repair/maintenance machinery.
Methods
Patients and samples
Heparin-anticoagulated PB samples from patients with monoclonal TCR-αβ+/CD4+ T-LGL lymphocytosis (n = 12), TCR-αβ+/CD8+ T-LGL leukemia (n = 3), TCR-γδ+ T-LGL leukemia (n = 5), and CD4+ T-cell chronic lymphoproliferative disorders other than LGL (n = 10) were collected in the Cytometry Service of the University Hospital of Salamanca following the recommendations and institutional review board approval of the University Hospital Ethics Committee and the tenets of the Declaration of Helsinki, and studied at diagnosis. In all cases, serologic studies were carried out in cell-free plasma samples from each patient to measure the presence of anti-CMV and anti-EBV specific IgG and IgM antibodies (LIAISON IgG and IgM, DiaSorin, Saluggia, Italy). In addition, a quantitative CMV-DNA polymerase chain reaction assay was used to monitor CMV viral load (COBAS AMPLICOR CMV, Roche Molecular Systems, Branchburg, NJ) in half of all TCR-αβ+/CD4+ T-LGL patients, the dynamic range of the test being of CMV 4 × 102 to 105 copies/mL. For the serologic studies, a group of 17 hCMV-seropositive normal healthy persons was studied in parallel.
Stimulation of PB samples with hCMV and hEBV
Two aliquots of RPMI 1640-diluted PB samples (1:1 vol:vol) were cultured for 6, 24, and 48 hours at 37°C in a 5% CO2-humidified atmosphere in the presence of 30 μM of TAPI-2 (a tumor necrosis factor alpha protease inhibitor; Cytognos SL, Salamanca, Spain), the anti-hCD28 and anti-hCD49d costimulatory monoclonal antibodies (mAbs), 1 μg/mL (BD Biosciences PharMingen, San Diego, CA), and either hCMV or hEBV whole viral lysates (5 μg/mL; Advanced Biotechnologies, Columbia, MD). Another aliquot of each PB sample was processed in parallel under the same conditions but in the absence of viral lysate, as an unstimulated sample (negative control).
Identification and functional characterization of hCMV-specific CD4+ T cells using flow cytometry
After stimulation, PB samples (100 μL) were stained with the following combination: fluorescein isothiocyanate (FITC)/phycoerythrin (PE)/peridinin chlorophyll protein-cyanin 5.5 (PerCPCy5.5)/allophycocyanin (APC), of mAb: CD3-FITC (clone 2A3; ImmunoSTEP SL, Salamanca, Spain), anti-tumor necrosis factor-α (TNF-α)-PE (clone mAb11; BD Biosciences, San Jose, CA), CD4 PerCPCy5.5 (clone SK3; BD Biosciences), anti-TCR-γδ-APC (clone B1; BD Biosciences PharMingen), CD8-APC (clone B9.11; Immunotech, Marseille, France), CD56-APC (NCAM 16.2; BD Biosciences), or CD7-APC (clone eBio124-1D1; eBioscience, San Diego, CA), chosen on the basis of the phenotypic features of the clonal T cells identified in each patient. Expression of the CD69-FITC (clone L78; BD Biosciences) and CD25-PE (clone 2A3; BD Biosciences) activation markers was also assayed in combination with CD4 PerCPCy5.5 and the APC-conjugated reagents listed. Information approximately between 0.5 and 1.5 × 105 cellular events/sample aliquot was collected using a FACSCalibur flow cytometer and the CellQUEST software program (BD Biosciences). For data analysis, the Infinicyt software program (Cytognos SL) was used.
Quantification of secreted cytokines using flow cytometry
Soluble levels of multiple secreted cytokines (interferon-γ ([IFN-γ]), TNF-α, LT-α, interleukin-2 ([IL-2]), IL-3, IL-4, IL-5, IL-6, IL-9, IL-10, IL-12p70, IL-13, granulocyte-macrophage colony-stimulating factor) were quantified in the supernatants of PB samples cultured for 6 hours with whole viral lysates, using the Cytometric Bead Array system (BD Biosciences) and a FACSCanto II flow cytometer (BD Biosciences), following the recommendations of the manufacturer.
Immune response to the “MQLIPDDYSNTHSTRYVTVK” hCMV peptide complementary of the HLA-DRB1*0701 allele
Based on Swiss-Prot database search, a single hCMV peptide (“MQLIPDDYSNTHSTRYVTVK”), derived from hCMV glycoprotein B, was identified that could be loaded in HLADRB1*0701. PB samples from patients with TCR-Vβ13.1+/CD4+ T-LGL lymphocytosis and an HLA-DRB1*0701 genotype (n = 4) as well as CD4+ T-LGL cases expressing TCRVβ families other than TCR-Vβ13.1 and cases diagnosed of other T-cell neoplasias (n = 4) were cultured for 6 hours with 0.5 μg/mL of the hCMV peptide (Inbios Srl, Napoli, Italy) in the presence of TAPI-2 (30 μM) and the anti-hCD28 and anti-hCD49d costimulatory mAb (1 μg/mL). Assessment of TNF-α+-activated cells as well as quantification of multiple secreted cytokines (IFN-γ, TNF-α, IL-2, IL-4, IL-6, and IL-10) were performed after stimulation with the peptide on a FACSCalibur flow cytometer, according to procedures that have been previously described in detail.16
Genome-wide expression profiling
Total RNA was isolated from magnetic-activated cell sorter-freshly purified hCMV-stimulated CD69+, hCMV-stimulated CD69−, and unstimulated monoclonal CD4+ T-LGL lymphocytes from PB samples from 4 TCR-αβ+/CD4+ T-LGL lymphocytosis patients (purity of ≥ 98%). Briefly, 100 ng of total RNA from each of the 12 purified cell fractions was amplified and labeled using the GeneChip 2 cycle cDNA synthesis kit and the GeneChip IVT labeling kit (Affymetrix, Santa Clara, CA), respectively. Then it was hybridized to the Human Genome U133 Plus 2.0 Array (Affymetrix).
In parallel, total RNA was also isolated from highly purified (≥ 98% purity) hCMV-stimulated (specific) CD69+ CD4+ T-lymphocytes isolated from PB samples from hCMV-seropositive healthy donors (n = 5, mean age, 36 ± 3 years) using a FACSAria flow cytometer (BD Biosciences). To get pure and highly concentrated RNA, the silica membrane technology NucleoSpin RNA XS (Macherey-Nagel, Düren, Germany) was used. Total RNA was then amplified, labeled, and hybridized to the Human Genome U133 Plus 2.0 Array (Affymetrix) as described in the previous paragraph.
Statistical methods
Mean values, standard deviation, median, and range were calculated for each variable under study using the SPSS program (version 12.0; SPSS, Chicago, IL). To establish the statistical significance of differences observed between groups, either the Pearson χ2 test or the Fisher exact test was used for categorical variables; the nonparametric Friedman and Wilcoxon tests and the Mann-Whitney U test were used for continuous variables corresponding to paired and unpaired samples, respectively (version 12.0; SPSS). P values less than .05 were considered to be associated with statistical significance.
For DNA oligonucleotide microarray-based data analysis, RNA expression values for each probe set were calculated using the Robust Multi-array Average algorithm17 ; further statistical analyses were performed using the R18 and Bioconductor19 software tools. First, a prospective unsupervised exploratory analysis was performed to ascertain whether samples were associated with an identifiable structure of the data at the gene expression level. For this purpose, both multidimensional scaling20 and hierarchical clustering were performed on those 1000 genes (of the original 54 675 probes) showing the highest level of variation among paired samples (P < .001). In a second step, a Significant Analysis of Microarrays algorithm was used to identify those genes with a statistically significantly different expression between paired cell fractions. Genes showing significantly different patterns of expression were selected based on a multiclass analysis, as those showing the lowest (< 1% in all comparisons) false discovery ratio with a q-value control.21 Finally, based on the expression of those genes showing the highest variation between the different fractions of purified monoclonal CD4+ T-LGL cells, a hierarchical clustering analysis was performed using complete linkage and euclidean distances. All microarray data have been deposited in Gene Expression Omnibus under accession number GSE12488.22
Results
Cellular and soluble responses to hCMV in patients with monoclonal TCR-αβ+/CD4+ T-LGL lymphocytosis
All CD4+ T-LGL patients analyzed were seropositive for hCMV with slightly higher levels of anti-hCMV-specific IgG antibodies (12 ± 7 IU/mL) compared with both hCMV-seropositive patients with other T-cell chronic lymphoproliferative disorders (7 ± 6 IU/mL; P = .1) and healthy donors (7 ± 4 IU/mL; P = .05). None of the CD4+ T-LGL patients analyzed showed detectable hCMV viral load in plasma (< 400 copies/mL). In turn, CD4+ T-LGL patients who were seropositive for hEBV (10 of 12 cases; 83%) also showed similar levels of anti-EBV Epstein-Barr virus-associated nuclear antigen and anti-EBV viral capsid antigen (VCA) specific IgG antibodies with respect to hEBV-seropositive patients with other T-cell chronic lymphoproliferative disorders (anti–Epstein Barr virus–associated nuclear antigen IgG of 249 ± 240 vs 247 ± 228 U/mL, respectively; P > .05; anti-EBV VCA IgG of 574 ± 307 vs 497 ± 295 U/mL, respectively; P > .05).
Most patients with monoclonal TCR-αβ+/CD4+ T-LGL lymphocytosis (n = 11 of 12; 92%) showed activated, hCMV-specific TNF-α+/CD4+ clonal T cells after 6 hours of culture in the presence of a whole hCMV viral lysate, whereas this response was found at a much lower frequency among the other T-CLPDs analyzed (Figure 1A). In addition, the specificity of the immunologic response found “in vitro” among patients with monoclonal TCR-αβ+/CD4+ T-LGL lymphocytosis was also confirmed by the demonstration of an increased secretion of both TNF-α (498 ± 731 pg/mL) and IFN-γ (761 ± 648 pg/mL) in the presence of hCMV but not of hEBV (IFN-γ, 43 ± 82 pg/mL; TNF-α, 50 ± 89 pg/mL; Figure 1B). In contrast, other T-CLPD patients analyzed showed an almost negligible response to both hCMV (IFN-γ, 55 ± 108 pg/mL; TNF-α, 124 ± 391 pg/mL) and hEBV (IFN-γ, 7 ± 19 pg/mL; TNF-α, 47 ± 116 pg/mL) whole viral lysates (Figure 1B).
Functional response to hCMV in patients with monoclonal CD4+ T-LGL vs other T-cell chronic lymphoproliferative disorders (T-CLPDs). (A) Most patients with monoclonal TCR-βα+/CD4+ T-LGL lymphocytosis (n = 11 of 12; 92%) showed activated hCMV-specific TNF-α+ clonal T cells in response to the viral lysate, whereas this response was found at much lower frequencies (P < .001) among other T-CLPDs analyzed (n = 2 of 18; 11%). The hCMV-specific response was also reflected by increased secretion of both IFN-γ (P < .001) and TNF-α (P < .01) into the culture supernatant in response to hCMV, but not hEBV (B), as well as by an increased expression of both the CD69 and CD25 activation-associated markers (P < .05) (C). In turn, extremely high levels of soluble IFN-γ were found after stimulation with the MQLIPDDYSNTHSTRYVTVK hCMV peptide, complementary of the HLA-DRB1*0701 allele, in patients with monoclonal TCRVβ13.1+/CD4+ T-LGL lymphocytosis compared with other T-CLPD cases carrying this HLA haplotype (P < .05) (D). Boxes in panels B to D extend from the 25th to the 75th percentiles; the line in the middle and vertical lines correspond to the median value and both the 10th and 90th percentiles, respectively.
Functional response to hCMV in patients with monoclonal CD4+ T-LGL vs other T-cell chronic lymphoproliferative disorders (T-CLPDs). (A) Most patients with monoclonal TCR-βα+/CD4+ T-LGL lymphocytosis (n = 11 of 12; 92%) showed activated hCMV-specific TNF-α+ clonal T cells in response to the viral lysate, whereas this response was found at much lower frequencies (P < .001) among other T-CLPDs analyzed (n = 2 of 18; 11%). The hCMV-specific response was also reflected by increased secretion of both IFN-γ (P < .001) and TNF-α (P < .01) into the culture supernatant in response to hCMV, but not hEBV (B), as well as by an increased expression of both the CD69 and CD25 activation-associated markers (P < .05) (C). In turn, extremely high levels of soluble IFN-γ were found after stimulation with the MQLIPDDYSNTHSTRYVTVK hCMV peptide, complementary of the HLA-DRB1*0701 allele, in patients with monoclonal TCRVβ13.1+/CD4+ T-LGL lymphocytosis compared with other T-CLPD cases carrying this HLA haplotype (P < .05) (D). Boxes in panels B to D extend from the 25th to the 75th percentiles; the line in the middle and vertical lines correspond to the median value and both the 10th and 90th percentiles, respectively.
This functional hCMV-specific T-cell response was also associated with up-regulation of the expression of both the CD69 and CD25 activation-associated markers in monoclonal TCR-αβ+/CD4+ T-LGL patients after longer stimulatory periods (24-48 hours) with hCMV but not hEBV whole viral lysate (Figure 1C).
On looking at the CD8 response to hCMV, a slightly lower response was observed among HLA-A2 patients vs all other CD4+ T-LGL cases (0.2% ± 0.2% TNF-α+/CD8+ T cells vs 0.7% ± 0.7% TNF-α+/CD8+ T cells; P = .02). In contrast, HLA-A2 healthy donors showed a similar CD8 T-cell response to hCMV than patients displaying other HLA-I haplotypes (0.2% ± 0.2% TNF-α+/CD8+ T cells vs 0.1% ± 0.2% TNF-α+/CD8+ T cells, respectively; P > .05).
Immunologic response to the “MQLIPDDYSNTHSTRYVTVK” hCMV peptide in monoclonal TCRVβ13.1+/CD4+ T-LGL patients
Stimulation of monoclonal T cells from patients with monoclonal TCR-Vβ13.1+/CD4+ T-LGL lymphocytosis and an HLA-DRB1*0701 genotype with the “MQLIPDDYSNTHSTRYVTVK” hCMV peptide, whose recognition is restricted by the HLA-DR7 allele (Swiss-Prot database), showed a clear response by the expanded monoclonal TCR-Vβ13.1+/CD4+ T-LGL, as reflected by an increased percentage of hCMV-specific TNF-α+ monoclonal T cells and a surprisingly high secretion of IFN-γ (13 875 ± 1377 pg/mL) into the extracellular medium. Of note, IFN-γ secretion was even higher than that obtained for the same patients in response to the hCMV whole viral lysate (Figure 1D). In contrast, none of 5 different hCMV-seropositive patients, diagnosed with monoclonal TCR-Vβ5.1+/CD4+ and TCR-Vβ22+/CD4+ T-LGL lymphocytosis, a TCR-Vβ13.1+/CD4+ T-cell Sezary syndrome, TCR-γδ+ T-LGL leukemia, and peripheral CD4+ T-cell lymphoma not otherwise specified, associated with an HLA-DRB1*0701 genotype, showed a specific response to this hCMV peptide, except for the CD4+ T cell lymphoma patient who displayed mild secretion of IFN-γ (1890 pg/mL; Figure 1D).
Impact of “in vitro” stimulation with hCMV in the gene expression profile of monoclonal TCR-αβ+/CD4+ T-LGL
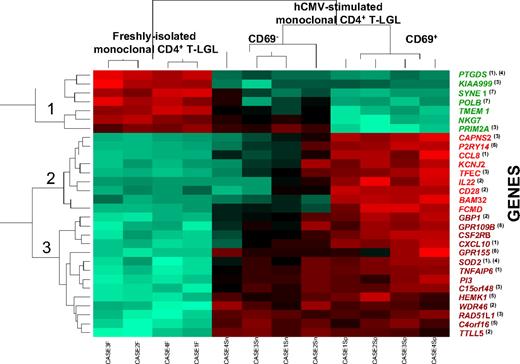
Supervised analysis of the gene expression profiles of purified monoclonal TCR-αβ+/CD4+ T cells using the Significant Analysis of Microarrays algorithm showed that 97 of 19 000 genes (54 675 probes) analyzed were significantly down- or up-regulated during hCMV stimulation (Table 1). Subsequent hierarchical clustering analysis (Figure 2) showed that 3 groups of samples corresponding to the 3 different cellular fractions subjected to analysis (unstimulated, CD69+-stimulated, and CD69−-stimulated clonal CD4+ T cells), could be clearly discriminated on the basis of the expression found for those 30 genes showing the highest variability on their expression among these samples. From the functional viewpoint, those 30 differentially expressed genes included genes involved in inflammatory and immune responses, cell proliferation and cell-cycle progression, apoptosis, protein synthesis, G-protein receptor-mediated signaling pathways, and the DNA repair and maintenance cellular machinery (Figure 2).
Genes whose expression in monoclonal CD4+ T-LGL cells significantly (P < .006) changed after short-term in vitro hCMV stimulation
| Probe set . | Gene symbol . | Score . | Gene description . |
|---|---|---|---|
| 211748_x_at | PTGDS | 10,13416371 | Prostaglandin D2 synthase 21 kDa (brain) |
| 244813_at | RAD51L1 | 7,676334816 | RAD51-like 1 (S cerevisiae) |
| 213915_at | NKG7 | 7,649233005 | Natural killer cell group 7 sequence |
| 215898_at | TTLL5 | 7,324824305 | Tubulin tyrosine ligase-like family, member 5 |
| 206026_s_at | TNFAIP6 | 7,270728576 | Tumor necrosis factor, alpha-induced protein 6 |
| 219023_at | C4orf16 | 7,237962005 | Chromosome 4 open reading frame 16 |
| 222858_s_at | DAPP1 | 6,933745384 | Dual adaptor of phosphotyrosine and 3-phosphoinositides |
| 223484_at | C15orf48 | 6,916996754 | Chromosome 15 open reading frame 48 |
| 208184_s_at | TMEM1 | 6,71864243 | Transmembrane protein 1 |
| 214038_at | CCL8 | 6,110026344 | Chemokine (C-C motif) ligand 8 |
| 209799_at | PRKAA1 | 5,895332958 | Protein kinase, AMP-activated, alpha 1 catalytic subunit |
| 231577_s_at | GBP1 | 5,876037622 | Quanylate binding protein 1, interferon-inducible, 67 kDa |
| 213002_at | MARCKS | 5,841826371 | Myristoylated alanine-rich protein kinase C substrate |
| 205159_at | CSF2RB | 5,826684303 | Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| 230012_at | C17orf44 | 5,810899214 | Chromosome 17 open reading frame 44 |
| 205267_at | POU2AF1 | 5,7541994 | POU domain, class 2, associating factor 1 |
| 222388_s_at | VPS35 | 5,750463855 | Vacuolar protein sorting 35 (yeast) |
| 1554885_a_at | PRIM2A | 5,734193947 | Primase, polypeptide 2A, 58 kDa |
| 223382_s_at | ZNRF1 | 5,700054082 | Zinc and ring finger 1 |
| 41469_at | PI3 | 5,667143829 | Peptidase inhibitor 3, skin-derived (SKALP) |
| 229344_x_at | FAM80B | 5,615111734 | Family with sequence similarity 80, member B |
| 218096_at | AGPAT5 | 5,548067875 | 1-Acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| 241917_at | FCHSD2 | 5,544997567 | FCH and double SH3 domains 2 |
| 1555884_at | PSMD6 | 5,462286485 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| 243512_x_at | IMMP2L | 5,43440168 | IMP2 inner mitochondrial membrane peptidase-like (S cerevisiae) |
| 222833_at | CAPNS2 | 5,404156833 | Calpain, small subunit 2 |
| 204533_at | CXCL10 | 5,397907696 | Chemokine (C-X-C motif) ligand 10 |
| 230434_at | KLHL23 | 5,367623661 | Kelch-like 23 (Drosophila) |
| 222845_x_at | TMBIM4 | 5,36183547 | Transmembrane BAX inhibitor motif containing 4 |
| 205283_at | FCMD | 5,330492853 | Fukuyama-type congenital muscular dystrophy (fukutin) |
| 1554929_at | KIAA0999 | 5,297695496 | NA |
| 52159_at | HEMK1 | 5,293319745 | HemK methyltransferase family member 1 |
| 224827_at | DC-UbP | 5,210263204 | NA |
| 205641_s_at | TRADD | 5,19931747 | TNFRSF1A-associated via death domain |
| 201295_s_at | WSB1 | 5,191526754 | WD repeat and SOCS box-containing 1 |
| 218507_at | HIG2 | 5,175505974 | NA |
| 222386_s_at | COPZ1 | 5,174500241 | Coatomer protein complex, subunit zeta 1 |
| 204057_at | IRF8 | 5,165308859 | Interferon regulatory factor 8 |
| 218562_s_at | TMEM57 | 5,163304991 | Transmembrane protein 57 |
| 221059_s_at | CHST6 | 5,152533315 | Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| 209969_s_at | STAT1 | 5,083186457 | Signal transducer and activator of transcription 1, 91 kDa |
| 232027_at | SYNE1 | 5,031819366 | Spectrin repeat containing, nuclear envelope 1 |
| 204633_s_at | RPS6KA5 | 5,023419317 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 5 |
| 216841_s_at | SOD2 | 5,017952939 | Superoxide dismutase 2, mitochondrial |
| 222691_at | SLC35B3 | 5,009110445 | Solute carrier family 35, member B3 |
| 207700_s_at | NCOA3 | 4,974277498 | Nuclear receptor coactivator 3 |
| 235574_at | GBP4 | 4,96365421 | Guanylate binding protein 4 |
| 222045_s_at | C20orf67 | 4,935840305 | Chromosome 20 open reading frame 67 |
| 204500_s_at | AGTPBP1 | 4,934205697 | ATP/GTP binding protein 1 |
| 1556967_at | ZDHHC14 | 4,92755238 | Zinc finger, DHHC-type containing 14 |
| 204613_at | PLCG2 | 4,924269359 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| 212378_at | GART | 4,924160286 | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| 210050_at | TPI1 | 4,920823757 | Triosephosphate isomerase 1 |
| 204125_at | NDUFAF1 | 4,920466452 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 |
| 204068_at | STK3 | 4,894787293 | Serine/threonine kinase 3 (STE20 homolog, yeast) |
| 201900_s_at | AKR1A1 | 4,890038496 | Aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| 242939_at | TFDP1 | 4,889767365 | Transcription factor Dp-1 |
| 226511_at | MCART1 | 4,864612596 | Mitochondrial carrier triple repeat 1 |
| 201921_at | LOC552891 | 4,824421802 | NA |
| 223767_at | GPR84 | 4,819947478 | G protein-coupled receptor 84 |
| 204565_at | THEM2 | 4,79843566 | Thioesterase superfamily member 2 |
| 229398_at | RAB18 | 4,788964099 | RAB18, member RAS oncogene family |
| 227458_at | C9orf46 | 4,769116802 | Chromosome 9 open reading frame 46 |
| 206584_at | LY96 | 4,766515038 | Lymphocyte antigen 96 |
| 213246_at | C14orf130 | 4,765949714 | Chromosome 14 open reading frame 130 |
| 213246_at | C14orf130 | 4,765949714 | Chromosome 14 open reading frame 130 |
| 203518_at | LYST | 4,755336993 | Lysosomal trafficking regulador |
| 236717_at | LOC165186 | 4,713732588 | NA |
| 226170_at | EYA3 | 4,699440582 | Eyes absent homolog 3 (Drosophila) |
| 212442_s_at | LASS6 | 4,673946958 | LAG1 longevity assurance homolog 6 (S. cerevisiae) |
| 227247_at | PLEKHA8 | 4,640609475 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| 201463_s_at | TALDO1 | 4,623771225 | Transaldolase 1 |
| 204702_s_at | NFE2L3 | 4,608324388 | Nuclear factor (erythroid-derived 2)-like 3 |
| 224445_s_at | ZFYVE21 | 4,60690338 | Zinc finger, FYVE domain containing 21 |
| 204446_s_at | ALOX5 | 4,606635061 | Arachidonate 5-lipoxygenase |
| 206478_at | KIAA0125 | 4,604412442 | KIAA0125 |
| 215380_s_at | C7orf24 | 4,59977998 | Chromosome 7 open reading frame 24 |
| 218230_at | ARFIP1 | 4,584558778 | ADP-ribosylation factor interacting protein 1 (arfaptin 1) |
| 229500_at | SLC30A9 | 4,576485193 | Solute carrier family 30 (zinc transporter), member 9 |
| 209222_s_at | OSBPL2 | 4,567763881 | Oxysterol binding protein-like 2 |
| 206267_s_at | MATK | 4,558282396 | Megakaryocyte-associated tyrosine kinase |
| 214681_at | GK | 4,538965602 | Glycerol kinase |
| 228106_at | C4orf30 | 4,537274088 | Chromosome 4 open reading frame 30 |
| 218095_s_at | TMEM165 | 4,527525331 | Transmembrane protein 165 |
| 212961_x_at | CXorf40B | 4,521246647 | Chromosome X open reading frame 40B |
| 227438_at | ALPK1 | 4,516024032 | Alpha-kinase 1 |
| 210347_s_at | BCL11A | 4,513152919 | B-cell CLL/lymphoma 11A (zinc finger protein) |
| 206637_at | P2RY14 | 4,513063383 | Purinergic receptor P2Y, G-protein coupled, 14 |
| 1561167_at | ETV6 | 4,500052398 | ets variant gene 6 (TEL oncogene) |
| 204254_s_at | VDR | 4,498636497 | Vitamin D (1,25- dihydroxyvitamin D3) receptor |
| 206995_x_at | SCARF1 | 4,491153208 | Scavenger receptor class F, member 1 |
| 201328_at | ETS2 | 4,487520459 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) |
| 221843_s_at | KIAA1609 | 4,485591378 | KIAA1609 |
| 207426_s_at | TNFSF4 | 4,483796011 | Tumor necrosis factor (ligand) superfamily, member 4 (tax-transcriptionally activated glycoprotein 1, 34 kDa) |
| 236995_x_at | TFEC | 4,482801297 | Transcription factor EC |
| 209238_at | STX3 | 4,468215629 | Syntaxin 3 |
| 40016_g_at | MAST4 | 4,464348703 | Microtubule associated serine/threonine kinase family member 4 |
| 222357_at | ZBTB20 | 4,452512078 | Zinc finger and BTB domain containing 20 |
| Probe set . | Gene symbol . | Score . | Gene description . |
|---|---|---|---|
| 211748_x_at | PTGDS | 10,13416371 | Prostaglandin D2 synthase 21 kDa (brain) |
| 244813_at | RAD51L1 | 7,676334816 | RAD51-like 1 (S cerevisiae) |
| 213915_at | NKG7 | 7,649233005 | Natural killer cell group 7 sequence |
| 215898_at | TTLL5 | 7,324824305 | Tubulin tyrosine ligase-like family, member 5 |
| 206026_s_at | TNFAIP6 | 7,270728576 | Tumor necrosis factor, alpha-induced protein 6 |
| 219023_at | C4orf16 | 7,237962005 | Chromosome 4 open reading frame 16 |
| 222858_s_at | DAPP1 | 6,933745384 | Dual adaptor of phosphotyrosine and 3-phosphoinositides |
| 223484_at | C15orf48 | 6,916996754 | Chromosome 15 open reading frame 48 |
| 208184_s_at | TMEM1 | 6,71864243 | Transmembrane protein 1 |
| 214038_at | CCL8 | 6,110026344 | Chemokine (C-C motif) ligand 8 |
| 209799_at | PRKAA1 | 5,895332958 | Protein kinase, AMP-activated, alpha 1 catalytic subunit |
| 231577_s_at | GBP1 | 5,876037622 | Quanylate binding protein 1, interferon-inducible, 67 kDa |
| 213002_at | MARCKS | 5,841826371 | Myristoylated alanine-rich protein kinase C substrate |
| 205159_at | CSF2RB | 5,826684303 | Colony stimulating factor 2 receptor, beta, low-affinity (granulocyte-macrophage) |
| 230012_at | C17orf44 | 5,810899214 | Chromosome 17 open reading frame 44 |
| 205267_at | POU2AF1 | 5,7541994 | POU domain, class 2, associating factor 1 |
| 222388_s_at | VPS35 | 5,750463855 | Vacuolar protein sorting 35 (yeast) |
| 1554885_a_at | PRIM2A | 5,734193947 | Primase, polypeptide 2A, 58 kDa |
| 223382_s_at | ZNRF1 | 5,700054082 | Zinc and ring finger 1 |
| 41469_at | PI3 | 5,667143829 | Peptidase inhibitor 3, skin-derived (SKALP) |
| 229344_x_at | FAM80B | 5,615111734 | Family with sequence similarity 80, member B |
| 218096_at | AGPAT5 | 5,548067875 | 1-Acylglycerol-3-phosphate O-acyltransferase 5 (lysophosphatidic acid acyltransferase, epsilon) |
| 241917_at | FCHSD2 | 5,544997567 | FCH and double SH3 domains 2 |
| 1555884_at | PSMD6 | 5,462286485 | Proteasome (prosome, macropain) 26S subunit, non-ATPase, 6 |
| 243512_x_at | IMMP2L | 5,43440168 | IMP2 inner mitochondrial membrane peptidase-like (S cerevisiae) |
| 222833_at | CAPNS2 | 5,404156833 | Calpain, small subunit 2 |
| 204533_at | CXCL10 | 5,397907696 | Chemokine (C-X-C motif) ligand 10 |
| 230434_at | KLHL23 | 5,367623661 | Kelch-like 23 (Drosophila) |
| 222845_x_at | TMBIM4 | 5,36183547 | Transmembrane BAX inhibitor motif containing 4 |
| 205283_at | FCMD | 5,330492853 | Fukuyama-type congenital muscular dystrophy (fukutin) |
| 1554929_at | KIAA0999 | 5,297695496 | NA |
| 52159_at | HEMK1 | 5,293319745 | HemK methyltransferase family member 1 |
| 224827_at | DC-UbP | 5,210263204 | NA |
| 205641_s_at | TRADD | 5,19931747 | TNFRSF1A-associated via death domain |
| 201295_s_at | WSB1 | 5,191526754 | WD repeat and SOCS box-containing 1 |
| 218507_at | HIG2 | 5,175505974 | NA |
| 222386_s_at | COPZ1 | 5,174500241 | Coatomer protein complex, subunit zeta 1 |
| 204057_at | IRF8 | 5,165308859 | Interferon regulatory factor 8 |
| 218562_s_at | TMEM57 | 5,163304991 | Transmembrane protein 57 |
| 221059_s_at | CHST6 | 5,152533315 | Carbohydrate (N-acetylglucosamine 6-O) sulfotransferase 6 |
| 209969_s_at | STAT1 | 5,083186457 | Signal transducer and activator of transcription 1, 91 kDa |
| 232027_at | SYNE1 | 5,031819366 | Spectrin repeat containing, nuclear envelope 1 |
| 204633_s_at | RPS6KA5 | 5,023419317 | Ribosomal protein S6 kinase, 90 kDa, polypeptide 5 |
| 216841_s_at | SOD2 | 5,017952939 | Superoxide dismutase 2, mitochondrial |
| 222691_at | SLC35B3 | 5,009110445 | Solute carrier family 35, member B3 |
| 207700_s_at | NCOA3 | 4,974277498 | Nuclear receptor coactivator 3 |
| 235574_at | GBP4 | 4,96365421 | Guanylate binding protein 4 |
| 222045_s_at | C20orf67 | 4,935840305 | Chromosome 20 open reading frame 67 |
| 204500_s_at | AGTPBP1 | 4,934205697 | ATP/GTP binding protein 1 |
| 1556967_at | ZDHHC14 | 4,92755238 | Zinc finger, DHHC-type containing 14 |
| 204613_at | PLCG2 | 4,924269359 | Phospholipase C, gamma 2 (phosphatidylinositol-specific) |
| 212378_at | GART | 4,924160286 | Phosphoribosylglycinamide formyltransferase, phosphoribosylglycinamide synthetase, phosphoribosylaminoimidazole synthetase |
| 210050_at | TPI1 | 4,920823757 | Triosephosphate isomerase 1 |
| 204125_at | NDUFAF1 | 4,920466452 | NADH dehydrogenase (ubiquinone) 1 alpha subcomplex, assembly factor 1 |
| 204068_at | STK3 | 4,894787293 | Serine/threonine kinase 3 (STE20 homolog, yeast) |
| 201900_s_at | AKR1A1 | 4,890038496 | Aldo-keto reductase family 1, member A1 (aldehyde reductase) |
| 242939_at | TFDP1 | 4,889767365 | Transcription factor Dp-1 |
| 226511_at | MCART1 | 4,864612596 | Mitochondrial carrier triple repeat 1 |
| 201921_at | LOC552891 | 4,824421802 | NA |
| 223767_at | GPR84 | 4,819947478 | G protein-coupled receptor 84 |
| 204565_at | THEM2 | 4,79843566 | Thioesterase superfamily member 2 |
| 229398_at | RAB18 | 4,788964099 | RAB18, member RAS oncogene family |
| 227458_at | C9orf46 | 4,769116802 | Chromosome 9 open reading frame 46 |
| 206584_at | LY96 | 4,766515038 | Lymphocyte antigen 96 |
| 213246_at | C14orf130 | 4,765949714 | Chromosome 14 open reading frame 130 |
| 213246_at | C14orf130 | 4,765949714 | Chromosome 14 open reading frame 130 |
| 203518_at | LYST | 4,755336993 | Lysosomal trafficking regulador |
| 236717_at | LOC165186 | 4,713732588 | NA |
| 226170_at | EYA3 | 4,699440582 | Eyes absent homolog 3 (Drosophila) |
| 212442_s_at | LASS6 | 4,673946958 | LAG1 longevity assurance homolog 6 (S. cerevisiae) |
| 227247_at | PLEKHA8 | 4,640609475 | Pleckstrin homology domain containing, family A (phosphoinositide binding specific) member 8 |
| 201463_s_at | TALDO1 | 4,623771225 | Transaldolase 1 |
| 204702_s_at | NFE2L3 | 4,608324388 | Nuclear factor (erythroid-derived 2)-like 3 |
| 224445_s_at | ZFYVE21 | 4,60690338 | Zinc finger, FYVE domain containing 21 |
| 204446_s_at | ALOX5 | 4,606635061 | Arachidonate 5-lipoxygenase |
| 206478_at | KIAA0125 | 4,604412442 | KIAA0125 |
| 215380_s_at | C7orf24 | 4,59977998 | Chromosome 7 open reading frame 24 |
| 218230_at | ARFIP1 | 4,584558778 | ADP-ribosylation factor interacting protein 1 (arfaptin 1) |
| 229500_at | SLC30A9 | 4,576485193 | Solute carrier family 30 (zinc transporter), member 9 |
| 209222_s_at | OSBPL2 | 4,567763881 | Oxysterol binding protein-like 2 |
| 206267_s_at | MATK | 4,558282396 | Megakaryocyte-associated tyrosine kinase |
| 214681_at | GK | 4,538965602 | Glycerol kinase |
| 228106_at | C4orf30 | 4,537274088 | Chromosome 4 open reading frame 30 |
| 218095_s_at | TMEM165 | 4,527525331 | Transmembrane protein 165 |
| 212961_x_at | CXorf40B | 4,521246647 | Chromosome X open reading frame 40B |
| 227438_at | ALPK1 | 4,516024032 | Alpha-kinase 1 |
| 210347_s_at | BCL11A | 4,513152919 | B-cell CLL/lymphoma 11A (zinc finger protein) |
| 206637_at | P2RY14 | 4,513063383 | Purinergic receptor P2Y, G-protein coupled, 14 |
| 1561167_at | ETV6 | 4,500052398 | ets variant gene 6 (TEL oncogene) |
| 204254_s_at | VDR | 4,498636497 | Vitamin D (1,25- dihydroxyvitamin D3) receptor |
| 206995_x_at | SCARF1 | 4,491153208 | Scavenger receptor class F, member 1 |
| 201328_at | ETS2 | 4,487520459 | v-ets erythroblastosis virus E26 oncogene homolog 2 (avian) |
| 221843_s_at | KIAA1609 | 4,485591378 | KIAA1609 |
| 207426_s_at | TNFSF4 | 4,483796011 | Tumor necrosis factor (ligand) superfamily, member 4 (tax-transcriptionally activated glycoprotein 1, 34 kDa) |
| 236995_x_at | TFEC | 4,482801297 | Transcription factor EC |
| 209238_at | STX3 | 4,468215629 | Syntaxin 3 |
| 40016_g_at | MAST4 | 4,464348703 | Microtubule associated serine/threonine kinase family member 4 |
| 222357_at | ZBTB20 | 4,452512078 | Zinc finger and BTB domain containing 20 |
NA indicates not applicable.
Hierarchical clustering analysis dendrogram of those 30 genes showing the highest differences between freshly isolated and both purified CD69+ and CD69− hCMV-stimulated monoclonal CD4+ T-LGL (n = 12 paired cell samples). The relative level of expression of each gene is represented by a color code where red represents an expression higher than the mean and green represents an expression below the mean values. Those genes showing different levels of expression in the distinct cell fractions analyzed are known to be involved in inflammatory (1) and immune (2) responses, cell proliferation and/or cell-cycle progression (3), apoptosis (4), protein synthesis (5), G-protein receptor-mediated cell signaling (6), and the DNA repair and maintenance machinery (7). Of note, both CD69+ and CD69− hCMV-stimulated monoclonal CD4+ T-LGLs showed a high similarity in their levels of expression for most of the genes displayed in the dendrogram, except for NKG7, PRIM2A, CAPNS2, P2RY14, CCL8, KCNJ2, TFEC, IL22, CD28, BAM32, and FCMD.
Hierarchical clustering analysis dendrogram of those 30 genes showing the highest differences between freshly isolated and both purified CD69+ and CD69− hCMV-stimulated monoclonal CD4+ T-LGL (n = 12 paired cell samples). The relative level of expression of each gene is represented by a color code where red represents an expression higher than the mean and green represents an expression below the mean values. Those genes showing different levels of expression in the distinct cell fractions analyzed are known to be involved in inflammatory (1) and immune (2) responses, cell proliferation and/or cell-cycle progression (3), apoptosis (4), protein synthesis (5), G-protein receptor-mediated cell signaling (6), and the DNA repair and maintenance machinery (7). Of note, both CD69+ and CD69− hCMV-stimulated monoclonal CD4+ T-LGLs showed a high similarity in their levels of expression for most of the genes displayed in the dendrogram, except for NKG7, PRIM2A, CAPNS2, P2RY14, CCL8, KCNJ2, TFEC, IL22, CD28, BAM32, and FCMD.
Interestingly, all genes involved in repression of inflammation, progression of cell cycle, and maintenance of DNA integrity (PTGDS, KIAA999, SYNE1, POLB, TMEM1, NKG7, PRIM2A) were down-regulated in both CD69+ and CD69− hCMV-stimulated monoclonal CD4+ T-LGL, compared with the paired freshly isolated monoclonal CD4+ T-LGL cell fraction. HCMV-stimulated CD69+ and CD69− monoclonal CD4+ T-LGL showed typical traits of an inflammatory response associated with an increased expression of the CCL8, CXCL10, TNFAIP6, and SOD2 genes involved in chemotaxis, cell-cell interactions, and the oxidative stress during inflammation23-26 (Figure 2). Likewise, these cells acquired a quiescent phenotype as reflected by a decreased expression of the KIAA999 and PRIM2A enzymes in association with overexpression of the RAD51L1, CAPNS2, TFEC, and C15orf48 genes, a gene expression profile presumably reflecting an arrest or a delay in cell-cycle progression27-31 (Figure 2). In addition, hCMV stimulation of monoclonal CD4+ T cells could also lead to an increased resistance to apoptosis resulting from down-regulation of the proapoptotic PTGDS gene and overexpression of the antiapoptotic SOD2 gene.32,33 In turn, increased expression of both the HEMK1 and C4orf16 genes, involved in protein translation and trafficking between the trans-Golgi network34 and endosomes,35 respectively, could just reflect an increase in protein synthesis associated with hCMV stimulation as confirmed by the evaluation of cytokine secretion by those cells. Furthermore, hCMV-specific monoclonal CD4+ T-LGL cells also showed up-regulation of several G-protein receptors (eg, P2RY14, GPR155, and GPR109B) that mediate signals provided by secreted cytokines and chemokines.36 Finally, hCMV stimulation induced down-regulation of both the SYNE1 and POLB enzymes, which are required for the organization of the nucleus as well as for the maintenance of DNA integrity and DNA repair,37,38 supporting the induction of an increased nuclear instability on hCMV-stimulated CD4+ T-LGL.
In contrast to hCMV-stimulated CD4+ T-LGL cells, CD69+/CD4+ T-cells from hCMV-seropositive healthy persons showed patterns of gene expression in response to hCMV associated with increased cell proliferation and apoptosis, more than 200 genes being differentially expressed (P < .001) in CD69+/CD4+ hCMV-activated T cells from healthy subjects versus T-LGL patients (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Tumoral versus normal hCMV-specific CD69+/CD4+ T cells showed down-regulation of genes involved in apoptosis (ie, FAF1 (Fas (TNFRSF6) associated factor 1), TNFRSF25 (tumor necrosis factor receptor superfamily member 25)) and cell cycle (ie, CCND2 (cyclin D2), CDC37 (cell division cycle 37 homolog), GSPT1 (G1- to S-phase transition 1)) among other genes; in addition, genes involved in preventing apoptosis, such as FAIM2 (Fas apoptotic inhibitory molecule 2) and BIRC7 (livin inhibitor of apoptosis), as well as the LIN37 gene, which is mainly expressed in quiescent cells,39 were also increased in the patient CD69+/CD4+ T-LGL cells.
Discussion
In a recent study, we have shown that monoclonal TCRVβ13.1+/CD4+ T-LGL patients show highly conserved TCRVβ CDR3 sequences in association with the HLA-DRB1*0701 allele.2 These findings suggest that the expansion of (mono)clonal T cells in these patients might result from antigenic stimulation through a common peptide. In contrast to cytotoxic CD8+ T lymphocytes, the exact role of CD4+ T cells bearing a cytotoxic immunophenotype in the clearance of viral infections has remained unclear for decades. However, recent results indicate that, although antigen recognition by CD4+ T lymphocytes usually requires antigen uptake by antigen-presenting cells, direct presentation of hCMV and hEBV peptides on HLA class II also occurs after viral infection.40,41 These findings, together with the low frequency of autoimmune disorders observed among CD4+ T-LGL patients,1,2 suggest that, in these patients, antigenic stimulation, rather than being associated with endogenous proteins, as proposed for CD8+ T-LGL, could most probably have an exogenous viral origin. Here we demonstrate, for the first time, that hCMV could be involved in the ontogeny of monoclonal CD4+/TCRVαβ+ T-LGL. Our results clearly show that, independently of the exact TCRVβ used, monoclonal CD4+ T cells from these patients display a strong response to hCMV whole viral lysate, whereas such hCMV-specific response could only be observed in a minor proportion of other T-CLPDs. Interestingly, the “MQLIPDDYSNTHSTRYVTVK” peptide from hCMV gB protein was able to reproduce the response of clonal CD4+ LGL cells to hCMV in patients in which the expanded T cells expressed TCRVβ13.1 in association with the HLA-DRB1*0701 allele. Previous studies have identified the “DYSNTHSTRYV” epitope contained in our peptide as the only hCMV peptide sequence that can be loaded in HLA-DRB1*0701 molecules.42,43 Of note, this peptide has also been shown to be directly expressed on HLA-II molecules from hCMV-infected cells, allowing for their direct recognition by hCMV-specific CD4+ T cells.42 However, to the best of our knowledge, no study has been reported so far in which it has been clearly demonstrated that this peptide could induce stimulation of expanded monoclonal CD4+ T cells from T-LGL patients.
Through the use of different markers, our results clearly show the ability of hCMV-stimulated monoclonal CD4+ T-LGL cells to both up-regulate the expression of early cell surface activation markers, such as CD69 and CD25, and to secrete both TNF-α and IFN-γ. The surprisingly high concentration of IFN-γ secreted into the extracellular medium in the subgroup of monoclonal TCRVβ13.1+/CD4+ T-LGL patients in response to a single hCMV peptide complementary to the HLA-DRB1*0701 allele is particularly enlightening; this is particularly true if we consider that the observed response could represent only part of the response actually occurring in vivo because antigen presentation developed in vitro by PB cells could be considerably less efficient as it is limited to circulating cells (dendritic cells and B lymphocytes) that are considered to be precursors of professional antigen-presenting cells44 and because of the potential interference of plasma proteins with both antigen uptake and HLA loading.45
Immunologic T-cell responses against viruses and other antigens are typically associated with clonal selection of T cells with restricted antigen specificities.46 This is particularly evident among CD8+ cytotoxic/effector T cells46 ; in turn, CD4+ T cells typically show a broader recognition of peptide epitopes probably the result of the existence of less stringent anchor positions in HLA class II vs HLA class I molecules. Despite this, oligoclonal expansions of CD4+ T cells bearing a relatively heterogeneous memory/effector cytotoxic phenotype have long been reported in association with hCMV.13-15,47 Interestingly, this also includes oligoclonal expansions of hCMV-specific TCRVβ13.1+/CD4+ T lymphocytes in HLA-DRB1*0701 healthy volunteers,15 further supporting the involvement of hCMV in the clonal expansion of CD4+ T-LGL patients.
Altogether, these observations provide unequivocal evidence about the specificity of the expanded monoclonal T cells in CD4+ T-LGL patients; however, they do not directly demonstrate the involvement of hCMV on the abnormally increased numbers of clonal T cells observed in these patients. To better understand the mechanisms leading to the abnormally increased (mono)clonal expansion of hCMV-specific CD4+ T cells in T-LGL patients, we investigated the changes occurring in the gene expression profiles (GEPs) of clonal CD4+ T cells after in vitro hCMV stimulation. Accordingly, our results show that hCMV-specific monoclonal CD4+ T-LGL cells producing increased levels of IFN-γ and TNF-α display GEPs associated with down-regulation of genes involved in cell cycle progression and maintenance of DNA integrity, together with up-regulation of genes involved in both inflammatory and immune responses and an increased resistance to apoptosis; this was in contrast to the response observed for hCMV-specific normal CD69+/CD4+ T cells from healthy subjects, which showed GEPs associated with increased cell proliferation and higher susceptibility to apoptosis. In line with these observations, Appay et al have recently shown that increased T-cell differentiation resulting from persistent immune activation under conditions of inflammation may lead to immunosenescence.48 In this sense, hCMV-specific monoclonal CD4+ T-LGL cells could represent an accumulation of a monoclonal population of aged lymphocytes with typical traits of replicative senescence, such as cell-cycle arrest, limited ability to proliferate, resistance to apoptosis, and absence of CD28.49 Likewise, the patients with monoclonal TCR-βα+/CD4+ T-LGL lymphocytosis here analyzed showed a high percentage of monoclonal CD27−/CD4+ T-LGL cells expressing CD57 (data not shown), a marker of terminal differentiation used to identify CMV-specific CD4+ T cells that have achieved replicative incompetence14 ; of note, the only CD4+ T-LGL patient who did not respond to hCMV displayed the highest percentage of monoclonal CD57+CD27-CD4+ T-LGL cells (data not shown). In this regard, it should be noted that in vivo accumulation of replicative incompetent monoclonal hCMV-specific CD4+ T-LGL cells does not seem to be an indicator of an inability of the immune system to remove such terminally differentiated effector population (CD27−) because no additional immunologic abnormalities in the distribution of the other major subsets of peripheral blood B, T (including total CD8+ and CD8+/CD57+/CD3+ T cells), and NK cells were found in these patients (data not shown). However, accumulation of CD4+ T-LGL cells could result from chronic hCMV stimulation of “stem cell–like,” self-renewing monoclonal memory lymphocytes with a preserved replicative potential,50 leading to the clonal expansion of terminally differentiated effector cells.
Altogether, these findings would reinforce the hypothesis suggested by Appay et al indicating that immune exhaustion as consequence of successive rounds of antigen-driven T-cell activation can deteriorate T-cell competence through the decline of T-cell renewal capacities in parallel to the loss of other T-cell clones relevant to the control and clearance of a specific virus.48 By contrast, they shed little light on the role of hCMV on the development of T-LGL, unless a minor population of hCMV-specific CD4+ T-LGL precursors exists, which retains the ability of self-renewal, at the same time it is responsible for generating an increasingly more numerous population of terminally differentiated senescent hCMV-specific CD4+ T-LGL cells. This concept could be of particularly high relevance because GEP studies showed an association of the referred quiescent state of hCMV-activated CD4+ T-LGL cells with a phenotype reflecting an increased genomic instability resulting from down-regulation of the SYNE1 and POLB enzymes involved in maintaining DNA integrity. In such cases, in vivo hCMV stimulation of T-LGL could increase the risk of accumulation of genetic changes and lesions in the expanded cells and favor their neoplastic and even malignant transformation.
In conclusion, in the present study, we demonstrate, for the first time, the involvement of hCMV in the ontogeny of CD4+ T-LGL, indicating that the antigenic hCMV stimulus could be responsible for the initiation and maintenance of the disease.
An Inside Blood analysis of this article appears at the front of this issue.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
This work was supported in part by the following grants: Fondo Investigacion Sanitaria (05/0399) from the Ministerio de Sanidad y Consumo, Madrid, Spain; Red Tematica de Investigacion Cooperativa de Cancer (RD06/0020/0035) from the Instituto de Salud Carlos III, Ministerio de Sanidad y Consumo, Madrid, Spain; and the Consejería de Salud, Junta de Andalucía (05/287), Sevilla, Spain. A.C.G.-M. is supported by Fondo Investigacion Sanitaria (grant CP03/00 035).
Authorship
Contribution: A.R.-C. designed the research, performed experiments, analyzed/interpreted results, made the figures, and wrote the paper; A.C.G.-M. designed the research, performed some experiments, and critically reviewed the paper; P.B. performed experiments; J.A. designed the research and critically reviewed the paper; F. R.-C. and P.G. contributed essential reagents; M.D.T. analyzed results of genome-wide expression; S.M.-C. determined the hCMV serologic status of patients and controls and monitored hCMV viral load in patients; Y.S. and A.W.L. contributed with technical support and critically review the paper; M.G. and A.B. performed the HLA typing of patients and healthy controls; and A.O. designed the research and wrote the paper.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Alberto Orfao, Centro de Investigación del Cáncer, Avenida Universidad de Coimbra s/n, Campus Miguel de Unamuno, 37007, Salamanca, Spain; e-mail: orfao@usal.es.