Abstract
Atypical hemolytic uremic syndrome (aHUS) is a disease of complement dysregulation. In approximately 50% of patients, mutations have been described in the genes encoding the complement regulators factor H, MCP, and factor I or the activator factor B. We report here mutations in the central component of the complement cascade, C3, in association with aHUS. We describe 9 novel C3 mutations in 14 aHUS patients with a persistently low serum C3 level. We have demonstrated that 5 of these mutations are gain-of-function and 2 are inactivating. This establishes C3 as a susceptibility factor for aHUS.
Introduction
Mutations in the genes encoding the complement regulators factor H,1-6 factor I,7,8 and membrane cofactor protein (MCP; CD46),9,10 as well as in the activating component factor B,11 have been detected in approximately 50% of patients with atypical hemolytic uremic syndrome (aHUS).12 A proportion of the remaining patients have persistently low serum levels of C3. In this study we have examined the hypothesis that mutations in the gene encoding C3 could be associated with aHUS in these patients
C3 is the pivotal component of the complement system.13 Activation of the classical, lectin, and alternative pathways results in cleavage of C3 to generate C3b and the anaphylatoxin C3a. When C3b is produced, the thioester is cleaved, and then this highly reactive species may bind covalently to targets. Interaction of the zymogen factor B with C3b and subsequent cleavage of factor B by factor D results in formation of the alternative pathway C3 convertase C3bBb. This set of reactions represents an amplification loop.
A series of complement regulators including factor H and MCP prevent feedback via this loop by increasing the rate of dissociation of C3bBb and/or by serving as cofactors for the serine protease factor I to cleave C3b. Mutations in the gene encoding factor B were recently found to enhance formation of C3bBb or increase resistance to inactivation.11
The importance of C3 as a susceptibility factor for human disease has been emphasized by recent studies documenting that a common nonsynonymous coding change in C3 (rs2230199, Arg80Gly, corresponding to C3S and C3F) is both a susceptibility factor for age-related macular degeneration14 and associated with long-term renal allograft survival.15
Methods
Subjects
In 2 independent cohorts of aHUS patients (Paris, France and Newcastle upon Tyne, United Kingdom), 26 patients (17 Paris, 9 Newcastle) with a serum C3 level persistently below the lower end of the normal range of 680 to 1380 mg/L were identified. In these patients functionally significant mutations in CFH, MCP, CFI, and CFB had not previously been detected. Mutation screening of C3 was undertaken in these patients.
Approval for this study was obtained from the Departement de la Rechereche Clinique et du Developement, DRRC Ile de France, France and the Northern and Yorkshire Multi-Center Research Ethics Committee, United Kingdom. Informed consent was obtained in accordance with the Declaration of Helsinki.
Mutation screening
The coding sequence of C3 was amplified with flanking primers (Table S1, available on the Blood website; see the Supplemental Materials link at the top of the online article). Direct sequencing was undertaken using a 96-capillary Sequencer 3700 (Applied Biosystems, Courtaboeuf, France) using the dye terminator method. For the genomic DNA sequence the first nucleotide A of the initiator ATG codon is denoted as nucleotide +1. The amino acid numbering does not include the 22 residues of the signal peptide.
Functional studies
C3 cDNA (gift of David Isenman, University of Toronto, Toronto, ON)16 was sequenced and compared with the sequence published for the C3 crystal structure.17 Two single basepair changes were found in the cDNA and altered (QuikChange Multi Site-Directed Mutagenesis Kit; Stratagene, La Jolla, CA) to match the published sequence of the C3 used in the structural analysis.17 Mutant clones were produced (QuikChange XL Site-Directed Mutagenesis Kit; Stratagene) and sequenced in both directions.
The mutant and WT C3 DNAs were transiently transfected into either 293T or COS-1 cells using Lipofectamine transfection reagent (Invitrogen, Carlsbad, CA). Two to 3 days after transfection, the supernatants were collected and concentrated. C3 was quantitated by ELISA and examined by Western blotting (see Document S1). Conversion of C3 to iC3 was accomplished by storage at 4°C or repetitive freeze thawing and monitored by autolytic cleavage18 (see Document S1). C3b ligand-binding assays were undertaken using recombinant MCP (see Document S1 for protocol), factor H (Complement Technologies, Tyler, TX), soluble CR1 (gift of H. Marsh, Avant Immunotherapeutics, Needham, MA) and factor B (Complement Technologies). Cofactor assays were undertaken using wild-type and the mutant C3 proteins, human factor I (Complement Technologies) and the above noted cofactor proteins.
See Document S1 for methodology relating to assays for cofactor protein binding to C3 and cofactor activity.
Results and discussion
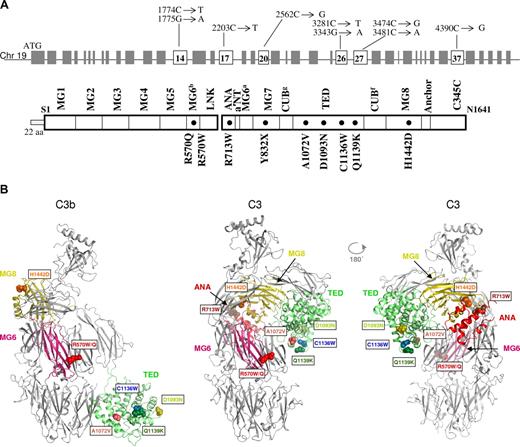
In 11 patients, we identified a heterozygous C3 mutation. Three additional family members were also affected by HUS, 2 from a pedigree (Figure S1) in which an unaffected individual also carried the same change, and another from an affected sibling pair. Therefore, 14 affected persons were found to harbor a mutation. There were a total of 9 distinct mutations identified in the initial 11 patients, the same mutation was carried by 2 unrelated persons. There were 8 heterozygous missense mutations (R570W, R570Q, R713W, A1072V, D1093N, C1136W, Q1139K, and H1142D) and one heterozygous nonsense mutation (Y832X). The positions of these are shown in Figure 1. None of these variants was present in 200 chromosomes from healthy persons.
Location of C3 mutations associated with aHUS. (A) Gene structure of C3, domains of the encoded protein, and position of the mutations. Note that the genomic structure numbering begins with the start site ATG, while the protein structures begin with the first amino acid of the mature protein. (B) Structures of complement component C3 and C3b showing the locations of mutations identified in atypical hemolytic uremic syndrome (aHUS) patients. Ribbon representation of C3b and C3 (2 views) with the domains containing mutations (labeled spheres).
Location of C3 mutations associated with aHUS. (A) Gene structure of C3, domains of the encoded protein, and position of the mutations. Note that the genomic structure numbering begins with the start site ATG, while the protein structures begin with the first amino acid of the mature protein. (B) Structures of complement component C3 and C3b showing the locations of mutations identified in atypical hemolytic uremic syndrome (aHUS) patients. Ribbon representation of C3b and C3 (2 views) with the domains containing mutations (labeled spheres).
Clinical and laboratory data are presented in Table S2. Median age at presentation was 6.5 years (range, 8 months to 40 years). Seven of the 14 recovered renal function after their initial presentation, and 4 of these had recurrences. Among the 14 patients there have been 12 renal transplantations (9 cadaveric and 3 live-related), 5 of which have been affected by recurrent disease.
Next, 8 of 9 C3 mutant constructs (except for the missense mutation) were prepared and transfected into mammalian cells. The C1163W mutant was either minimally or not detectably expressed. Of the remaining 7, each was expressed in approximately comparable amounts, and protein bands in gels migrated identically to wild-type C3 (Figure S2).
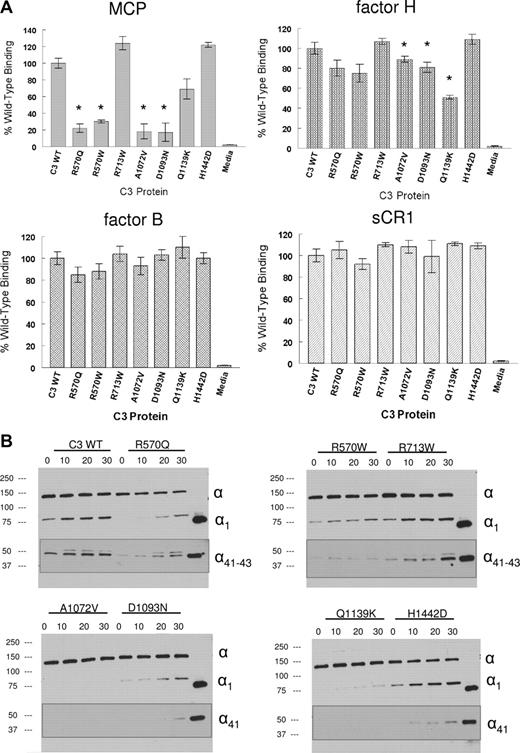
Assays were performed to assess the interaction of the 7 secreted C3 mutants with MCP.19 Binding of R570Q, R570W, A1072V, D1093N, and Q1139K was 22%, 30%, 18%, 17%, and 69% of wild-type, respectively (Figure 2A). Binding of R713W and H1442D was not statistically different from wild-type. The results of the cofactor assays paralleled the ligand binding data (Figure 2B). The 2 mutants with normal MCP binding also had normal cofactor activity, whereas the 5 mutants with decreased MCP binding had decreased cofactor activity. Specifically, there were no detectable cleavage fragments in 4 and modest cleavage by mutant R570Q. Based on the loss of the α-chain and appearance of major α1 and α41 kDa cleavage fragments, there was more than 90% reduction in cofactor activity for these 5 mutants. Thus, the reduced interaction of 5 of the 7 secreted C3 mutant proteins with MCP (the major membrane cofactor regulator of C3b) is likely to lead to a gain of function relative to complement activation.
Ligand binding and cofactor activity of the C3 proteins. Proteins were transiently expressed in 293T cells, concentrated and quantified before analysis (see Document S1). (A) Binding to MCP, factor H, factor B, and soluble CR1 (sCR1) in ELISA. For MCP and factor H, C3 protein was incubated at 15 ng/mL and for factor B, 200 ng/mL. Detection was made with chicken anti–human C3 and HRP-linked donkey anti–chicken IgY. * indicates a significant reduction in binding (P < .05). (B) Kinetic analysis of cofactor activity. C3 preparations were incubated for 0 to 30 minutes at 37°C with factor I and a cofactor protein (MCP, factor H, or sCR1). The zero control is before the addition of factor I. The last lane is an iC3b control. Samples were reduced and analyzed by Western blotting using either chicken anti–human C3 or goat anti–human C3 (insets) followed by an HRP-linked antiglobulin (see Document S1). Cofactor activity is assessed by the loss of the α chain and appearance of the α1 and α41 kDa major cleavage fragments. The minor α43 kDa cleavage fragment is more variable but no pattern was observed that was specific for a mutant. Data are representative of 5 similar experiments.
Ligand binding and cofactor activity of the C3 proteins. Proteins were transiently expressed in 293T cells, concentrated and quantified before analysis (see Document S1). (A) Binding to MCP, factor H, factor B, and soluble CR1 (sCR1) in ELISA. For MCP and factor H, C3 protein was incubated at 15 ng/mL and for factor B, 200 ng/mL. Detection was made with chicken anti–human C3 and HRP-linked donkey anti–chicken IgY. * indicates a significant reduction in binding (P < .05). (B) Kinetic analysis of cofactor activity. C3 preparations were incubated for 0 to 30 minutes at 37°C with factor I and a cofactor protein (MCP, factor H, or sCR1). The zero control is before the addition of factor I. The last lane is an iC3b control. Samples were reduced and analyzed by Western blotting using either chicken anti–human C3 or goat anti–human C3 (insets) followed by an HRP-linked antiglobulin (see Document S1). Cofactor activity is assessed by the loss of the α chain and appearance of the α1 and α41 kDa major cleavage fragments. The minor α43 kDa cleavage fragment is more variable but no pattern was observed that was specific for a mutant. Data are representative of 5 similar experiments.
We next evaluated the interaction of the 7 secreted mutants with complement receptor 1 (soluble CR1, sCR1) and the plasma regulatory protein factor H. Both of these inhibitors also bind C3b and are cofactors for its cleavage by factor I. There were no statistically significant differences between binding of sCR1 to wild-type C3 versus that of the 7 mutants. For factor H, however, the binding profile generally resembled that of MCP except that the magnitude of the decrease in binding was less. However, there was a statistically significant decrease in iC3 binding of A1072V, D1093N, and Q1139K, while R570Q and R570W were modestly (10%-20%) but not statistically significantly decreased. Cofactor activity for Q1139K was also decreased compared with native C3 (Figure S3).
We also assessed binding to factor B. Factor B interacts with C3b or iC3 to begin the formation of the alternative pathway C3 convertase. There were no statistically significant alterations in binding of these mutants to factor B. Taken together, these results further suggest that it is a modification in interactions with regulators that leads to a secondary gain of function in these C3 mutants.
Homozygous C3 deficiency in association with recurrent bacterial infections has been recognized for many years.20 Following the description of the genomic structure of C3 in 1990,21 a molecular basis for C3 deficiency was first described.22 Subsequently, inherited C3 deficiency has been identified in 19 families (reviewed in Reis et al23 ) and the underlying molecular mechanism established in 8 of these. In this study we have identified 9 heterozygous mutations in C3. Five (R570Q, R570W, A1072V, D1093N, and Q1139K) lead to impaired binding to the regulator MCP and, thus, resistance to cleavage by factor I.
Recent structural studies of human C3 have provided unique insights into the functional domains of the molecule.17,24 Putative binding sites for factor H, complement receptors 1 and 2 (CR2; CD21), properdin, and factor B have been mapped to the structure of C3.13 The 5 mutations associated with impaired binding to MCP also provide additional information on potential interactive sites for this regulator as well as possibly for factor I.
The functional significance of R713W and H1442D mutations remains to be elucidated as they had normal binding to MCP and subsequent cleavage by factor I. While the C3 mutants with a resistance to regulator binding fit with current concepts of aHUS pathogenesis, the mutations Y832X and C1136W lead to impaired secretion of C3 and haploinsufficiency. The latter is more difficult to understand relative to the concept of increased complement activation for a given degree of cellular injury as the fundamental predisposing event for aHUS.
In summary, this study establishes a new association between heterozygous mutations in C3 and aHUS.
The online version of this article contains a data supplement.
The publication costs of this article were defrayed in part by page charge payment. Therefore, and solely to indicate this fact, this article is hereby marked “advertisement” in accordance with 18 USC section 1734.
Acknowledgments
We thank Florence Marliot for expert technical support and Paula Bertram for production and purification of recombinant soluble MCP.
T.H.J.G. was supported by the Robin Davies Trust, the Foundation for Children with atypical HUS, and the Medical Research Council (grant G0701325). V.F.B. was supported by the Délégation Régionale à la Recherche Clinique, Assistance Publique–Hôpitaux de Paris (Grants PHRC AOM 05 130). This work was supported in part by funding from the National Institutes of Health (R01 AI037618, R01 AI041592, and T32 HL07317 to E.C.M., M.K.L., R.H., and J.P.A.). R.G. was supported by a National Institutes of Health training grant (T32 AI007163).
National Institutes of Health
Authorship
Contribution: V.F.-B., T.H.J.G., and J.P.A. designed the research, analyzed the data, and wrote the paper. V.F.-B., E.C.M., M.K.L., M.-A.D.-D., L.S., R.G., and R.H. performed the research. J.B., A.L.B., N.M., B.S.K., R.A.W., K.L., G.K., T.M., H.N., W.W., S.G., B.H. de L., M.F., V.M., C.L., and W.H.F. contributed clinical information. B.J.C.J. undertook the structural analysis. E.C.M., M.K.L., R.G., and R.H. prepared the mutant proteins and performed the ligand binding and cofactor assays.
Conflict-of-interest disclosure: The authors declare no competing financial interests.
Correspondence: Veronique Frémeaux-Bacchi, Service d'Immunologie Biologique, Hôpital Européen Georges Pompidou, 20 rue Leblanc, 75908, Paris, Cedex 15, France; e-mail: veronique.fremeaux-bacchi@egp.aphp.fr; or John P. Atkinson, Washington University School of Medicine, Division of Rheumatology, 660 South Euclid Avenue, Campus Box 8045, St Louis, MO 63110; e-mail: jatkinso@im.wustl.edu; or Prof Timothy H. J. Goodship, Institute of Human Genetics, Newcastle University, Central Parkway, Newcastle upon Tyne, United Kingdom NE1 3BZ; e-mail: t.h.j.goodship@ncl.ac.uk.
References
Author notes
*V.F.-B., T.H.J.G., and J.P.A. contributed equally to this work.

This feature is available to Subscribers Only
Sign In or Create an Account Close Modal